Abstract
Plantago major (Plantaginaceae) is a medicinal plant in Chinese folk culture, known for its famous medicinal components such as plantagomain. In this study, we conducted genome sequencing of P. major using Illumina sequencing technology. The assembled complete chloroplast genome had a length of 165,044 bp, comprising a large single-copy regions (82,963 bp), a small single-copy regions (4,633 bp), and a pair of inverted repeat regions (38,724 bp). A total of 140 genes were detected, including 94 CDS, 38 tRNA, and 8 rRNA. Phylogenetic analysis revealed a close genetic relationship between P. major and P. rigida. These findings provide valuable data for a comprehensive understanding of the biological characteristics of P. major.
Introduction
Plantago major (Linaeus 1753) is an accepted species in the genus Plantago (family Plantaginaceae) distributed in the temperate regions of the Eurasian continent, with the type specimen collected from Europe. However, it has now become naturalized in most areas of the world (Iwanycki Ahlstrand et al. Citation2022). Due to the existence of a large number of transitional morphologies, the deep evolutionary relationships among different species still need further clarification, which requires more samples and DNA data to be supplemented (Bagheri et al. Citation2022; Iwanycki Ahlstrand et al. Citation2022). Combining the released chloroplast genomes of other plants in this genus, we will be able to more clearly reveal the complex genetic relationships of this genus.
In China, P. major is renowned as a medicinal plant with abundant nutritional components (Lukova et al. Citation2020; Zhang et al. Citation2021). Its young shoots and tender stems are edible and can be prepared in various ways, including blanching in boiling water, pickling, stir-frying, and stewing. Moreover, this plant contains bioactive constituents like plantamajoside, demonstrating significant pharmacological activities (Samuelsen Citation2000; Li et al. Citation2014; Wu et al. Citation2016; Liu et al. Citation2019; Zeng et al. Citation2022), which have sparked great interest among researchers. The effects of its defatted methanol extract are similar to the commercially-used drug silymarin, indicating its potential as an affordable dietary supplement or nutritional medicinal formulation, offering liver support to patients with hepatic dysfunction (Eldesoky et al. Citation2018). Additionally, seed extracts can be utilized for meat preservation (Noshad et al. Citation2021). Currently, the lack of data on the chloroplast genome of this species hinders in-depth research in genetic field. In this study, the whole chloroplast genome of P. major was characterized and assembled for the first time to reveal the genetic characteristics of this species at the molecular level and to deepen our understanding of the evolutionary relationships of P. major within the Plantago genus.
Materials and methods
In this study, we utilized Illumina sequencing technology to assemble and analyze the complete chloroplast genome of P. major plant, which was grown in the organic environments in Mashan county, Nanning, China (22°51′34″N, 108°22′10″E; ; identified by Yingying Liu with contact information: [email protected]). Fresh leaves were collected and voucher specimen (No. LYY20230515002) was deposited in the herbarium of Guangxi Botanical Garden of Medicinal Plants (abbr. GBGMPP).
Figure 1. The morphological characteristics of P. major in our research. It is a perennial herb with basal leaves. Photoed by Yingying Liu in Guangxi Botanical Garden of Medicinal Plants, Nanning city, China.

Genome sequencing was performed using the Illumina platform (HiseqPE150). The clean data was assembled into the chloroplast genome using NOVOPlasty (Dierckxsens et al. Citation2017). The assembled sequence was annotated using PGA-master (Qu et al. Citation2019), with reference to the chloroplast genome annotation of P. media (NC_028520), and manually corrected using Genious 10.2 (Kearse et al. Citation2012).
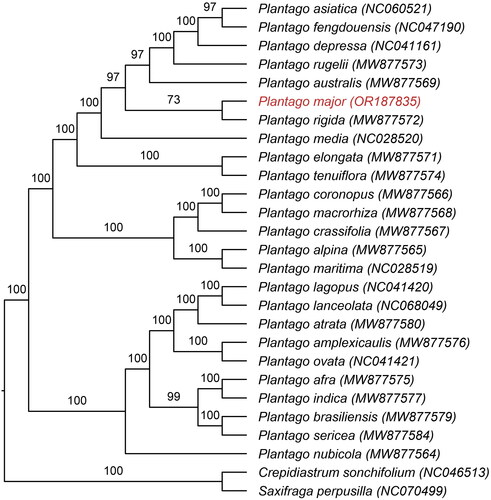
Twenty-four chloroplast genomes of the genus Plantago were obtained from the National Center for Biotechnology Information (NCBI), using the chloroplast genomes of Crepidiastrum sonchifolium (NC_046513) and Saxifraga perpusilla (NC_070499) as outgroup for phylogenetic analysis, for these two species are representative taxa of closely related clades. A total of 79 shared CDS were selected and aligned using MAFFT (Katoh and Standley Citation2013). ModelFinder v2.2.0 (Kalyaanamoorthy et al. Citation2017) was used to select the best-fit model using bayesian information criterion (BIC TVM + F + I + I+R2) criterion. A maximum likelihood (ML) tree was constructed using IQtree (Nguyen et al. Citation2015) with 10,000 ultrafast bootstraps.
Results
As evidence of correct genome assembly, the reads obtained from sequencing exhibit a high coverage depth (Figure S1). The results of the study revealed that the chloroplast genome of P. major (GenBank accession OR187835) is organized into four regions, with a total length of 165,044 bp (). Number of cis-splicing genes and trans-splicing genes are shown in Supplemental Figure S2. The chloroplast genome consists of a large single-copy regions (LSC; 82,963 bp), a small single-copy regions (SSC; 4,633 bp), and a pair of inverted repeats (IRs, 38,724 bp). A total of 140 genes were detected, including 94 coding sequence (CDS), 38 transfer RNA (tRNA), and 8 ribosomal RNA (rRNA). The overall GC content of this chloroplast genome was found to be 38.1%. The phylogenetic analysis revealed that P. major was taxonomically classified within the genus Plantago. Furthermore, it was observed that P. major formed a monophyletic clade together with P. rigida, while P. asiatica, P. fengouensis, P. depressa, P. rugelii, P. australis were found to be their sister taxon ().
Figure 2. Circular map of the complete chloroplast genome of P. major generated by CPGview. The map contains six tracks in default. From the center outward, the first track shows the dispersed repeats. The dispersed repeats consist of direct (D) and palindromic (P) repeats, connected with red and green arcs. The second track shows the long tandem repeats as short blue bars. The third track shows the short tandem repeats or microsatellite sequences as short bars with different colors. The colors, the type of repeat they represent, and the description of the repeat types are as follows. Black: c (complex repeat); green: p1 (repeat unit size = 1); yellow: p2 (repeat unit size = 2); purple: p3 (repeat unit size = 3); blue: p4 (repeat unit size = 4); orange: p5 (repeat unit size = 5); red: p6 (repeat unit size = 6). The small single-copy (SSC), inverted repeat (IRa and IRb), and large single-copy (LSC) regions are shown on the fourth track. The GC content along the genome is plotted on the fifth track. The genes are shown on the sixth track. The optional codon usage bias is displayed in the parenthesis after the gene name. Genes are color-coded by their functional classification. The transcription directions for the inner and outer genes are clockwise and anticlockwise, respectively. The functional classification of the genes is shown in the bottom left corner.
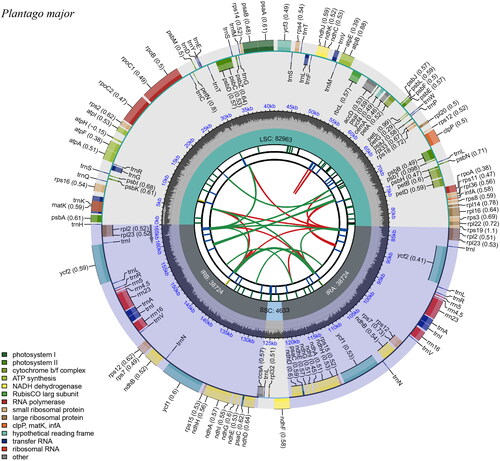
Figure 3. Maximum-likelihood phylogenetic tree of 25 plantago species was constructed based on 76 CDS sequences from chloroplast genomes of P. major and its closely related specie, with Crepidiastrum sonchifolium and Saxifraga perpusilla as outgroups. The number on each branch indicates the boot support value. The following sequences were used: P. asiatica NC 060521 (Si et al. Citation2022), P. fengdouensis NC 047190 (Wang et al. Citation2020), P. depressa NC 041161 (Kwon et al. Citation2019), P. rugelii MW 877573, P. australis MW 877569, P. rigida MW 877572, P. elongata MW 877571, P. tenuiflora MW 877574, P. coronopus MW 877566, P. macrorhiza MW 877568, P. crassifolia MW 877567, P. alpina MW 877565, P. atrata MW77580, P. amplexicaulis MW 877576, P. indica MW 877577, P. afra MW 877575, P. brasiliensis MW 877579, P. nubicola MW 877564, P. sericea MW 877584 (Mower et al. Citation2021), P. media NC 028520, P. maritima NC 028519 (Zhu et al. Citation2016), P. lagopus NC 041420, P. ovata NC 041421 (Sun et al. Citation2019), P. lanceolata NC 068049 (Zhao et al. Citation2023), C. sonchifolium NC 046513 (Cho et al. Citation2020), and S. perpusilla NC 070499 (Yuan et al. Citation2023).
Discussion and conclusion
In this study, we first utilized Illumina sequencing technology to assemble and analyze the complete chloroplast genome of P. major and found that P. major formed a monophyletic clade together with P. rigida. It is interesting to note that P. rigida is primarily distributed in Ecuador and exhibits significant morphological differences from P. major, such as greatly reduced plant size and leaves. Whereas, the close proximity of the two species on the phylogenetic tree indicates a close genetic relationship between P. major and P. rigida. Chloroplasts play a crucial role in the biological characteristics and distribution of plants. The unveiling of the chloroplast genome sequence of P. major will provide meaningful information for the phylogeny and plant molecular research of the genus Plantago.
Ethical approval
P. major L. is not a threatened or endangered species in the Red list of the International Union for Conservation of Nature (IUCN). This Chloroplast Genome research on P. major L., including the collection of plant and DNA material (cultivated) and field work, were permitted and granted by GBGMP. No ethical items were involved in the research.
Authors’ contributions
L. Jiang & L. Chen designed the research and gave financial support. D. Hu, W. Zeng, Z. Yan, Y. Liu & L. Gui. collected plant and DNA materials. Y. Liu & L. Gui conducted the research and wrote the manuscript.
Figure 3.jpg
Download JPEG Image (1.5 MB)Figure 2.jpg
Download JPEG Image (2 MB)Figure 1.jpg
Download JPEG Image (4.2 MB)Manuscript clean.docx
Download MS Word (755.2 KB)Supplementary figures.docx
Download MS Word (196.2 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data of this study are openly accessible in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession number OR187835. The associated BioProject, SRA, and BioSample numbers are PRJNA987869, SRR25033257, and SAMN35994512, respectively.
Additional information
Funding
References
- Bagheri M, Heidari B, Dadkhodaie A, Heidari Z, Daneshnia N, Richards CM. 2022. Analysis of genetic diversity in a collection of Plantago species: application of ISSR markers. J Crop Sci Biotechnol. 25(1):1–8. doi:10.1007/s12892-021-00107-3.
- Cho MS, Kim SH, Yang J, Crawford DJ, Stuessy TF, López-Sepúlveda P, Kim SC. 2020. Plastid phylogenomics of Dendroseris (Cichorieae; Asteraceae): insights into structural organization and molecular evolution of an endemic lineage from the Juan Fernández Islands. Front Plant Sci. 11:594272. doi:10.3389/fpls.2020.594272.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18–e18. doi:10.1093/nar/gkw955.
- Eldesoky AH, Abdel Rahman RF, Ahmed OK, Soliman GA, Saeedan AS, Elzorba HY, Elansary AA, Hattori M. 2018. Antioxidant and hepatoprotective potential of Plantago major growing in Egypt and its major phenylethanoid glycoside, acteoside. J Food Biochem. 42(5):e12567. doi:10.1111/jfbc.12567.
- Iwanycki Ahlstrand N, Gopalakrishnan S, Vieira FG, Bieker VC, Meudt HM, Dunbar-Co S, Rothfels CJ, Martinez-Swatson KA, Maldonado C, Hassemer G, et al. 2022. Travel tales of a worldwide weed: genomic signatures of Plantago major L. reveal distinct genotypic groups with links to colonial trade routes. Front Plant Sci. 13:838166. doi:10.3389/fpls.2022.838166.
- Kalyaanamoorthy S, Minh BQ, Wong TK, Von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. doi:10.1038/nmeth.4285.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi:10.1093/molbev/mst010.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649. doi:10.1093/bioinformatics/bts199.
- Kwon W, Kim Y, Park CH, Park J. 2019. The complete chloroplast genome sequence of traditional medical herb, Plantago depressa Willd. (Plantaginaceae). Mitochondrial DNA Part B Resour. 4(1):437–438. doi:10.1080/23802359.2018.1553530.
- Li Y, Gan L, Li GQ, Deng L, Zhang X, Deng Y. 2014. Pharmacokinetics of plantamajoside and acteoside from Plantago asiatica in rats by liquid chromatography–mass spectrometry. J Pharm Biomed Anal. 89:251–256. doi:10.1016/j.jpba.2013.11.014.
- Liu F, Huang X, He J, Song C, Peng L, Chen T, Wu BL. 2019. Plantamajoside attenuates inflammatory response in LPS-stimulated human gingival fibroblasts by inhibiting PI3K/AKT signaling pathway. Microb Pathog. 127:208–211. doi:10.1016/j.micpath.2018.11.034.
- Lukova P, Nikolova M, Petit E, Elboutachfaiti R, Vasileva T, Katsarov P, Manev H, Gardarin C, Pierre G, Michaud P, et al. 2020. Prebiotic activity of poly-and oligosaccharides obtained from Plantago major L. leaves. Appl Sci. 10(8):2648. doi:10.3390/app10082648.
- Mower JP, Guo W, Partha R, Fan W, Levsen N, Wolff K, Nugent JM, Pabón-Mora N, González F. 2021. Plastomes from tribe Plantagineae (Plantaginaceae) reveal infrageneric structural synapormorphies and localized hypermutation for Plantago and functional loss of ndh genes from Littorella. Mol Phylogenet Evol. 162:107217. doi:10.1016/j.ympev.2021.107217.
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. doi:10.1093/molbev/msu300.
- Noshad M, Alizadeh Behbahani B, Jooyandeh H, Rahmati Joneidabad M, Hemmati Kaykha ME, Ghodsi Sheikhjan M. 2021. Utilization of Plantago major seed mucilage containing Citrus limon essential oil as an edible coating to improve shelf‐life of buffalo meat under refrigeration conditions. Food Sci Nutr. 9(3):1625–1639. doi:10.1002/fsn3.2137.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):50. doi:10.1186/s13007-019-0435-7.
- Samuelsen AB. 2000. The traditional uses, chemical constituents and biological activities of Plantago major L. A review. J Ethnopharmacol. 71(1–2):1–21. doi:10.1016/s0378-8741(00)00212-9.
- Si H, Li R, Zhang Q, Liu L. 2022. Complete chloroplast genome of Plantago asiatica and its phylogenetic position in Plantaginaceae. Mitochondrial DNA Part B Resour. 7(5):819–821. doi:10.1080/23802359.2022.2073838.
- Sun H, Li S, Wang K. 2019. The complete chloroplast genome sequence of Plantago lagopus L. Mitochondrial DNA Part B. 4(1):382–383. doi:10.1080/23802359.2018.1547141.
- Wang Q, Mao LH, Ding B, Li MT, Fu ZX, Deng HP. 2020. The complete chloroplast genome of Plantago fengdouensis (Plantaginaceae): An endemic and endangered species from China. Mitochondrial DNA B Resour. 5(1):51–52. doi:10.1080/23802359.2019.1694851.
- Wu H, Zhao G, Jiang K, Chen X, Zhu Z, Qiu C, Li C, Deng G. 2016. Plantamajoside ameliorates lipopolysaccharide-induced acute lung injury via suppressing NF-κB and MAPK activation. Int Immunopharmacol. 35:315–322. doi:10.1016/j.intimp.2016.04.013.
- Yuan R, Ma X, Zhang Z, Gornall RJ, Wang Y, Chen S, Gao Q. 2023. Chloroplast phylogenomics and the taxonomy of Saxifraga section Ciliatae (Saxifragaceae). Ecol Evol. 13(1):e9694. doi:10.1002/ece3.9694.
- Zeng G, An H, Fang D, Wang W, Han Y, Lian C. 2022. Plantamajoside protects H9c2 cells against hypoxia/reoxygenation-induced injury through regulating the akt/Nrf2/HO-1 and NF-κB signaling pathways. J Recept Signal Transduct Res. 42(2):125–132. doi:10.1080/10799893.2020.1859534.
- Zhao F, Liu B, Liu S, Min D-Z, Zhang T, Cai J, Zhou X-X, Chen B, Olmstead RG, Xiang C-L, et al. 2023. Disentangling a 40-year-old taxonomic puzzle: the phylogenetic position of Mimulicalyx (Lamiales). Bot J Linn Soc. 201(2):135–153. doi:10.1093/botlinnean/boac033.
- Zhang S, Hu J, Sun Y, Tan H, Yin J, Geng F, Nie S. 2021. Review of structure and bioactivity of the Plantago (Plantaginaceae) polysaccharides. Food Chem X. 12:100158. doi:10.1016/j.fochx.2021.100158.
- Zhu A, Guo W, Gupta S, Fan W, Mower JP. 2016. Evolutionary dynamics of the plastid inverted repeat: the effects of expansion, contraction, and loss on substitution rates. New Phytol. 209(4):1747–1756. doi:10.1111/nph.13743.
