Abstract
Described originally from Heilongjiang, China, Odontothrips phaseoli is a potential pest of threatening bean plant in northern China. The complete mitochondrial genome of O. phaseoli was sequenced and assembled, with a total length of 15,540 bp. Within this genome, 37 genes have been identified: 13 PCGs, 22 tRNAs, two rRNAs, and two putative control regions. Most PCGs terminate with TAA, while four genes (atp8, nad1, nad2 and nad4) use an incomplete ‘T’ and nad6 employs TAG as the stop codon. Compared to the mitogenome of the ancestral insect, O. phaseoli displays significant gene rearrangement. However, it retains three conserved gene blocks in common with its related species, Megalurothrips usitatus, both of which belong to the Megalurothrips genus-group. The phylogenetic tree, constructed based on the entire mitogenome dataset of all thrips species available in NCBI, shows that the two species cluster closely together. This alignment might underscore the close link between gene arrangements and the phylogeny relationships.
Introduction
The second most species-rich subfamily in Thysanoptera, Thripinae includes 1785 extant species in 231 genera (ThripsWiki Citation2023), of which many members are most of pest thrips and all of the tospovirus vectors. The species of flower-living genus Odontothrips are known as damaging alfalfa and vegetables of Fabaceae. Especially, Odontothrips phaseoli, described from Heilongjiang, China by Kurosawa (Citation1941), is now considered as an important pest of bean plant of Phaseolus in northern China. In addition, in the eight genera of the Megalurothrips genus-group, Megalurothrips and Odontothrips are particularly closely related and confused (Mound and Palmer Citation1981; Xie et al. Citation2010). In morphology, the species of these genera share a pair of dorso-apical setae on the first antennal segment which is unusual in Thripinae (Hakimara and Minaei Citation2019), but it is difficult to distinguish them by morphology. Currently, the mitogenome of insect, as a molecular marker, is helpful to analyze the relationships of tribes and genera in many insects (Chen et al. Citation2019; Zhou et al. Citation2020; Chen et al. Citation2021; Wu et al. Citation2022). In Thysanoptera, some attempts were made to reveal phylogenetic and evolutionary relationships among genera and species with mitogenome data (Chakraborty et al. Citation2018; Kumar et al. Citation2019; Liu et al. Citation2022; Pakrashi et al. Citation2023). In NCBI, only one mitogenome of Megalurothrips, M. usitatus, is available but none of Odontothrips (Xing-Ming et al. Citation2023). To help understanding the relationship among them, the complete mitochondrial genome of O. phaseoli was sequenced, and the phylogenetic tree was constructed.
Materials and methods
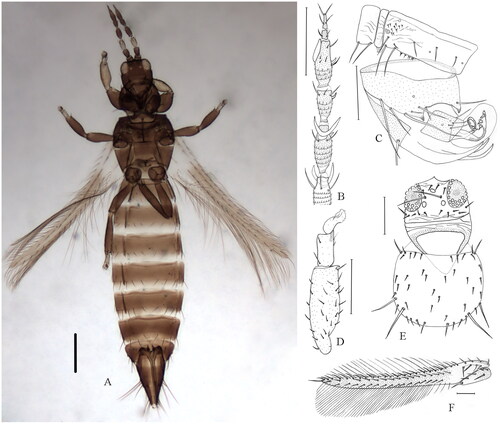
The samples of O. phaseoli were collected from flowers of Phaseolus in Shaanxi Province of China in 2021 (33°86′88″N, 109°94′74″E), and were preserved in 95% ethanol and stored at −20 °C for DNA extraction. The voucher specimens (No. JM2021030) were deposited at the School of Biological Science and Engineering, Shaanxi University of Technology (L.H. Dang, [email protected]). The diagnosis of O. phaseoli is as follows (): ocellar setae S1 present, S2 much elongate; antennal segment VI with inner sense cone having a wide base at least one-third as long as the length of inner margin of this segment; pronotum with two pairs of major setae; fore tarsi without apical tubercle on inner surface; forewing brown with pale at base, first vein with 4 + 11–17 + 2 setae; fore tibiae with a large apical claw ventrally; antennal segment only III pale, other segments uniform brown; abdominal sternites without discal setae, IV with three pairs of posteromarginal setae arising at margin, VII setal pair S1 arise in front of margin, S2 and S3 arise at margin; male with 3 endothecal spines at and near apex of each canaliculus.
Figure 1. Dorsal view of O. phaseoli. (A) Body, female; (B) Antennal segments III–VIII; (C) Male tergite VIII–X with three endothecal spines at and near the apex of each canaliculus; (D) Fore tibiae and tarsal; (E) Head and pronotum; (F) Forewing. Scale bars = 100μm. Identified and photographed by Lihong Dang.
Total genomic DNA was extracted from a single thrips sample using the Biomarker Micro Cell/Tissue DNA Kit. And sent to BerryGenomics (Beijing, China) forsequencing, which was conducted using the Illumina NovaSeq 6000 platform (San Diego, CA, USA) with 150 bp paired-end reads. Finally, a total of 4.24 Gb clean data was used to assembly by NOVOPlasty (Dierckxsens et al. Citation2017) with default parameters. PCGs and RNAs of the complete mitogenome was firstly predicted using MITOS2 web server (Bernt et al. Citation2013), and then manually annotated and corrected using Geneious v. R9 software (Kearse et al. Citation2012). The AT-rich control regions were identified via boundaries of adjacent genes. Nucleotide composition, codon usage was estimated in PhyloSuite v1.2.2 (Zhang et al. Citation2020). Circular map of the mitogenome was drawn with CGView Server (Grant and Stothard Citation2008).
The whole mitogenome dataset (13PCG + 22tRNA + 2rRNA) of 34 thrips species including the target species were aligned by MAFFT v7.313 (Katoh and Standley Citation2013) in PhyloSuite v1.2.2 (Zhang et al. Citation2020), with Aphis gossypii (Zhang et al. Citation2016) and Alloeorhynchus bakeri (Li et al. Citation2012) as outgroups. MACSE (Ranwez et al. Citation2018) was utilized to optimize the alignments of protein-coding genes, while the protein-coding genes and RNA genes were trimmed by Gblocks (Talavera and Castresana Citation2007). BI (Bayesian Inference) tree was reconstructed by MrBayes (Ronquist et al. Citation2012). The resultant tree was visualized in iTOL (https://itol.embl.de/).
Results
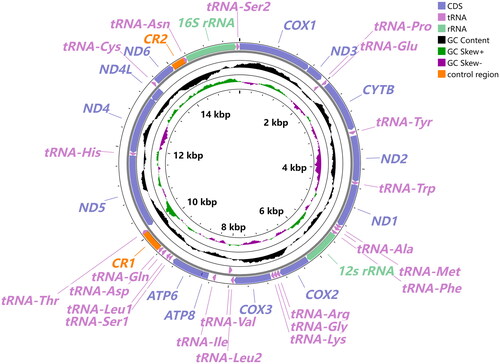
The whole mitochondrial genome of O. phaseoli has a total length of 15,540 bp (GenBank: OR593754; NCBI Reference Sequence: NC084197), including 37 genes and two putative control regions (CR1 and CR2) (). Six genes (nad5, nad4, nad4L, trnH, trnV and trnP) are encoded in the L-strand, while the others were encoded in the H-strand (Table S1). There are 18 intergenic spaces and six pairs of genes overlap in the entire mitogenome (Table S1). The nucleotide composition of this mitogenome was 78.8% A + T content. The total length of the 13 PCGs, ranging from 160 bp (atp8) to 1695 bp (nad5), was 10,953bp, in which all PCGs used ATN start codons, but most of them used TAA or TAG as a stop codon while four genes (atp8, nad1, nad2 and nad4) have an incomplete ‘T’. All 22 tRNAs were found in this mitogenome, with a total length of 1,435bp ranging from 56 bp (trnS1) to 80 bp (trnV) in size (Table S1). Most tRNAs have a typical clover-leaf secondary structure with one exception, trnS1, lacking the DHU arm (Figure S2). Two rRNA genes have a full length of 775 bp in rrnS which is located between trnF and cox2, and of 1,088bp in rrnL that is located between trnN and trnS2. The coverage depth of O. phaseoli is shown in Figure S1.
Figure 2. The circular representation of the complete mitogenome of O. phaseoli. The innermost and Middle circles depict the GC-skew and GC content, respectively. The outermost circle indicates the arrangements of genes: inner genes from the reverse strand, and outer genes from the forward strand, with PCGs in bluish violet, rRNAs in light green, and tRNAs in light purple. Different colors are used to show different functional categories, as shown in the upper right of the picture.
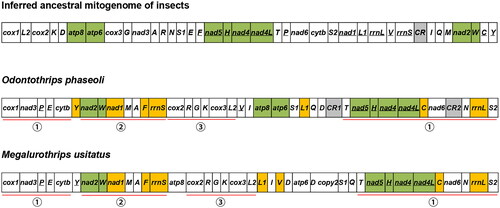
When comparing the mitochondrial gene order of O. phaseoli to the inferred insect ancestral arrangement it displayed significant rearrangement differences (Clary and Wolstenholme Citation1985) (). But three gene blocks (atp8-atp6, nad5-trnH-nad4-nad4L and nad2-trnW) are conserved in O. phaseoli (). A comparison between the mitochondrial gene arrangements of O. phaseoli and its related species, Megalurothrips usitatus, shows that they both share three conserved gene blocks ( - ①, ②, ③).
Figure 3. Mitochondrial gene arrangements in the ancestor insect, O. phaseoli, and M. usitatus. Conserved gene blocks are highlighted in green, and the inverted genes in orange.
The phylogeny confirms the monophyly of two suborders, Tubulifera and Terebrantia. Moreover, O. phaseoli was clustered to Megalurothrips usitatus with a high support value (PP = 1), which is consistent with previous phylogenetic studies () (Mound and Palmer Citation1981). Especially, the results of DNA barcoding studies supported that these two species were also close related (Rebijith et al. Citation2014). The two genera that belong to the Megalurothrips genus-group share the morphological characteristics: antennae 8-segmented, ocellar setae pair I present, median metanotal setae at anterior margin, metanotal spinula absent, tergite VIII with posteromarginal comb usually interrupted, and sternal discal setae absent, which might be a synapomorphy for this group (Hakimara and Minaei Citation2019).
Figure 4. BI phylogenetic tree inferred from the whole mitogenome. The tree was constructed based on concatenated nucleotide sequences of the whole mitogenome dataset (13PCG + 22tRNA + 2rRNA) of 36 species in 6 families. The numbers under the internodes represent Bayesian inference (BI) posterior probabilities (PP). The scale bar refers to 0.1 nucleotide substitutions per character. The used sequences and their references were listed in (Table S3): Alloeorhynchus bakeri (HM235722; Li et al. Citation2012), Aphis gossypii (KJ669654; Zhang et al. Citation2016), Acaciothrips ebneri (MZ645927; not available), Bactrothrips quadrituberculatus (MW233591; not available), Elaphrothrips spiniceps (OP919351; not available), Megathrips lativentris (OP723481; not available), Gynaikothrips ficorum (MT892761; Dang et al. Citation2021), Gynaikothrips uzeli (MK940484; Tyagi et al. Citation2020), Haplothrips aculeatus (KP198620; not available), Psephenothrips eriobotryae (MW793907; Dang et al. Citation2024), Holarthrothrips indicus (MN072397; Tyagi et al. Citation2020), Aeolothrips indicus (MW899051; Pakrashi et al. Citation2021), Aeolothrips xinjiangensis (MW376485; Liu et al. Citation2022), Franklinothrips vespiformis (MN072395; Tyagi et al. Citation2020), Anaphothrips obscurus (KY498001; Liu et al. Citation2017), Frankliniella panamensis (OR060662; not available), Dendrothrips minowai (MF582634; Chen et al. Citation2017), Frankliniella intonsa (JQ917403; Yan et al. Citation2014), Frankliniella occidentalis (JN835456; Yan et al. Citation2012), Scirtothrips hansoni (OR044712; not available), Neohydatothrips samayunkur (MF991901; Kumar et al. Citation2019), Mycterothrips gongshanensis (MZ913437; not available), Rhipiphorothrips cruentatus (MN072396;Tyagi et al. Citation2020), Scirtothrips dorsalis EA1 (KM349826; Dickey et al. Citation2015), Scirtothrips dorsalis SA1 (KM349827, KM349828; Dickey et al. Citation2015), Odontothrips phaseoli (OR593754; this study), Stenchaetothrips biformis (ON653412; Hu et al. Citation2023), Taeniothrips tigris (MW751816; Pakrashi et al. Citation2021), Thrips imagines (AF335993; Shao and Barker Citation2003), Thrips hawaiiensis (MW582621; Wang et al. Citation2021), Thrips palmi (MH253898; Chakraborty et al. Citation2018), Thrips setosus (MN148452; not available), Opimothrips tubulatus (MN787503; not available), Megalurothrips usitatus (ON815612; Xing-Ming et al. Citation2023), Aptinothrips stylifer (OQ559124; Li et al. Citation2024), Pseudodendrothrips mori (MN167468; not available).

Discussion and conclusion
In this study, the complete mitochondrial genome of Odontothrips phaseoli was characterized, in which 37 genes and two putative CRs are recognized in a total length of 15,540bp. Compared to the primitive ancestral mitogenome of arthropods, O. phaseoli displayed significant gene rearrangement differences. In addition, there is a noticeable similarity in the occurrence of gene transpositions between O. phaseoli and M. usitatus. The phylogenetic tree showed that O. phaseoli was robustly clustered with M. usitatus, affirming its affiliation within the Megalurothrips genus-group. Furthermore, this alignment might underscore the close link between gene arrangements and the phylogeny relationships.
Authors contributions
D-X W, C-W L wrote the original manuscript, and analyzed the data; L-H D revised and edited the manuscript, conceived and designed the project, and approved the version to be published. All authors agree to be accountable for all aspects of the work.
Ethics statement
No special permits were required for the insect specimens collected for this study. No endangered or protected species were involved in the field study. The insect species sequenced is a common thrips in China, which is not included in the List of Protected Animals in China.
Table S1.docx
Download MS Word (26.9 KB)Figure S2.docx
Download MS Word (9.1 MB)Table S3.docx
Download MS Word (27.2 KB)Table S2.docx
Download MS Word (23.8 KB)Figure S1.docx
Download MS Word (128.6 KB)Data availability statement
The data that support the findings of this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov/genbank/ under the accession no. OR593754. The associated BioProject, Bio-Sample numbers, and SRA are PRJNA1021028, SAMN37542142, and SRR26222581, respectively.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. doi:10.1016/j.ympev.2012.08.023.
- Chakraborty R, Tyagi K, Kundu S, Rahaman I, Singha D, Chandra K, Patnaik S, Kumar V. 2018. The complete mitochondrial genome of Melon thrips, Thrips palmi (Thripinae): comparative analysis. PLOS One. 13(10):e0199404. doi:10.1371/journal.pone.0199404.
- Chen J, Wang Y, Qin M, Jiang LY, Qiao GX. 2019. The mitochondrial genome of Green-idea psidii van der Goot (Hemiptera: Aphididae: Greenideinae) and comparisons with other Aphididae aphids. Int J Biol Macromol. 122:824–832. doi:10.1016/j.ijbiomac.2018.10.209.
- Chen SC, Jiang HY, Peng P, Hu X, Lin Q, Wang XQ. 2017. The complete mitochondrial genome of the stick tea thrips Dendrothrips minowai (Thysanoptera: Thripidae). Mitochondrial DNA B Resour. 3(1):58–59. doi:10.1080/23802359.2017.1419099.
- Chen X, Li C, Song Y. 2021. The complete mitochondrial genomes of two erythroneurine leafhoppers (Hemiptera, Cicadellidae, Typhlocybinae, Erythroneurini) with assessment of the phylogenetic status and relationships of tribes of Typhlocybinae. Zookeys. 1037:137–159. doi:10.3897/zookeys.1037.63671.
- Clary DO, Wolstenholme DR. 1985. The mitochondrial DNA molecular of Drosophila yakuba: nucleotide sequence, gene organization, and genetic code. J Mol Evol. 22(3):252–271. doi:10.1007/BF02099755.
- Dang LH, Li CW, Wang DX, Gao YX, Zhao LP, Zhang B. 2024. Characterizing the complete mitochondrial genome of Psephenothrips eriobotryae Dang & Qiao (Thysanoptera: Phlaeothripidae) with massive gene arrangement in Phlaeothripidae. Zool Syst. 49(2):116–128. doi:10.11865/zs.2024112.
- Dang LH, Wang X, Xie DL, Gao YX, Zhao LP. 2021. Complete mitochondrial genome of Gynaikothrips ficorum (Thysanoptera: Phlaeothripidae). Mitochondrial DNA B Resour. 6(7):2033–2034. doi:10.1080/23802359.2021.1923412.
- Dickey AM, Kumar V, Morgan JK, Jara-Cavieres A, Shatters RG, McKenzie CL, OSBorne LS. 2015. A novel mitochondrial genome architecture in thrips (Insecta: thysanoptera): extreme size asymmetry among chromosomes and possible recent control region duplication. BMC Genomics. 16(1):439. doi:10.1186/s12864-015-1672-4.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18. doi:10.1093/nar/gkw955.
- Grant JR, Stothard P. 2008. The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 36:W181–W184. doi:10.1093/nar/gkn179.
- Hakimara M, Minaei K. 2019. A new species of Odontothrips (Thysanoptera, Thripidae) from Fars province, Iran. Zootaxa. 4674(1):zootaxa.4674.1.9. doi:10.11646/zootaxa.4674.1.9.
- Hu QL, Ye ZX, Zhang CX. 2023. High-throughput sequencing yields a complete mitochondrial genome of the rice thrips, Stenchaetothrips biformis (Thysanoptera: thripidae). Mitochondrial DNA B Resour. 8(2):204–206. doi:10.1080/23802359.2023.2169572.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi:10.1093/molbev/mst010.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649. doi:10.1093/bioinformatics/bts199.
- Kumar V, Tyagi K, Kundu S, Chakraborty R, Singha D, Chandra K. 2019. The first complete mitochondrial genome of marigold pest thrips, Neohydatothrips samayunkur (Sericothripinae) and comparative analysis. Sci Rep. 9(1):191. doi:10.1038/s41598-018-37889-6.
- Kurosawa M. 1941. Thysanoptera of Manchuria. Kontyu. 15(3):35–45.
- Li C, Gao Y, Wang D, Dang L. 2024. The extraordinary rearrangement of mitochondrial genome of the wheat pest, Aptinothrips stylifer and the mitochondrial phylogeny of Thripidae (Thysanoptera). Arch Insect Biochem Physiol. 115(1):e22086. doi:10.1002/arch.22086.
- Li H, Liu H, Cao L, Shi A, Yang H, Cai W. 2012. The complete mitochondrial genome of the damsel bug Alloeorhynchus bakeri (Hemiptera: Nabidae). Int J Biol Sci. 8(1):93–107. doi:10.7150/ijbs.8.93.
- Liu H, Li H, Song F, Gu W, Feng J, Cai W, Shao R. 2017. Novel insights into mitochondrial gene rearrangement in thrips (Insecta: Thysanoptera) from the grass thrips, Anaphothrips obscurus. Sci Rep. 7(1):4284. doi:10.1038/s41598-017-04617-5.
- Liu Q, He J, Song F, Tian L, Cai W, Li H. 2022. Positive correlation of the gene rearrangements and evolutionary rates in the mitochondrial genomes of thrips (Insecta: Thysanoptera). Insects. 13(7):585. doi:10.3390/insects13070585.
- Mound LA, Palmer JM. 1981. Phylogenetic relationships between some genera of Thripidae (Thysanoptera). Entomologica Scandinavica. 15:153–170.
- Pakrashi A, Patidar A, Singha D, Kumar V, Tyagi K. 2023. Comparative analysis of the two suborders of Thysanoptera and characterization of the complete mitochondrial genome of Thrips parvispinus. Arch Insect Biochem Physiol. 114(2):1–15. doi:10.1002/arch.22010.
- Pakrashi A, Tyagi K, Kumar V. 2021. The complete mitochondrial genome of Aeolothrips indicus Bhatti, 1964 (Thysanoptera: Thripidae). Mitochondrial DNA B Resour. 6(10):2808–2809. doi:10.1080/23802359.2021.1970647.
- Pakrashi A, Tyagi K, Kumar V. 2021. The complete mitochondrial genome of Taeniothrips tigris Bhatti, 1995 (Thysanoptera: Thripidae). Mitochondrial DNA B Resour. 6(8):2256–2257. doi:10.1080/23802359.2021.1947916.
- Ranwez V, Douzery EJP, Cambon C, Chantret N, Delsuc F. 2018. MACSE v2: toolkit for the alignment of coding sequences accounting for frameshifts and stop codons. Mol Biol Evol. 35(10):2582–2584. doi:10.1093/molbev/msy159.
- Rebijith KB, Asokan R, Krishna V, Ranjitha HH, Kumar NKK, Ramamurthy VV. 2014. DNA barcoding and elucidation of cryptic diversity in thrips (Thysanoptera). The Florida Entomologist. 97(4):1328–1347. doi:10.1653/024.097.0407.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. doi:10.1093/sysbio/sys029.
- Shao R, Barker SC. 2003. The highly rearranged mitochondrial genome of the plague thrips, Thrips imaginis (Insecta: thysanoptera): convergence of two novel gene boundaries and an extraordinary arrangement of rRNA genes. Mol Biol Evol. 20(3):362–370. doi:10.1093/molbev/msg045.
- Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 56(4):564–577. doi:10.1080/10635150701472164.
- ThripsWiki. 2023. ThripsWiki-providing information on the World’s thrips. [accessed 2023 Sep 30]. http://thrips.info/wiki/Main_Page
- Tyagi K, Chakraborty R, Cameron SL, Sweet AD, Chandra K, Kumar V. 2020. Rearrangement and evolution of mitochondrial genomes in Thysanoptera (Insecta). Sci Rep. 10(1):695. doi:10.1038/s41598-020-57705-4.
- Wang Y, Wu C, Xie Y, Liu H, Zhang H. 2021. Complete mitochondrial genome sequence for the Thrips hawaiiensis (Thysanoptera: Thripidae). Mitochondrial DNA B Resour. 6(7):2092–2094. doi:10.1080/23802359.2021.1942268.
- Wu C, Zhou Y, Tian T, Li TJ, Chen B. 2022. First report of complete mitochondrial genome in the subfamily Alleculinae and mitochondrial genome-based phylogenetics in Tenebrionidae (Coleoptera: Tenebrionoidea). Insect Sci. 29(4):1226–1238. doi:10.1111/1744-7917.12983.
- Xie Y, Zhang H, Mound LA. 2010. A new species from southwestern China of the holarctic genus Odontothrips (Thysanoptera: Thripidae). Zootaxa. 2729(1):53–57. doi:10.11646/zootaxa.2729.1.4.
- Xing-Ming L, Pan L, Du-Cheng C, Jin-Hua L. 2023. Characterization of the complete mitochondrial genome and phylogenetic analysis of bean thrips Megalurothrips usitatus (Bagnall, 1913) (Thysanoptera: Thripidae). Mitochondrial DNA B Resour. 8(10):1032–1035. doi:10.1080/23802359.2023.2261644.
- Yan D, Tang Y, Hu M, Liu F, Zhang D, Fan J. 2014. The mitochondrial genome of Frankliniella intonsa: insights into the evolution of mitochondrial genomes at lower taxonomic levels in Thysanoptera. Genomics. 104(4):306–312. doi:10.1016/j.ygeno.2014.08.003.
- Yan D, Tang Y, Xue X, Wang M, Liu F, Fan J. 2012. The complete mitochondrial genome sequence of the western flower thrips Frankliniella occidentalis (Thysanoptera: Thripidae) contains triplicate putative control regions. Gene. 506(1):117–124. doi:10.1016/j.gene.2012.06.022.
- Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355. doi:10.1111/1755-0998.13096.
- Zhang S, Luo J, Wang C, Lv L, Li C, Jiang W, Cui J, Rajput LB. 2016. Complete mitochondrial genome of Aphis gossypii Glover (Hemiptera: Aphididae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(2):854–855. doi:10.3109/19401736.2014.919474.
- Zhou X, Dietrich CH, Huang M. 2020. Characterization of the complete mitochondrial genomes of two species with preliminary investigation on phylogenetic status of Zyginellini (Hemiptera: Cicadellidae: Typhlocybinae). Insects. 11(10):684. doi:10.3390/insects11100684.
