Abstract
Singerocybe alboinfundibuliformis (Seok et al.) Yang, Qin & Takah 2014 is an edible mushroom distributed in several East or Southeast Asian countries. Herein, we report the mitochondrial genome of S. alboinfundibuliformis based on Illumina sequencing data. The overall length of the mitochondrial genome is 64,279 bp, with a GC content of 29.0%. It contains 14 typical protein-coding genes, 27 tRNA genes, two rRNA genes, and 13 intergenic ORFs. Most of these genes (39 out of 56) are transcribed at the forward strand, and few (17 out of 56) are transcribed at the reverse strand. Among these genes, only the rnl gene is invaded by an intron, and all other genes are intron-free. Phylogenetic analysis based on mitochondrial amino acid sequences supports the phylogenetic position of S. alboinfundibuliformis in Clitocybaceae, being close to Lepista sordida (Schumach.) Singer 1951. This study serves as a springboard for future investigation on fungal evolution in Clitocybaceae.
1. Introduction
Singerocybe is a fungal genus whose members are saprotrophic and usually found in habitats like soils rich in humus, dead wood, or rotting leaves (Kumla et al. Citation2016). To date, only seven species are recognized in the genus with most being geographically restricted in their distributions (Qin et al. Citation2014). Singerocybe alboinfundibuliformis (Seok et al.) Yang, Qin & Takah (2014) was first described as a new species under the name Clitocybe alboinfundibulliforme Seok, Kim, Park, Kim, Yoo & Park (Citation2009) from Korea (Seok et al. Citation2009). Based on molecular phylogenetic analysis, it was later transferred to the genus Singerocybe (Qin et al. Citation2014). The fungus is currently known from several different countries in East or Southeast Asia, including China, Japan, Korea, and Thailand (Seok et al. Citation2009; Qin et al. Citation2014; Kumla et al. Citation2016). In China, the fungus has been known from several different provinces, including Guizhou, Yunnan, Shanxi, Shandong, Hubei, Jilin, Liaoning, and Beijing (Qin et al. Citation2014; Deng Citation2016; Liu et al. Citation2022; He et al. Citation2023), and it is often eaten by local mycophagists in Guizhou and Yunnan Provinces, China (Deng Citation2016; Yang et al. Citation2021).
The genus Singerocybe has been the subject of controversy since its proposal in 1988. For a long time, its family-level affiliation has been either in Tricholomataceae or undetermined (Qin et al. Citation2014). A recent study suggested Singerocybe as a member of the family Clitocybaceae based on molecular phylogenetic analyses (He et al. Citation2023). Currently, there is no available genome information for species in Singerocybe. There are a total of 192 nucleotide records in GenBank (as of 20 March 2024), mainly focusing on nuclear ribosomal DNA fragments and protein-coding genes that are commonly used as markers in molecular phylogeny.
Mitochondrial DNA can provide insights into fungal evolution and phylogeny (Bullerwell and Lang Citation2005). In this study, we assembled the mitochondrial genome (mitogenome) of Singerocybe alboinfundibuliformis and deduced its phylogenetic position within Agaricales based on mitochondrial amino acid sequences.
2. Materials and methods
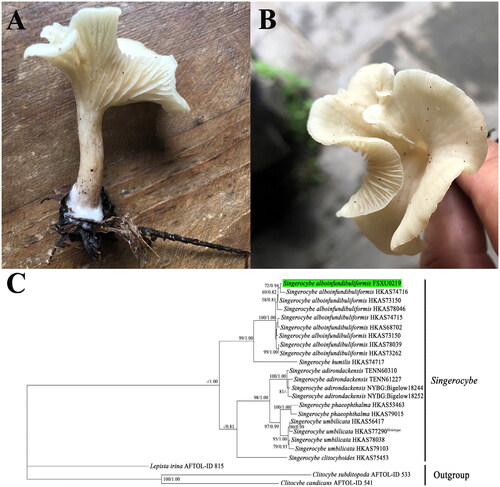
The specimen of S. alboinfundibuliformis used in this study was collected from Mountain Taishan (37°44′57″N, 112°26′26″E) located in Taiyuan, Shanxi Province, China. A voucher specimen was deposited in the herbarium at Shanxi University (Deposit No. FSXU0219; contact person: Yongjie Zhang, [email protected]). Identification of the specimen was verified by both morphology and molecular phylogeny ().
Figure 1. Morphology and phylogenetic identification of S. alboinfundibuliformis. (A, B) Side and top view of the S. alboinfundibuliformis fruiting body. The photograph was taken by Yongjie Zhang from Mountain Taishan in Taiyuan (37°44′57″N, 112°26′26″E), Shanxi Province, China. (C) ML and BI phylogenetic tree of Singerocybe species based on the concatenated dataset of four nuclear DNA fragments, namely nrDNA ITS, nrDNA LSU, RPB2, and TEF1, with Clitocybe subditopoda, Clitocybe candicans, and Lepista irina as outgroups. Bootstrap values (BP) ≥50% from ML analysis and Bayesian posterior probabilities (PP) ≥0.80 from BI analysis are shown at internal nodes. The specimen from this study is highlighted. Among the seven known species in Singerocybe, the type species of the genus, Singerocybe viscida, has no available DNA sequence and therefore is not included in the phylogenetic analysis.
Total DNA was extracted from the specimen using the CTAB (cetyl trimethyl ammonium bromide) method (Zhang et al. Citation2010). DNA library of 350-bp insert size was constructed and then used for sequencing using an Illumina NovaSeq 6000 platform at Novogene Co. Ltd. (Beijing, China). Mitochondrial sequences were de novo assembled using GetOrganelle v1.7.5 (Jin et al. Citation2020) and NOVOPlasty v4.3.1 (Dierckxsens et al. Citation2017). For GetOrganelle, default parameters were used. For NOVOPlasty, we set a genome range of 2000–100,000 and used the output sequences resulting from GetOrganelle as the seed input. Mitogenome sequence was annotated by referring to those described previously (Zhang et al. Citation2017; Ren et al. Citation2021). The circular map of the mitogenome was visualized using OGDRAW (Greiner et al. Citation2019).
In order to know the phylogenetic position of S. alboinfundibuliformis in Agaricales, amino acids of the 14 typical protein-coding genes (atp6, atp8, atp9; cob, cox1-3; nad1-6, and nad4L) were used for phylogenetic analysis. Except for the sequences of S. alboinfundibuliformis reported in the study, sequences of 36 additional fungal species in Agaricales were used. Two species in Tremellales, namely Cryptococcus gattii (Vanbreus. & Takashio) Kwon-Chung & Boekhout 2002 and Cryptococcus neoformans (San Felice) Vuill. 1901, were employed as outgroups. Phylogenetic relationships were estimated using both maximum-likelihood (ML) and Bayesian (BI) approaches as implemented in IQ-TREE v1.6.12 (Nguyen et al. Citation2015) and MrBayes v3.2.7 (Ronquist et al. Citation2012), respectively, with identical settings described in our previous publication (Ren et al. Citation2021).
3. Results
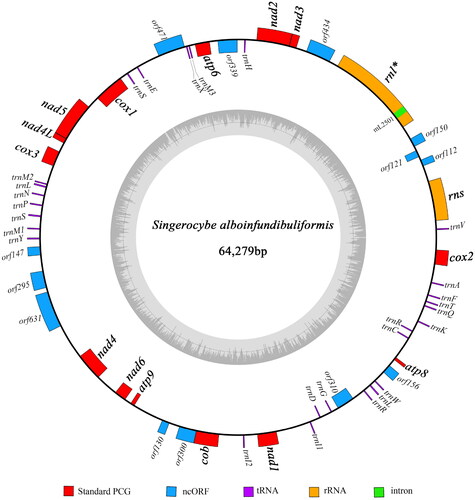
We employed both NOVOPlasty and GetOrganelle to assemble the mitogenome of S. alboinfundibuliformis, and they generated identical sequences of 64,279 bp in length, with a GC content of 29.0% and an average sequencing depth of 2558× (; Fig. S1). The S. alboinfundibuliformis mitogenome contains 14 protein-coding genes of the oxidative phosphorylation system (atp6, atp8, atp9, cob, cox1, cox2, cox3, nad1, nad2, nad3, nad4, nad4L, nad5, and nad6), two rRNA genes (rns and rnl), 27 tRNA genes, and 13 intergenic open reading frames (encoding ribosomal protein S3, DNA polymerase, or hypothetical protein). Most of these genes (39 out of 56 genes) are transcribed at the forward strand, and others (17 out of 56) are transcribed at the reverse strand (). Among these genes, there is only one intron-containing gene (i.e. rnl), and other genes are not invaded by an intron. The rnl intron (designated as mL2501) is short in length (only 350 bp) and does not encode an intronic ORF (Fig. S2). It is noteworthy that intergenic region (31,722 bp, 49.3%) accounts for half of the mitogenome.
Figure 2. Circular map of the S. alboinfundibuliformis mitogenome. The outer ring marks relative positions of different genes, and the inner ring represents GC contents. Blocks outside the ring represent forward strand genes, and blocks inside the ring represent reverse strand genes. Different kinds of genes are shown in different colors. The 14 protein-coding genes typically found in fungal mitogenomes and the two rRNA genes are shown in bold. Intron-containing genes are followed by asterisks after gene names.
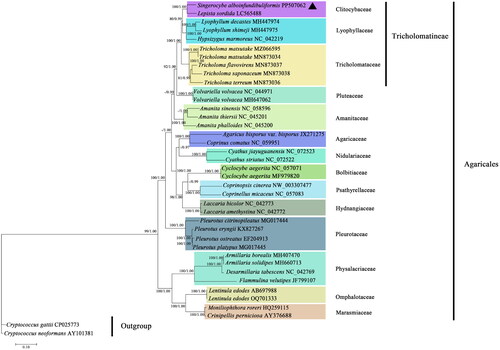
Phylogenetic analyses based on mitochondrial amino acid sequences showed that S. alboinfundibuliformis clustered together with Lepista sordida (Schumach.) Singer 1951 with 100% bootstrap value (), forming the Clitocybaceae clade. The families Clitocybaceae, Lyophyllaceae, and Tricholomataceae further clustered together to form the suborder Tricholomatineae in the order Agaricales. All other Agaricales species clustered according to their known family-level affiliations.
Figure 3. Phylogenetic tree constructed based on 14 core protein-coding genes. The phylogenetic tree shows the evolutionary relationships between S. alboinfundibuliformis and 36 other Agaricales species, with Cryptococcus gattii and Cryptococcus neoformans as the outgroups. The tree shown here was the single best topology recovered from ML, and the topology was identical to that recovered from BI. Support values from ML (before forward slash) and BI (after forward slash) analyses are given for nodes receiving strong supports (i.e. ML bootstrap values ≥70% or BI posterior probability ≥0.95). The sample resulting from this study is denoted in triangle.
4. Discussion and conclusions
In this study, the mitogenome of S. alboinfundibuliformis was sequenced and annotated for the first time. This study represents the first mitogenome in the genus Singerocybe. Our phylogenetic analyses based on mitochondrial sequences support the placement of the fungus within the family Clitocybaceae. This clustering is consistent with that reported in a recent study (He et al. Citation2023).
As for the family Clitocybaceae, six genera are currently recognized, namely Clitocybe, Collybia, Dendrocollybia, Lepista, Pseudolyophyllum, and Singerocybe (He et al. Citation2023). Currently, Lepista and Singerocybe have available mitogenome information. Other four genera still lack an available mitogenome. In addition, the taxonomy of the suborder Tricholomatineae has been recently revised to include 12 families (Vizzini et al. Citation2024); however, only three of these families have available mitogenome information. To gain evolutionary insights into fungal evolution in Clitocybaceae and Tricholomatineae, mitogenomes from more species are expected in the future.
Ethical approval
Specimen collection conformed to international ethical requirements, and did not cause damage to the local environment. Both the process and purpose of this experimental research were in line with rules and regulations of our institute. There were no ethical issues or other conflicts of interest related to this study.
Author contributions
Y.J.Z. and S.Z. collected the sample and designed research; N.X. and Y.J.Z. performed research and analyzed data; N.X., S.Z., and Y.J.Z. wrote the paper. All authors have read and agreed to the published version of the manuscript.
Supplemental Material
Download MS Word (1.3 MB)Acknowledgements
We thank the High-Performance Simulation Platform of Shanxi University for providing computing resource.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
BioProject, BioSample, and SRA accession numbers related to the sample of this study are PRJNA1090458, SAMN40566090, and SRR28409254, respectively. The mitochondrial genome data that support the findings of this study are openly available in the Nation Center for Biotechnology Information (NCBI) at https://www.ncbi.nlm.nih.gov, with an accession number PP507062.
Additional information
Funding
References
- Bullerwell CE, Lang BF. 2005. Fungal evolution: the case of the vanishing mitochondrion. Curr Opin Microbiol. 8(4):362–369. doi:10.1016/j.mib.2005.06.009.
- Deng CY. 2016. A new genus recorded to Guizhou—Singerocybe. Guizhou Sci. 34(1):29–31.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18. doi:10.1093/nar/gkw955.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64. doi:10.1093/nar/gkz238.
- He Z, Chen Z, Bau T, Wang G, Yang Z. 2023. Systematic arrangement within the family Clitocybaceae (Tricholomatineae, Agaricales): phylogenetic and phylogenomic evidence, morphological data and muscarine-producing innovation. Fungal Divers. 123(1):1–47. doi:10.1007/s13225-023-00527-2.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241. doi:10.1186/s13059-020-02154-5.
- Kumla J, Suwannarach N, Vadthanarat S, Raspé O, Lumyong S. 2016. First report of Singerocybe in Thailand. Mycotaxon. 131(1):205–209. doi:10.5248/131.205.
- Liu H, Zhang Z, Dong S, Bai Y. 2022. New records of mushrooms in Shanxi: Descolea quercina and Singerocybe alboinfundibuliformis. Edible Fungi. 44(6):21–23, 26.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. doi:10.1093/molbev/msu300.
- Qin J, Feng B, Yang ZL, Li YC, Ratkowsky D, Gates G, Takahashi H, Rexer KH, Kost GW, Karunarathna SC. 2014. The taxonomic foundation, species circumscription and continental endemisms of Singerocybe: evidence from morphological and molecular data. Mycologia. 106(5):1015–1026. doi:10.3852/13-338.
- Ren LY, Zhang S, Zhang YJ. 2021. Comparative mitogenomics of fungal species in Stachybotryaceae provides evolutionary insights into Hypocreales. Int J Mol Sci. 22(24):13341. doi:10.3390/ijms222413341.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. doi:10.1093/sysbio/sys029.
- Seok SJ, Kim YS, Park KM, Kim WG, Yoo KH, Park IC. 2009. New species of Agaricales. Mycobiology. 37(4):295–299. doi:10.4489/MYCO.2009.37.4.295.
- Vizzini A, Alvarado P, Consiglio G, Marchetti M, Xu J. 2024. Family matters inside the order Agaricales: systematic reorganization and classification of Incertae sedis clitocyboid, pleurotoid and tricholomatoid taxa based on an updated 6-gene phylogeny. Stud Mycol. 107(1):67–148. doi:10.3114/sim.2024.107.02.
- Yang ZL, Wu G, Li YC, Wang XH, Cai Q. 2021. Common edible and poisonous mushrooms of Southwestern China. Beijing: Science Press.
- Zhang YJ, Yang XQ, Zhang S, Humber RA, Xu J. 2017. Genomic analyses reveal low mitochondrial and high nuclear diversity in the cyclosporin-producing fungus Tolypocladium inflatum. Appl Microbiol Biotechnol. 101(23–24):8517–8531. doi:10.1007/s00253-017-8574-0.
- Zhang YJ, Zhang S, Liu XZ, Wen HA, Wang M. 2010. A simple method of genomic DNA extraction suitable for analysis of bulk fungal strains. Lett Appl Microbiol. 51(1):114–118. doi:10.1111/j.1472-765X.2010.02867.x.
