Abstract
Bird’s nest fungi involve six different genera, but only one of these genera (i.e. Cyathus) have available mitochondrial genomes (mitogenomes) to date. In this study, we report the first mitogenome in the genus Nidula with Nidula shingbaensis K. Das & R.L. Zhao 2013 as a representative. The mitogenome is a circular molecule of 65,793 bp with a GC content of 26.2%. There are a total of 43 genes, including 14 typical protein-coding genes, 26 tRNA genes, two rRNA genes, and one free-standing intergenic open reading frame (ORF). Three introns (two in cox1 and one in cob) are present in the mitogenome, with each containing an ORF encoding for a LAGLIDADG endonuclease. Phylogenetic analysis based on mitochondrial amino acid sequences confirms the phylogenetic placement of N. shingbaensis in Nidulariaceae in Agaricales. This study serves as a springboard for future investigation on fungal evolution in Nidulariaceae.
1. Introduction
Bird’s nest fungi are charismatic mushrooms resembling tiny egg-filled bird’s nests. As they are saprobic, feeding on decomposing substrates, they are often seen growing on mulch, woody chips, fallen logs, leaf litter, or animal dung (Kraisitudomsook and Smith Citation2024). All Bird’s nest fungi are currently classified in the family Nidulariaceae, including six genera, Cyathus, Crucibulum, Mycocalia, Nidula, Nidularia, and Retiperidiolia (Kraisitudomsook et al. Citation2022). These genera distinguished from each other by differences in morphology and peridiole structure. To date, there is no evidence to suggest that bird’s nest fungi are pathogenic to plants or toxic to humans or other animals. On the contrary, some species show significant neurotrophic and neuroprotective activities due to their production of cyathane diterpenoids (Qi et al. Citation2023).
The genus Nidula was originally described in 1902 and currently contains seven accepted species according to statistics in the Fungal Names database (Wang et al. Citation2023). Different from species in Cyathus or Crucibulum, the egg-like spore mass (known as peridiole) of Nidula does not connect to fruiting bodies (known as peridia) by special cords (known as funiculi) (Brodie Citation1975). Among Nidula species, Nidula shingbaensis K. Das & R.L. Zhao 2013 was described as new to science in 2013 (Das and Zhao Citation2013). It is currently known to be distributed in India and Thailand (Das and Zhao Citation2013; Kraisitudomsook et al. Citation2021).
So far, there have been four species in Nidulariaceae with available mitogenomes, and they are all from the genus Cyathus (Li et al. Citation2023). In this study, we report the first mitogenome in Nidula with N. shingbaensis as a representative. This study will provide a reference for future evolutionary studies among different genera in Nidulariaceae.
2. Materials and methods
2.1. Fungal materials
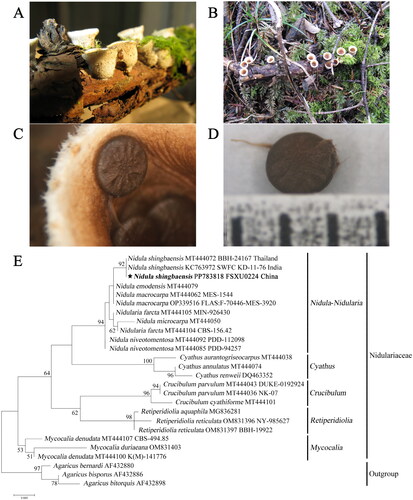
Specimens used in this study were derived from unknown fallen twigs from Shennongjia Forestry District (N 31.48°, E 110.31°), Hubei, China. A specimen was deposited at the Herbarium of Shanxi University (http://life.sxu.edu.cn/, contact person: Yongjie Zhang, [email protected]) under the voucher number FSXU0224. Both morphology and phylogenetic analysis based on nrDNA ITS sequences identified it as Nidula shingbaensis (). A culture was obtained, showing identical nrDNA ITS sequence with the fruiting body. The culture was cultivated on potato dextrose agar medium at 28 °C for 15 days, and mycelia were collected and used for DNA extraction.
Figure 1. Morphology and phylogenetic identification of the N. shingbaensis specimen used in this study. (A, B) Mature basidiocarps. (C, D) Small packets (called peridioles) within nest-shaped fruiting bodies. Photos of the specimen were taken by Yongjie Zhang. (E) Phylogenetic tree based on nrDNA ITS sequences, as implemented with the maximum-likelihood method in MEGA. Bootstrap support values are shown for nodes receiving values larger than 50%. Each fungal taxon is followed by the GenBank accession number of its nrDNA ITS sequence. For species within more than two voucher specimens, the voucher number is also followed. For the focal species N. shingbaensis, the country where the specimen was collected is further followed. The short bar at the bottom of the tree is the distance scale (scale bar = 0.020).
2.2. DNA sequencing, mitogenome assembly, and annotation
Genomic DNA was extracted using the cetyl trimethyl ammonium bromide method (Zhang et al. Citation2010). High-throughput sequencing was performed using an Illumina NovaSeq 6000 sequencing platform in PE150 mode (Novogene, Tianjin, China). Mitogenome sequence was de novo assembled using two programs NOVOPlasty (https://github.com/ndierckx/NOVOPlasty) (Dierckxsens et al. Citation2017) and GetOrganelle (https://github.com/Kinggerm/GetOrganelle) (Jin et al. Citation2020).
The mitogenome sequence was primarily annotated using MFannot (https://megasun.bch.umontreal.ca/apps/mfannot/) based on the mold mitochondrial genetic code (i.e. Genetic Code 4), but necessary manual corrections were also needed according to previous publications (Zhang et al. Citation2017; Ren et al. Citation2021). Open reading frames (ORFs) at intronic and intergenic regions were only considered if they were longer than 300 bp. Introns in protein-coding genes (PCGs) were named according to established nomenclatures (Zhang and Zhang Citation2019). The circular map of the mitogenome was visualized using OGDRAW (https://chlorobox.mpimp-golm.mpg.de/OGDraw.html) (Greiner et al. Citation2019). Sequencing depth and coverage plot were drawn according to an online protocol (https://protocols.io/view/generating-sequencing-depth-and-coverage-map-for-o-cswxwffn.html) (Ni et al. Citation2023). Map of cis-splicing genes was drawn using PMGmap (http://www.1kmpg.cn/pmgmap).
2.3. Phylogenetic position of N. shingbaensis in Agaricales
In order to investigate the phylogenetic position of N. shingbaensis, 33 Agaricales species and two Boletales species were chosen as ingroups and outgroups, respectively. Amino acids of the 14 typical PCGs (atp6, atp8, atp9; cob, cox1-3; nad1-6, and nad4L) present in fungal mitogenomes were concatenated and used for phylogenetic analysis. Phylogenetic relationships were estimated using both maximum-likelihood (ML) and Bayesian (BI) approaches, with identical settings described in our previous publication (Ren et al. Citation2021).
3. Results
3.1. Basic features of the N. shingbaensis mitogenome
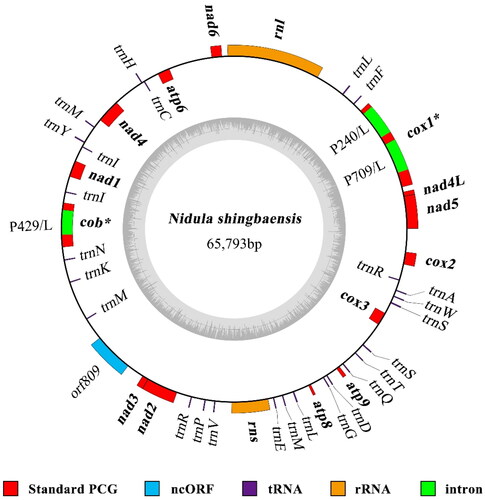
Both programs NOVOPlasty and GetOrganelle generated an identical circular mitogenome of 65,793 bp for N. shingbaensis, with an average sequencing depth of 1390× (Fig. S1). The mitogenome contains 43 genes, including two rRNA genes (rnl and rns), 26 tRNA genes, 14 typical PCGs associated with oxidative phosphorylation, and one intergenic ORF (orf809) encoding ribosomal protein S3 (). The 26 tRNAs encode all 20 canonical amino acids. The 14 typical PCGs encode seven subunits of NADH dehydrogenases (nad1-6 and nad4L), three cytochrome oxidase subunits (cox1-3), the cytochrome b (cob), and three ATP synthase subunits (atp6, atp8, and atp9). A total of three introns (all belonging to type I) are identified from the mitogenome, including two (named cox1P240 and cox1P709) in cox1 and one (named cobP429) in cob (Fig. S2). Each intron contains an intronic ORF coding for a LAGLIDADG endonuclease. It is noteworthy that intergenic regions (35,146 bp in total length, 53.4%) account for more than half of the mitogenome. No large-fragment duplications, introns, or plasmid-like sequences are present at these intergenic regions.
Figure 2. Circular map of the N. shingbaensis mitogenome. The outer ring indicates relative positions of different genes, and the inner ring indicates GC contents. Different types of genes/sequences are indicated by different colors as shown at the bottom of the figure. Blocks outside the ring represent forward strand genes, and blocks inside the ring represent reverse strand genes. The 14 standard PCGs and the two rRNA genes commonly found in fungal mitogenomes are shown in bold. Intron-containing genes are followed by an asterisk. For introns, standard intron names (with insertion site information) and functions of intronic ORFs (L, LAGLIDADG endonuclease) are given.
3.2. Phylogenetic position of N. shingbaensis in Agaricales
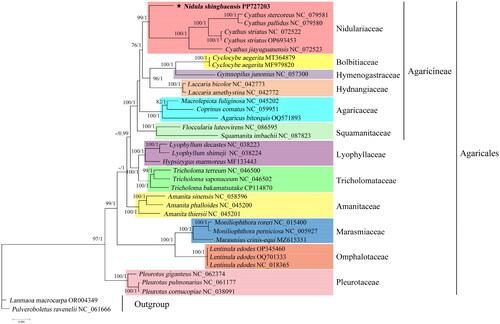
Based on the phylogenetic analysis, N. shingbaensis clustered with Cyathus stercoreus (Schwein.) De Toni 1888, Cyathus pallidus Berk. & M.A. Curtis 1869, Cyathus jiayuguanensis J. Yu, T.X. Zhou & L.Z. Zhao 2002 and Cyathus striatus Willd. 1787, forming the family Nidulariaceae with high support values (99% for ML and 1.00 for BI) (). Nidulariaceae further clustered with some other families like Bolbitiaceae and Hymenogastraceae to form the suborder Agaricineae in the order Agaricales. All other Agaricales species clustered according to their known family-level affiliations.
Figure 3. Phylogenetic tree constructed based on 14 core mitochondrial protein-coding genes. The phylogenetic tree shows the evolutionary relationship between N. shingbaensis and 33 other Agaricales species, with Lanmaoa macrocarpa and Pulveroboletus ravenelii as outgroups. This topology is the best topology constructed by Bayesian analysis. Two phylogeny-building methods (BI and ML) generate basically identical topologies. The node support value from ML is before the slash, and the node support value from BI is after the slash. Each fungal family is shown in different colors. The short bar at the bottom of the tree is the distance scale (scale bar = 0.050). Mitochondrial genomes used in the phylogenetic analysis were as follows, along with the corresponding references: Nidula shingbaensis PP727203 (this study), Cyathus stercoreus NC_079581, Cyathus pallidus NC_079580, Cyathus jiayuguanensis NC_072523, Cyathus striatus NC_072522, Cyathus striatus OP693453 (Li et al. Citation2023), Cyclocybe aegerita MT364879 (Liu et al. Citation2020), Cyclocybe aegerita MF979820 (Xu et al. Citation2017), Gymnopilus junonius NC_057300 (Cho et al. Citation2021), Laccaria bicolor NC_042773, Laccaria amethystina NC_042772 (Li et al. Citation2020), Macrolepiota fuliginosa NC_045202 (unpublished), Coprinus comatus NC_059951 (unpublished), Agaricus bitorquis OQ571893 (Zhao et al. Citation2023), Floccularia luteovirens NC_086595 (unpublished), Squamanita imbachii NC_087823 (unpublished), Lyophyllum decastes NC_038223, Lyophyllum shimeji NC_038224 (Li et al. Citation2019), Hypsizygus marmoreus MF133443 (unpublished), Tricholoma terreum NC_046500, Tricholoma saponaceum NC_046502 (Huang et al. Citation2021), Tricholoma bakamatsutake CP114870 (Ichida et al. Citation2023), Amanita sinensis NC_058596, Amanita phalloides NC_045200, Amanita thiersii NC_045201 (Li et al. Citation2023), Moniliophthora roreri NC_015400, Moniliophthora perniciosa NC_005927, Marasmius crinis-equi MZ615351 (Ali et al. Citation2021), Lentinula edodes OP345460 (Kim et al. Citation2022), Lentinula edodes OQ701333 (unpublished), Lentinula edodes NC_018365 (unpublished), Pleurotus giganteus NC_062374 (Liu et al. Citation2022), Pleurotus pulmonarius NC_061177 (unpublished), Pleurotus cornucopiae NC_038091 (Xu et al. Citation2018), Lanmaoa macrocarpa OR004349 (Zheng et al. Citation2023), and Pulveroboletus ravenelii NC_061666 (Cho et al. Citation2022).
4. Discussion and conclusions
The study represents the first documentation of N. shingbaensis in China and the first mitogenome in Nidula. The N. shingbaensis mitogenome at 65,793 bp is obviously smaller than those in Cyathus, which ranges from 114,236 bp in C. pallidus to 129,263 bp in C. stercoreus (Li et al. Citation2023). Phylogenetic analyses based on mitochondrial amino acid sequences confirm the placement of N. shingbaensis in Nidulariaceae (). Our results find that Nidulariaceae is sister to a clade of Bolbitiaceae, Hymenogastraceae, and Hydnangiaceae, with moderate to strong support (). This is different from previous studies, which showed that Nidulariaceae is sister to Squamanitaceae (Kraisitudomsook et al. Citation2021; Liu et al. Citation2021).
For inter-generic relationships of the six genera of bird’s nest fungi, there are inconsistencies among different studies (Kraisitudomsook et al. Citation2021, Citation2022, Citation2024). Together with this study, only two genera (Cyathia and Nidula) have available mitogenomes in Nidulariaceae. Mitogenomes from other genera of bird’s nest fungi need to be assembled in the future in order to provide evolutionary insights into inter-generic relationships in Nidulariaceae.
Ethics statement
The study did not involve humans or animals. In this study, samples can be collected without ethical approval or permission.
Author contributions
Y.J.Z. and S.Z. collected the sample and designed research; Q.Q. and Y.J.Z. performed research and analyzed data; Q.Q., S.Z., and Y.J.Z. wrote the paper. All authors agreed to be accountable for all aspects of the work.
Supplemental Material
Download PDF (315 KB)Acknowledgements
We thank the High-Performance Simulation Platform of Shanxi University for providing computing resource.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
BioProject, BioSample, and SRA accession numbers related to the sample of this study are PRJNA1104276, SAMN41074236, and SRR28795957, respectively. The mitochondrial genome and the nrDNA ITS sequence that support the findings of this study are openly available in the National Center for Biotechnology Information (NCBI) at https://www.ncbi.nlm.nih.gov, with accession numbers PP727203 and PP783818, respectively.
Additional information
Funding
References
- Ali SS, Amoako Attah I, Shao J, Kumi Asare E, Meinhardt LW, Bailey BA. 2021. Mitochondrial genomics of six cacao pathogens from the Basidiomycete family Marasmiaceae. Front Microbiol. 12:752094. doi:10.3389/fmicb.2021.752094.
- Brodie HJ. 1975. The bird’s nest fungi. Toronto and Buffalo: University of Toronto Press.
- Cho SE, Jo JW, Kwag YN, Lee H, Chung JW, Oh SH, Kim CS. 2021. Complete mitochondrial genome sequence of Gymnopilus junonius. Mitochondrial DNA B Resour. 6(3):1020–1021. doi:10.1080/23802359.2021.1895692.
- Cho SE, Kwag YN, Han SK, Lee DH, Kim CS. 2022. Complete mitochondrial genome sequence of Pulveroboletus ravenelii (Boletales, Basidiomycota). Mitochondrial DNA B Resour. 7(9):1581–1582. doi:10.1080/23802359.2022.2110006.
- Das K, Zhao RL. 2013. Nidula shingbaensis sp. nov., a new bird’s nest fungus from India. Mycotaxon. 125(1):53–58. doi:10.5248/125.53.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18. doi:10.1093/nar/gkw955.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64. doi:10.1093/nar/gkz238.
- Huang WL, Feng HY, Tu WY, Xiong C, Jin X, Li P, Wang X, Li Q. 2021. Comparative mitogenomic analysis reveals dynamics of intron within and between Tricholoma species and phylogeny of Basidiomycota. Front Genet. 12:534871. doi:10.3389/fgene.2021.534871.
- Ichida H, Murata H, Hatakeyama S, Yamada A, Ohta A. 2023. Near-complete de novo assembly of Tricholoma bakamatsutake chromosomes revealed the structural divergence and differentiation of Tricholoma genomes. G3. 13(11):jkad198. doi:10.1093/g3journal/jkad198.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241. doi:10.1186/s13059-020-02154-5.
- Kim S, Eom H, Nandre R, Choi YJ, Lee H, Ryu H, Ro HS. 2022. Comparative structural analysis on the mitochondrial DNAs from various strains of Lentinula edodes. Front Microbiol. 13:1034387. doi:10.3389/fmicb.2022.1034387.
- Kraisitudomsook N, Ahrendt S, Riley R, LaButti K, Lipzen A, Daum C, Barry K, Grigoriev IV, Rämä T, Martin F, et al. 2024. On the origin of bird’s nest fungi: phylogenomic analyses of fungi in the Nidulariaceae (Agaricales, Basidiomycota). Mol Phylogenet Evol. 193:108010. doi:10.1016/j.ympev.2024.108010.
- Kraisitudomsook N, Choeyklin R, Boonpratuang T, Pobkwamsuk M, Anaphon S, Smith ME. 2022. Hidden in the tropics: Retiperidiolia gen. nov., a new genus of bird’s nest fungi (Nidulariaceae), and a systematic study of the genus Mycocalia. Mycol Progress. 21(6):56. doi:10.1007/s11557-022-01807-y.
- Kraisitudomsook N, Healy RA, Smith ME. 2021. Molecular systematics and taxonomic overview of the bird’s nest fungi (Nidulariaceae). Fungal Biol. 125(9):693–703. doi:10.1016/j.funbio.2021.04.003.
- Kraisitudomsook N, Smith ME. 2024. Bird’s nest fungi: charismatic mushrooms in your garden. Gainesville: University of Florida Institute of Food and Agricultural Sciences; p. 361. doi:10.32473/edis-pp361-2020.
- Li Q, Luo YY, Sha AJ, Xiao WQ, Xiong Z, Chen XD, He J, Peng LX, Zou L. 2023. Analysis of synonymous codon usage patterns in mitochondrial genomes of nine Amanita species. Front Microbiol. 14:1134228. doi:10.3389/fmicb.2023.1134228.
- Li Q, Wang QF, Jin X, Chen ZQ, Xiong C, Li P, Zhao J, Huang WL. 2019. Characterization and comparison of the mitochondrial genomes from two Lyophyllum fungal species and insights into phylogeny of Agaricomycetes. Int J Biol Macromol. 121:364–372. doi:10.1016/j.ijbiomac.2018.10.037.
- Li Q, Yang LX, Xiang DB, Wan Y, Wu Q, Huang WL, Zhao G. 2020. The complete mitochondrial genomes of two model ectomycorrhizal fungi (Laccaria): features, intron dynamics and phylogenetic implications. Int J Biol Macromol. 145:974–984. doi:10.1016/j.ijbiomac.2019.09.188.
- Li ZC, Xie TC, Feng XL, Wang ZX, Lin C, Li GM, Li XZ, Qi JZ. 2023. The first five mitochondrial genomes for the family Nidulariaceae reveal novel gene rearrangements, intron dynamics, and phylogeny of Agaricales. Int J Mol Sci. 24(16):1–22. doi:10.3390/ijms241612599.
- Liu JW, Ge ZW, Horak E, Vizzini A, Halling RE, Pan CL, Yang ZL. 2021. Squamanitaceae and three new species of Squamanita parasitic on Amanita basidiomes. IMA Fungus. 12(1):4. doi:10.1186/s43008-021-00057-z.
- Liu XR, Wu XP, Tan H, Xie BG, Deng YJ. 2020. Large inverted repeats identified by intra-specific comparison of mitochondrial genomes provide insights into the evolution of Agrocybe aegerita. Comput Struct Biotechnol J. 18:2424–2437. doi:10.1016/j.csbj.2020.08.022.
- Liu ZL, Wu SJ, Chen XF, Zhang WL, Zhou SY, Wang XG. 2022. The complete mitochondrial genome of the edible mushroom Pleurotus giganteus (Agaricales, Pleurotus) and insights into its phylogeny. Mitochondrial DNA B Resour. 7(7):1313–1315. doi:10.1080/23802359.2022.2096418.
- Ni Y, Li JL, Zhang C, Liu C. 2023. Generating sequencing depth and coverage map for organelle genomes. protocols.io. doi:10.17504/protocols.io.4r3l27jkxg1y/v1.
- Qi JZ, Gao YQ, Kang SJ, Liu CW, Gao JM. 2023. Secondary metabolites of bird’s nest fungi: chemical structures and biological activities. J Agric Food Chem. 71(17):6513–6524. doi:10.1021/acs.jafc.3c00904.
- Ren LY, Zhang S, Zhang YJ. 2021. Comparative mitogenomics of fungal species in Stachybotryaceae provides evolutionary insights into Hypocreales. Int J Mol Sci. 22(24):13341. doi:10.3390/ijms222413341.
- Wang F, Wang K, Cai L, Zhao MJ, Kirk PM, Fan GM, Sun QL, Li B, Wang S, Yu ZF, et al. 2023. Fungal names: a comprehensive nomenclatural repository and knowledge base for fungal taxonomy. Nucleic Acids Res. 51(D1):D708–D716. doi:10.1093/nar/gkac926.
- Xu LM, Hinsinger DD, Jiang GF. 2017. The complete mitochondrial genome of the Agrocybe aegerita, an edible mushroom. Mitochondrial DNA B Resour. 2(2):791–792. doi:10.1080/23802359.2017.1398618.
- Xu LM, Hinsinger DD, Jiang GF. 2018. The complete mitochondrial genome of the Basidiomycete fungus Pleurotus cornucopiae (Paulet) Rolland. Mitochondrial DNA B Resour. 3(1):73–75. doi:10.1080/23802359.2017.1422405.
- Zhang S, Zhang YJ. 2019. Proposal of a new nomenclature for introns in protein-coding genes in fungal mitogenomes. IMA Fungus. 10(1):15. doi:10.1186/s43008-019-0015-5.
- Zhang YJ, Yang XQ, Zhang S, Humber RA, Xu J. 2017. Genomic analyses reveal low mitochondrial and high nuclear diversity in the cyclosporin-producing fungus Tolypocladium inflatum. Appl Microbiol Biotechnol. 101(23–24):8517–8531. doi:10.1007/s00253-017-8574-0.
- Zhang YJ, Zhang S, Liu XZ, Wen HA, Wang M. 2010. A simple method of genomic DNA extraction suitable for analysis of bulk fungal strains. Lett Appl Microbiol. 51(1):114–118. doi:10.1111/j.1472-765X.2010.02867.x.
- Zhao C, Feng XL, Wang ZX, Qi J. 2023. The first whole genome sequencing of Agaricus bitorquis and its metabolite profiling. J Fungi. 9(4):485. doi:10.3390/jof9040485.
- Zheng YT, Chen LL, Zhao K. 2023. Complete mitochondrial genome sequence of Lanmaoa macrocarpa (Boletales, Basidiomycota). Mitochondrial DNA B Resour. 8(10):1067–1070. doi:10.1080/23802359.2023.2266231.
