Abstract
Tropical savannas cover over 20% of land surface. They sustain a high diversity of mammalian herbivores and promote frequent fires, both of which are dependent on the underlying grass composition. These habitats are typically dominated by relatively few taxa, and the evolutionary origins of the dominant grass species are largely unknown. Here, we trace the origins of the genus Themeda, which contains a number of widespread grass species dominating tropical savannas. Complete chloroplast genomes were assembled for seven samples and supplemented with chloroplast and nuclear ITS markers for 71 samples representing 18 of the 27 Themeda species. Phylogenetic analysis supports a South Asian origin for both the genus and the widespread dominant T. triandra. This species emerged ~1.5 Ma from a group that had lived in the savannas of Asia for several million years. It migrated to Australia ~1.3 Ma and to mainland Africa ~0.5 Ma, where it rapidly spread in pre-existing savannas and displaced other species. Themeda quadrivalvis, the second most widespread Themeda species, is nested within T. triandra based on whole chloroplast genomes, and may represent a recent evolution of an annual growth form that is otherwise almost indistinguishable from T. triandra. The recent spread and modern-day dominance of T. triandra highlight the dynamism of tropical grassy biomes over millennial time-scales that has not been appreciated, with dramatic shifts in species dominance in recent evolutionary times. The ensuing species replacements likely had profound effects on fire and herbivore regimes across tropical savannas.
Introduction
Tropical savannas are among Earth’s most productive biomes and cover around 20% of global land area (Lehmann and Parr Citation2016). Throughout the tropics and subtropics, these grassy biomes are dominated by C4 grass species, which displaced C3 species across these regions during the Late Miocene and Pliocene as Earth’s environment warmed and dried (3–8 Ma; Cerling et al. Citation1997; Edwards et al. Citation2010). Consumer centred feedbacks related to fire and grazing have been invoked as underlying determinants of the globally synchronised rapid expansion of tropical grassy biomes (Beerling and Osborne Citation2006; Edwards et al. Citation2010; Scheiter et al. Citation2012). Indeed, there is evidence for the co-evolution of grazing mammals with the grass species they consume (Bouchenak-Khelladi et al. Citation2009; Sage and Stata Citation2015), just as there is palaeo-evidence of a global increase in fire with the expansion of C4 grass-dominated savannas (Edwards et al. Citation2010). Today, these tropical grassy biomes both sustain large numbers of grazing mammals and promote frequent fire (Archibald and Hempson Citation2016), and feedbacks between both fire and herbivory have been demonstrated as central to the modern dynamics of tropical grassy biomes over much of their range (Archibald and Hempson Citation2016; Lehmann et al. Citation2014). Similarly, at local scales, many grass species rely on fire and herbivory for their persistence and dispersal (Bond et al. Citation2003). The origins of these co-dependency feedbacks remain, however, poorly understood.
While the timing of the origin of C4 grass dominated biomes is relatively well resolved, many questions regarding the composing flora are unanswered. Today, a number of C4 grass species from these habitats have exceptional geographic ranges that span multiple continents. This is in stark contrast to the woody flora of savannas, which are typically derived from local forest ancestors in response to the novel ecological conditions generated by the development of a C4 grass ground layer (Maurin et al. Citation2014; Simon et al. Citation2009). Establishing why savannas are dominated by relatively few species requires considering the origins of the grass species that dominate these habitats, in terms of space, time and ancestral conditions. Among C4 grass species, variation in flammability is phylogenetically constrained, and the traits of the grassy ground layer have ecosystem-level impacts on fire frequency and intensity (Archibald et al. Citation2013; Simpson et al. Citation2016). A key question is therefore whether grass traits underlying savanna dynamics existed before the colonisation of these ecosystems, or whether they evolved in situ. This problem needs to be addressed by elucidating the history of groups that are nowadays involved in the fire/herbivore feedbacks in these systems.
In this study, we reconstruct the phylogeography of the genus Themeda Forssk. (Poaceae, Panicoideae, Andropogoneae). This genus includes one of world’s most widespread C4 grasses species, Themeda triandra Forssk., along with a modern invasive, Themeda quadrivalvis (L.) Kuntze, a grassy weed common in disturbed areas worldwide (Keir and Vogler Citation2006). In total, there are 27 recognised Themeda species with both annual and perennial lifeforms (Clayton et al. Citation2014). The range of the genus is defined by the range of T. triandra and all other species are either regionally or locally restricted. The highest concentration of species diversity is located in Asia, in particular India (Morales Citation2014). Despite ongoing phylogenetic work in Andropogoneae (Giussani et al. Citation2001; Mathews et al. Citation2002; Sánchez-Ken and Clark Citation2010; Burke, Wysocki et al. Citation2016; Arthan et al. Citation2017), the tribe that includes Themeda, detailed analyses of this ecologically important genus are lacking.
Today, T. triandra is central to the ecological dynamics of palaeotropical savannas (Gibbs Russell et al. Citation1991; Jessop, Dashorst, and James Citation2006; Snyman, Ingram, and Kirkman Citation2013), where the absence of this particular species has been used as an indicator of reduced ecosystem function and soil quality (Mills et al. Citation2005; du Preez and Snyman Citation1993). T. triandra is generally abundant where it is found, with both fire and grazing by mammals necessary for its persistence (Snyman, Ingram, and Kirkman Citation2013). Indeed, the ecological dominance of T. triandra relies on periodic burning and its ability to rapidly resprout post fire (Bond et al. Citation2003; Morgan and Lunt Citation1999). For example, in Serengeti region T. triandra comprises approximately 50% of grass cover in areas of light to moderate grazing (Vuorio et al. Citation2014), but is lost from a system where both fire and grazing are excluded (Danckwerts Citation1993). Themeda triandra is also a crucial food source for domestic livestock and wildlife in both Africa and Australia (McNaughton Citation1985; Morgan and Lunt Citation1999). Despite the ecological importance of T. triandra, the timing and rate of its geographic spread remain unknown.
Here, we investigate the evolutionary origins of the Themeda genus and T. triandra in particular via phylogenetic reconstructions. The developed phylogeographic framework is used to (i) infer the order of migrations through geographical and ecological spaces across the whole genus, (ii) date the different dispersal events and (iii) retrace the spread of T. triandra through space and time. Our analyses of this paleotropical savanna dominant sheds new light on the biogeographic factors underlying the assembly of new ecosystems during recent geological times.
Methods
Sampling and sequencing
Leaf samples for Themeda (n = 71; species = 18) and other closely related Andropogoneae (n = 7; species = 6) were collected from the field or obtained from the herbaria at the Royal Botanic Gardens Kew and Edinburgh (Table S1). The sampling strategy optimised the morphological, ecological and geographic diversity within the genus given the sample availability. DNA was extracted from silica dried leaves and herbarium material using the DNeasy Plant Mini Kit (Qiagen, Texas, USA), following the manufacturer’s protocol. PCR and Sanger sequencing of five plastid regions (trnK-matK, rpl16, ndhF, rpoC2 and trnL-trnF) and the nuclear-encoded ITS marker (i.e. internal transcribed spacers of the ribosomal DNA) were performed as described in Lundgren et al. (Citation2015). In brief, 25 μl PCR reactions were amplified with a 48°C annealing temperature, subsequently cleaned using the Exo-SAP treatment (Affymetric, High Wycombe, UK) and finally sequenced using the Big Dye 3.1 Terminator Cycle Sequencing chemistry (Applied Biosystems, California, USA). Multiple sets of PCR primers were used (Table S2). The older herbarium specimens yielded degraded DNA, and markers were consequently amplified in short overlapping fragments. The resultant chromatographs were manually corrected in Geneious v.5.3.6. All Sanger sequences were deposited in the NCBI GenBank database (KY991068-KY991366).
Four Themeda triandra, two Heteropogon Pers. and one Themeda quadrivalvis accessions were subjected to low-coverage whole-genome sequencing (genome-skimming), as described in Lundgren et al. (Citation2015). In brief, samples were individually barcoded, pooled and sequenced on an Illumina HiSeq-2500 or HiSeq-3000 at the Genopole platform of Toulouse. In total, 1/24th of a lane paired-end data were generated per sample, with all raw data deposited in the NCBI Sequence Read Archive (project identifier PRJNA377519; Table S3).
Assembly, alignment and phylogenetic inference
A complete chloroplast genome (hereafter plastome) was assembled de novo for one sample using the genome-walking method described in Besnard et al. (Citation2013). In brief, a combination of extractread2 (included in the OBITools package, http://metabarcoding.org/obitools) and velvet (Zerbino and Birney Citation2008) was used to identify overlapping chloroplast reads in the raw data and subsequently assemble them. For each sample, we then mapped the paired-end Illumina data back to this initial reference using bowtie2 v.2.2.9 (Langmead and Salzberg Citation2012). Indels and SNPs were called with SAMtools mpileup function v.1.2 (Li et al. Citation2009) and filtered using the accompanying vcfutils.pl script (parameters: d = 10, a = 5 and Q = 30). Plastome sequences for each sample were generated using vcftools v.0.1.11 (Danecek et al. Citation2011). To remove sequencing errors from SNP calls, we discarded minor alleles that had a within sample frequency of < 0.2. Plastome sequences were submitted to GenBank (accession numbers: KY707767-KY707773).
The newly generated plastomes were aligned with others publicly available for Andropogoneae and Arundinella Raddi using MAFFT v. 7.123b (Arthan et al. Citation2017; Katoh and Standley Citation2013). The alignment was manually refined and the inverted repeat region removed. The Sanger sequencing data, and additional data retrieved from GenBank, were aligned to the plastome matrix using MAFFT and then concatenated by sample. A phylogeny was obtained using Bayesian inference, as implemented in MrBayes version 3.2.0 (Huelsenbeck and Ronquist Citation2001) with the GTR + G + I substitution model. Two analyses, each composed of four chains, were run for 10 million generations, sampling a tree every thousand generations. Convergence was evaluated with Tracer v. 1.5.0 (Drummond and Rambaut Citation2007), and a consensus was computed using all the trees sampled after a relative burn-in period of 10%. To verify the effect of missing data on phylogenetic inference, we also constructed a phylogeny using a trimmed alignment containing five chloroplast genes (trnK-matK, rpl16, ndhF, rpoC2 and trnL-trnF) either obtained via Sanger sequencing or extracted from plastomes. The ITS sequences were similarly aligned with MAFFT, and a Bayesian phylogeny was inferred as above.
Molecular dating
Divergent groups within Themeda were identified from the plastid and ITS phylogenies, and the accessions from each clade with the most complete sequences were retained for molecular dating analysis as implemented in BEAST v. 1.8.4 (Drummond and Rambaut Citation2007). Arundinella deppeana Nees was used as the outgroup, based on the results of Grass Phylogeny Working Group II (Citation2012) that demonstrate the monophyly of the ingroup (Andropogoneae). Dating subgroups of grasses is complicated by the limited fossil record, and no informative fossil is available for the group studied here. We consequently adopted a secondary calibration approach, with a calibration point extracted from a previous, angiosperm-wide dating analysis (Christin et al. Citation2014). The age of grasses has been hotly debated, with controversial fossils potentially pushing the origin of the group back in time (Prasad et al. Citation2011; Burke, Lin et al. Citation2016), although their effect is reduced when the evidence for non-grass groups is incorporated (Christin et al. Citation2014). Because this debate affects only the scale of the dating analyses and not the ages relative to each other, we decided to fix the calibration point to values previously estimated and present the results of our dating analyses under two different scenarios; with only macrofossils and with macrofossils plus phytoliths, in both cases using the dates that take into account the evidence for groups outside grasses (Christin et al. Citation2014). Our approach has the advantage of showing the uncertainty resulting from the genetic data (confidence intervals around the age estimates) independently of the calibration density (uncertainty about the scale). The divergence of Zea mays L. and Sorghum bicolor was fixed at 15.26 Ma (or 22.38 when taking into account controversial microfossils; Christin et al. Citation2014), which was achieved with a normal distribution that had a mean of 15.26 and standard deviation of 0.0001. Two different analyses were run for 50,000,000 generations, sampling a tree every 1,000 generations with a GTR + G model, a Yule process speciation prior and a relaxed log-normal clock. After evaluating the convergence of the runs in Tracer v. 1.5.0 (Drummond and Rambaut Citation2007), the burn-in period was set to 10%, and the maximum credibility tree was identified from all trees sampled from both runs after the burn-in period, mapping median ages on nodes. To check the effect of missing data on our results, we repeated this analysis using only the samples with full chloroplast genomes.
Genome-wide phylogenetic tree
The seven genome-skimmed individuals were also genotyped across the nuclear genome as described in Olofsson et al. (Citation2016). In brief, the reads were cleaned using NGS QC Toolkit v.2.3.3 (Patel and Jain Citation2012) and mapped onto the chromosomes of the closest available model species (Sorghum bicolor (L.) Moench, version Sorbi1; Paterson et al. Citation2009) using Bowtie2 v. 2.2.3 (Langmead et al. Citation2009). Only reads uniquely aligned in pairs were subsequently used for SNP calling using SAMtools v.0.1.19 and previously published scripts (Olofsson et al. Citation2016). The low-coverage genome-skimming data mean that some alleles are likely missed, leading to an overestimation of homozygosity. However, no bias is expected in the missing allele, so that the low coverage is unlikely to lead to spurious groupings (Olofsson et al. Citation2016). A Bayesian phylogeny was inferred for the SNP data using MrBayes as described above, run for three million generations.
Results
Phylogenetic relationships within Themeda based on plastid markers
In this study, we sequenced and assembled seven plastomes, which were supplemented with a further 26 publicly available plastomes from Andropogoneae (including five Themeda accessions), and the plastome of Arundinella deppeana. Sanger-sequenced chloroplast data were obtained for a further 62 samples with a mean of 3,572 bp per sample (SD = 1623 bp; Table S1). These were aligned to the reference chloroplast genome, trimmed of the inverted repeat, and the phylogeny inferred from this 125,270 bp alignment included 18 of the 27 Themeda species (Table S1).
The Bayesian chloroplast tree identified seven major Themeda clades (clades A–H; Figure , Table ). A previously published chloroplast genome attributed to Themeda arguens (L.) Hack. may come from an incorrectly identified individual as it is nested within T. triandra (Figure ) and is subsequently treated as such. Themeda triandra then forms a monophyletic group with T. quadrivalvis (clade H; posterior probability = 1; Figure ). The T. triandra clade is sister to T. tremula (Nees ex Steud.) Hack. (clade F; Figure ), with the two being sister to clade G (Figure ), which includes species from Australia and Papua New Guinea (Figure ). Some of the other clades include multiple morphological species without variation in the genetic markers (clades C and D; Figure ). Within the T. triandra / T. quadrivalvis clade, there appears to be three groups, which match the geographic regions (H.1, H.2 and H.3; Figure and Fig. S1). Clade H.1 includes all the mainland African samples, in addition to those from Turkey and Yemen (Figure ). Clade H.2 contains all the T. quadrivalvis samples, whether they come from Madagascar or Asia (Figure ). Finally, clade H.3 encompasses all the Australian accessions, in addition to those from Bhutan, Nepal and Thailand (Figure ). In clade H.3, there is a division between some of the Australian samples and the rest. Note that the overall diversity among T. triandra plastomes (defined as clade H) is low. Across the plastomes of T. triandra (mean length with one inverted repeat = 116,090 bp, SD = 35 bp), 99.2% of sites were conserved, for a 99.8% pairwise identity between the eight T. triandra plastomes. There is some effect of reducing the data-set on the topology, but the relationships within T. triandra remain the same when only the four densely sampled markers are used (Fig. S1).
Figure 1. Bayesian chloroplast tree inferred with MrBayes. Posterior probabilities are indicated near nodes when greater than 0.8. The major clades within Themeda are delimited on the right (A–H). Within T. triandra, branches are coloured based on geographical origin. Scale bar shows the expected number of substitutions per site.
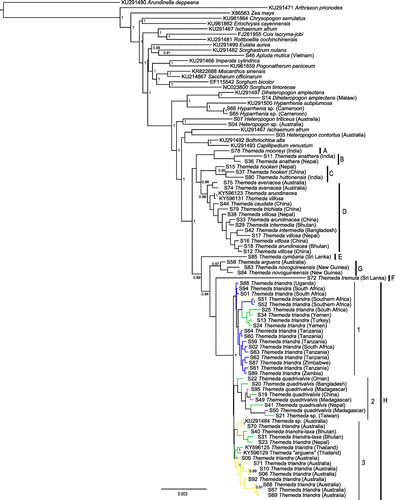
Table 1. Themeda species used in this study and their distribution. In total, we sequenced 18 out of the 27 known species. The clade assignment is based on chloroplast and nuclear markers, with the number of samples sequenced indicated (n), and ecological information derived from Morales (Citation2014).
Phylogenetic relationships based on the nuclear ITS
A majority of the clades from the plastid phylogeny were congruent with the phylogeny inferred from the 625-bp ITS alignment containing 71 samples (59 sequenced here) and 17 of the 27 Themeda species (Fig. S2). One notable exception is the division of T. novoguineensis (Reeder) Jansen from T. arguens, which was not achieved with the chloroplast markers. Based on ITS, T. triandra, T. quadrivalvis and T. australis (R.Br.) Stapf (not sampled for chloroplast markers and hypothesised as a synonym of T. triandra) form a clade with no distinction among the morphological species, nor the clades identified based on plastid markers (Figure ).
Molecular dating
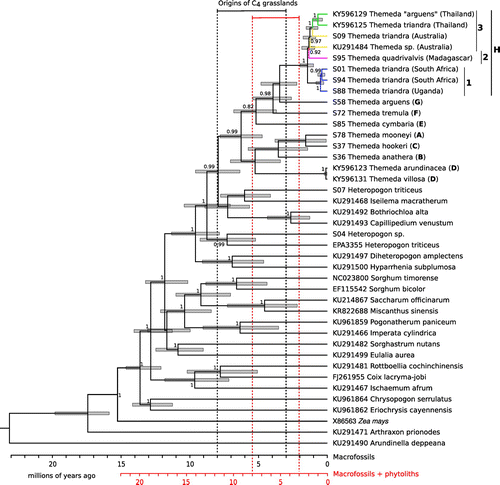
Based on our dating analyses, the divergence between Themeda and its sister group occurred at 7.98 Ma (95% HPD 5.01–12.78), which is largely congruent with previous estimates (Estep et al. Citation2014; Spriggs, Christin, and Edwards Citation2014). Within Themeda, the divergence of T. triandra from its sister group (T. arguens) occurred at 3.50 Ma (95% HPD 2.16–5.02 Ma) and the first divergence within T. triandra (crown node) at 1.48 Ma (95% HPD 0.79–3.45; Figure ). This first divergence within T. triandra corresponds to the divergence of mainland African accessions (H.1) from those from Madagascar (H.2) and Australia/Thailand (H.3; Figure ). The Madagascan samples split from the Australian and Thai ones at 1.37 Ma (95% HPD 0.73–3.07), and this represents the divergence of clades H.1 and H.2 (Figure ). Australia was colonised over 1 Ma, and the two Thailand samples diverged 0.69 Ma (95% HPD 0.21–1.22). The African lineage began diversifying relatively recently, with the Uganda sample splitting from the South African ones 0.52 Ma (95% HPD 0.19–1.30; Figure ). Note that the three African samples capture the earliest split within clade H.1 (Figure ). While all date estimates are older when considering microfossils, the diversification of T. triandra is still placed within the last two million years (Figure ). Missing data appeared to have little effect on the dates, since removing partially sequenced individuals does not alter the results (Fig. S3).
Figure 2. Dating the divergence within Themeda using chloroplast data. Posterior probabilities are indicated near nodes (>0.8), and bars represent the 95% HPD of the estimated dates. The clade that each Themeda species represents is indicated (A–H; Table ). NCBI GenBank accession numbers are given for samples sequenced in previous studies. Within T. triandra, branches are coloured based on geographical origin. The scale is given at the bottom for a scenario that incorporates or ignores the microfossils. The origins of C4 grasslands are plotted based on each scenario.
Genome-wide nuclear phylogeny
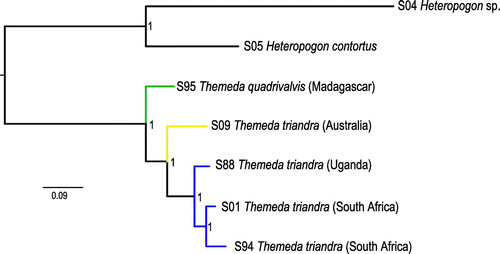
A nuclear phylogeny was inferred from the genome-skimming data generated here, using the methods described in Olofsson et al. (Citation2016). A total of 753,190 SNPs were used to infer a Bayesian phylogeny, with a mean of 11% missing data per samples. All nodes were fully supported (Figure ). The tree was rooted on the branch separating Heteropogon from Themeda. Unlike in the chloroplasts and ITS phylogenies (Figures , S2), T. quadrivalvis collected from Madagascar is strongly supported as sister to T. triandra in this tree (Figure ). Within T. triandra, the Australian sample separates first, with the three African samples forming a derived monophyletic group (Figure ).
Discussion
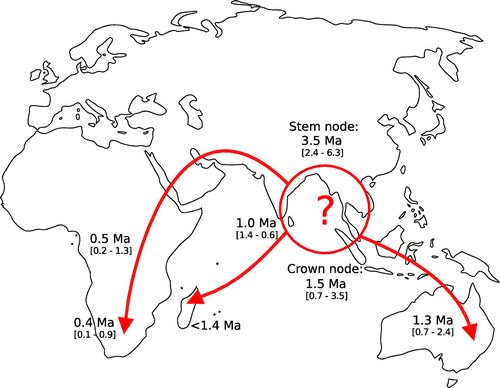
Based on the phylogenetic relationships and species distributions, Themeda triandra spread to its current cosmopolitan distribution surprisingly recently. This ecologically important species began to diversify approximately one and a half million years ago and has since been able to colonise the grassy biomes of Africa, Madagascar and Australia, long after these biomes initially assembled (Figure ). Therefore, T. triandra replaced other C4 grass species already established in these habitats, rather than being one of the founding species. Themeda trianda is a major component of modern day savannas and has rapidly become a key component of the ground flora of these ecosystems (Snyman, Ingram, and Kirkman Citation2013). This highlights the dynamic turn-over of tropical grassy biomes, with dramatic shifts in species dominance occurring in relatively recent evolutionary times
Figure 4. Inferred putative dispersal routes for Themeda. Dates are from a dated BEAST phylogeny based on chloroplast markers (Figure ).
Is Themeda quadrivalvis a synonym of Themeda triandra?
Both species are extremely similar and often dominate savannas. The key morphological difference between the two is that T. quadrivalvis is annual, and T. triandra is perennial. Previously, T. quadrivalvis has been considered as a potential synonym of T. triandra (Veldkamp Citation2016). However, if speciation is relatively recent, we would predict similar patterns for savanna species to what we observe in our phylogenies (Pennington and Lavin Citation2016) with a monophyletic daughter species (T. quadrivalvis) nested within a paraphyletic ancestor (T. triandra). This scenario is supported by the whole chloroplast genomes and ITS phylogenies (Figures , S1, S2, S3), and it may be concluded that T. quadrivalvis represents a recent evolution of an annual growth form from a perennial ancestor. Switching between these two life histories may be a surprisingly simple transition in grasses (Linder and Rudall Citation2005), being controlled by only two genes (Hu et al. Citation2003). However, the nuclear phylogeny identifies T. quadrivalvis as sister to T. triandra (Figure ). The inconsistencies between the plastome and nuclear phylogenies might stem from incomplete lineage sorting or hybridisation during the early diversification of the species complex, in which case T. quadrivalvis and T. triandra represent recently diverged sister taxa. Either way, both species are extremely closely related, and whether the two forms do interbreed in the wild or represent distinct species will require dedicated analyses in the future.
A long history in Asian savannas
Both the chloroplast and nuclear phylogenies place a series of Asian species as successive sister groups to the T. triandra clade (Figs. , S2), clearly suggesting Asia as the ancestral area for the group. The earliest division within the genus divides clades A–D from E–H (Figure ). Broadly speaking, this separates the predominately savanna species (clades E–H; including T. triandra and its close relatives) from the taller wetland species (clade D; e.g. T. villosa (Lam.) A.Camus and T. caudata (Nees ex Hook. & Arn.) A.Camus; Morales Citation2014; Veldkamp Citation2016) and those found in dry and/or high altitude environments (clade A–C; e.g. T. anathera (Nees ex Steud.) Hack.; Table ). The sister taxa of Themeda are also savanna species (e.g. Bothriochloa alta (Hitchc.) Henrard, Capillipedium venustum (Thwaites) Bor, Iseilema macratherum Domin, Heteropogon contortus (L.) P.Beauv. ex Roem. & Schult., Heteropogon triticeus (R.Br.) Stapf ex Craib, Diheteropogon amplectens (Nees) Clayton and Hyparrhenia subplumosa Stapf). We therefore conclude that the savanna ecology, with its suite of associated traits, predates the genus. The Andropogoneae tribe as a whole contains many of the species typical of grassy biomes in the paleotropics (Osborne Citation2008). The group evolved C4 photosynthesis more than 17 Ma (Christin et al. Citation2008), a trait that, in association with their capacity to quickly accumulate biomass between fires (Bond et al. Citation2003; Morgan and Lunt Citation1999), might have favoured their spread in novel grassy biomes as they expanded around the world. The savanna Themeda species themselves (clade E–H) began to diversify approximately 5 Ma (Figure ), during the global expansion of C4 grassy biomes (Cerling et al. Citation1997; Edwards et al. Citation2010). This means that the ancestors of T. triandra spent millions of years in the extensive and diverse grassy habitats of Asia (Dixon et al. Citation2014), and therefore T. triandra was probably well suited for the African savannas when it was eventually able to colonise this continent.
Paleotropical spread of Themeda triandra
From its Asian origin, T. triandra has spread to Australia and Africa (Figure ). It is likely that T. triandra arrived in Australia before mainland Africa, as supported by higher genetic diversity within Australia and to a lesser extent, the earlier divergence of Madagascan and Australian accessions compared to the divergence of the mainland African accessions sampled (South Africa and Uganda). Our dating analyses show that the lineage containing the mainland African accession diverged from those that remained in Asia approximately one and a half million years ago. Individuals from this group subsequently colonised mainland Africa, with the first split within this lineage estimated at half a million years ago (South Africa and Uganda), further supporting a recent and rapid spread throughout Africa (Figure ). The fact that this species was able to conquer multiple continents while its congeners remained geographic restricted is intriguing.
The capacity for wind dispersal of T. triandra seeds is poor, with most landing <1.75 m from a parent plant (Everson, Yeaton, and Everson Citation2009), but they can travel epizoochorously potentially over long distances (Agnew and Flux Citation1970; Milton Citation1993; Reynolds and Cumming Citation2016). Hydroscopic awns might aid their attachment to animal fur or feathers, but such awns are found in numerous Andropogoneae genera (Kellogg and Watson Citation1993) and in all Themeda species (Morales Citation2014). It is therefore possible that the impressive spread of T. triandra is not the result of a higher dispersal ability, but instead reflects an enhanced survival after occasional long distance dispersal.
Evolving from a genus already associated with tropical grassy biomes over a long period means that T. triandra is inherently adapted to these habitats. Its height and rapid growth enable T. triandra to out compete neighbouring species for light, water and nutrients. Fire also plays an important role in its dominance, with T. triandra being highly flammable relative to co-occurring grass species (Simpson et al. Citation2016) and its persistence reliant on burning (Danckwerts Citation1993). These characteristics are, however, at least partially shared across the genus (Morales Citation2014).
Polyploidy may also play a role in the success of T. triandra following dispersal, with the species recognised as having numerous populations of different ploidy levels (Birari Citation1980; Hayman Citation1960). Ploidy has been associated with the invasive potential of species, through improving the capacity to colonise new environments, via maintaining genetic diversity as new populations establish (te Beest et al. Citation2011). However, other species within the genus also include polyploids (Birari Citation1981). Furthermore, previous studies investigating allopolyploidy and diversification during Miocene grassland expansion showed that there is no correlation with the origin of novel morphological characters (Estep et al. Citation2014). We therefore conclude that the success of T. triandra cannot be attributed to a single characteristic, but probably reflects the combined action of multiple traits favouring dispersal and survival, but also potentially contingency. Indeed, a handful of chance dispersal events over long distances would have been sufficient to bring the species to the African continent, where its characteristics would have allowed a rapid colonisation of C4 grassy biomes that already existed there.
Conclusion
Themeda triandra is one of the species dominating savannas throughout Africa, Asia and Oceania. Interestingly, our dating analyses indicate that this species originated relatively recently in Asia, long after the expansion of C4 grassy biomes. It emerged from a group that had inhabited Asian savannas for millions of years. Therefore, characteristics favouring the dominance of African C4 grassy biomes predate the colonisation of these habitats and evolved in distinct ecosystems with similar properties. Interestingly, the spread of the species is independent from the expansion of the ecosystems, highlighting the importance of the biogeographic history, including random dispersal events, in the assemblage of large communities around the world. Following the colonisation of Africa, T. triandra likely displaced other savanna species that lived there, since it now represents the dominant grass in multiple systems across the continent. Overall, our results highlight the turn-over of tropical grassy biomes, which may have experienced many dramatic shifts in species dominance during recent evolutionary times, with a succession of dominant species. This is likely to have had a profound effect on fire and herbivore regimes across these globally important ecosystems.
Disclosure statement
No potential conflict of interest was reported by the authors.
Funding
This research was funded by NERC (grant NE/M00208X/1), the ERC (grant ERC-2014-STG-638333) and a Royal Society University Research Fellowship (URF120119) awarded to PAC. GB is supported by TULIP (ANR-10-LABX-0041).
Notes on contributors
Luke T. Dunning is a postdoctoral research associate at the University of Sheffield. His research focuses on the genomics of adaptation in grasses. Contribution: conceived the study, secured plant material, performed lab work and data analysis and wrote the paper with the help of all authors.
Anne-Lise Liabot was a research assistant at the University of Sheffield and is now an engineer research assistant at Institut Nationale de la Recherche Agronomique, INRA, Angers France. Contribution: performed lab work.
Jill K. Olofsson is a postdoctoral research associate at the University of Sheffield. Her research focuses on the population genomics of complex trait evolution. Contribution: data analysis.
Emma K. Smith is a PhD student at the University of Sheffield. Contribution: data analysis.
Maria S. Vorontsova is a research leader at The Royal Botanic Gardens, Kew. Contribution: secured plant material.
Guillaume Besnard is a researcher at the French Centre National de la Recherche Scientifique. His research focuses on the biogeography of various plant and animal groups. Contribution: secured plant material and performed lab work.
Kimberley J. Simpson is a PhD student at the University of Sheffield. Her research focuses on grass fire ecology. Contribution: secured plant material.
Marjorie R. Lundgren is a postdoctoral researcher at the University of Sheffield. Contribution: secured plant material.
Eda Addicott is a principal botanist with the Queensland Herbarium, Australia. Her research focuses on vegetation mapping and classification. Contribution: secured plant material.
Rachael V. Gallagher is a Research Fellow in plant ecology at Macquarie University, Sydney. Contribution: secured plant material.
Yingying Chu is a research assistant at the Center for Integrative Conservation, Xishuangbanna Tropical Botanical Garden. Contribution: secured plant material.
R. Toby Pennington is a tropical botanist at the University of Exeter and Royal Botanic Garden Edinburgh with research interests in the biogeographic history of tropical savannas and dry forests. Contribution: secured plant material.
Pascal-Antoine Christin is a Royal Society Research Fellow at the University of Sheffield. His research focuses on grass evolution. Contribution: conceived the study, secured plant material, performed data analysis and co-wrote the paper.
Caroline E. R. Lehmann is a lecturer in biogeography at the University of Edinburgh. Her research focuses on savanna ecology and evolution. Contribution: conceived the study, secured plant material and co-wrote the paper.
Supplemental data
Supplemental data for this article can be accessed here. https://doi.org/10.1080/23818107.2017.1391120
TABG_1391120_Supplementary_Material.pdf
Download PDF (2.7 MB)Acknowledgements
The authors would like to thank all those who provided material, including: The Royal Botanic Gardens, Kew, The Royal Botanic Gardens, Edinburgh, and Tricia Handasyde from the DBCA, Western Australia.
References
- Agnew, A. D. Q., and J. E. C. Flux. 1970. “Plant Dispersal by Hares (Lepus capensis L.) in Kenya.” Ecology 51 (4): 735–737.10.2307/1934057
- Archibald, S., and G. P. Hempson. 2016. “Competing Consumers: Contrasting the Patterns and Impacts of Fire and Mammalian Herbivory in Africa.” Philosophical Transactions of the Royal Society B 371 (1703): 20150309.10.1098/rstb.2015.0309
- Archibald, S., C. E. R. Lehmann, J. L. Gómez-Dans, and R. A. Bradstock. 2013. “Defining Pyromes and Global Syndromes of Fire Regimes.” Proceedings of the National Academy of Sciences 110 (16): 6442–6447.10.1073/pnas.1211466110
- Arthan, W., M. R. McKain, P. Traiperm, C. A. D. Welker, J. K. Teisher, and E. A. Kellogg. 2017. “Phylogenomics of Andropogoneae (Panicoideae: Poaceae) of Mainland Southeast Asia.” Systematic Botany 42 (3): 418–431.10.1600/036364417X696023
- Beerling, D. J., and C. P. Osborne. 2006. “The Origin of the Savanna Biome.” Global Change Biology 12 (11): 2023–2031.10.1111/gcb.2006.12.issue-11
- te Beest, M., J. J. Le Roux, D. M. Richardson, A. K. Brysting, J. Suda, M. Kubešová, and P. Pyšek. 2011. “The More the Better? The Role of Polyploidy in Facilitating Plant Invasions.” Annals of Botany 109 (1): 19–45.
- Besnard, G., P.-A. Christin, P.-J. G. Malé, E. Coissac, H. Ralimanana, and M. S. Vorontsova. 2013. “Phylogenomics and Taxonomy of Lecomtelleae (Poaceae), an Isolated Panicoid Lineage from Madagascar.” Annals of Botany 112 (6): 1057–1066.10.1093/aob/mct174
- Birari, S. P. 1980. “Apomixis and Sexuality in Themeda forssk. At Different Ploidy Levels (Gramineae).” Genetica 54 (2): 133–139.10.1007/BF00055982
- Birari, S. P. 1981. “Polyploidy in Species of Themeda Forsk.” Caryologia 34 (3): 301–310.10.1080/00087114.1981.10796896
- Bond, W. J., G. F. Midgley, F. I. Woodward, M. T. Hoffman, and R. M. Cowling. 2003. “What Controls South African Vegetation – Climate or Fire?” South African Journal of Botany 69 (1): 79–91.10.1016/S0254-6299(15)30362-8
- Bouchenak-Khelladi, Y. A., G. A. Verboom, T. R. Hodkinson, N. Salamin, O. Francois, G. Ni Chonghaile, and V. Savolainen. 2009. “The Origins and Diversification of C4 Grasses and Savanna-Adapted Ungulates.” Global Change Biology 15 (10): 2397–2417.10.1111/gcb.2009.15.issue-10
- Burke, S. V., W. P. Wysocki, F. O. Zuloaga, J. M. Craine, J. C. Pires, P. P. Edger, D. Mayfield-Jones, L. G. Clark, S. A. Kelchner, and M. R. Duvall. 2016. “Evolutionary Relationships in Panicoid Grasses Based on Plastome Phylogenomics (Panicoideae; Poaceae).” BMC Plant Biology 16 (1): 3247.10.1186/s12870-016-0823-3
- Burke, S. V., C.-S. Lin, W. P. Wysocki, L. G. Clark, and M. R. Duvall. 2016. “Phylogenomics and Plastome Evolution of Tropical Forest Grasses (Leptaspis, Streptochaeta: Poaceae).” Frontiers in plant science 7.
- Cerling, T. E., J. M. Harris, B. J. MacFadden, and M. G. Leakey. 1997. “Global Vegetation Change through the Miocene/Pliocene Boundary.” Nature 389 (6647): 153–158.10.1038/38229
- Christin, P.-A., G. Besnard, E. Samaritani, M. R. Duvall, T. R. Hodkinson, V. Savolainen, and N. Salamin. 2008. “Oligocene CO 2 Decline Promoted C4 Photosynthesis in Grasses.” Current Biology 18 (1): 37–43.10.1016/j.cub.2007.11.058
- Christin, P.-A., E. Spriggs, C. P. Osborne, C. A. E. Strömberg, N. Salamin, and E. J. Edwards. 2014. “Molecular Dating, Evolutionary Rates, and the Age of the Grasses.” Systematic Biology 63 (2): 153–165.10.1093/sysbio/syt072
- Clayton, W. D., M. S. Vorontsova, K. T. Harman, and H. Williamson. 2014. “GrassBase: The Online World Grass Flora.” Accessed July 20, 2014. http://www.kew.org/data/grasses–db
- Danckwerts, J. E. 1993. “Reserve Carbon and Photosynthesis: Their Role in Regrowth of Themeda Triandra, a Widely Distributed Subtropical Graminaceous Species.” Functional Ecology 7 (5): 634–641.10.2307/2390141
- Danecek, P., A. Auton, G. Abecasis, C. A. Albers, E. Banks, M. A. DePristo, R. E. Handsaker, et al. 2011. “The Variant Call Format and VCFtools.” Bioinformatics 27 (15): 2156–2158.10.1093/bioinformatics/btr330
- Dixon, A. P., D. Faber-Langendoen, C. Josse, J. Morrison, and C. J. Loucks. 2014. “Distribution Mapping of World Grassland Types.” Journal of Biogeography 41 (11): 2003–2019.10.1111/jbi.2014.41.issue-11
- Drummond, A. J., and A. Rambaut. 2007. “BEAST: Bayesian Evolutionary Analysis by Sampling Trees.” BMC Evolutionary Biology 7 (1): 214.10.1186/1471-2148-7-214
- Edwards, E. J., C. P. Osborne, C. A. E. Strömberg, S. A. Smith, and C4 Grasses Consortium. 2010. “The Origins of C4 Grasslands: Integrating Evolutionary and Ecosystem Science.” Science 328 (5978): 587–591.
- Estep, M. C., M. R. McKain, D. V. Diaz, J. Zhong, J. G. Hodge, T. R. Hodkinson, D. J. Layton, S. T. Malcomber, R. Pasquet, and E. A. Kellogg. 2014. “Allopolyploidy, Diversification, and the Miocene Grassland Expansion.” Proceedings of the National Academy of Sciences 111 (42): 15149–15154.10.1073/pnas.1404177111
- Everson, T. M., R. I. Yeaton, and C. S. Everson. 2009. “Seed Dynamics of Themeda triandra in the Montane Grasslands of South Africa.” African Journal of Range and Forage Science 26 (1): 19–26.10.2989/AJRFS.2009.26.1.3.698
- Gibbs Russell, G. E., L. Watson, M. Koekemoer, L. Smook, N. P. Barker, H. M. Anderson, and M. J. Dallwitz. 1991. Grasses of Southern Africa. Memoirs of the Botanical Survey of South Africa, No. 58. Pretoria: Botanical Research Institute.
- Giussani, L. M., J. H. Cota-Sanchez, F. O. Zuloaga, and E. A. Kellogg. 2001. “A Molecular Phylogeny of the Grass Subfamily Panicoideae (Poaceae) Shows Multiple Origins of C4 Photosynthesis.” American Journal of Botany 88 (11): 1993–2012.10.2307/3558427
- Grass Phylogeny Working Group II. 2012. “New Grass Phylogeny Resolves Deep Evolutionary Relationships and Discovers C4 Origins.” New Phytologist 193 (2): 304–312.
- Hayman, D. L. 1960. “The Distribution and Cytology of the Chromosome Races of Themeda australis in Southern Australia.” Australian Journal of Botany 8 (1): 58–68.10.1071/BT9600058
- Hu, F. Y., D. Y. Tao, E. Sacks, B. Y. Fu, P. Xu, J. Li, Y. Yang, et al. 2003. “Convergent Evolution of Perenniality in Rice and Sorghum.” Proceedings of the National Academy of Sciences 100 (7): 4050–4054.10.1073/pnas.0630531100
- Huelsenbeck, J. P., and F. Ronquist. 2001. “MrBayes: Bayesian Inference of Phylogenetic Trees.” Bioinformatics 17 (8): 754–755.
- Jessop, J., G. R. M. Dashorst, and F. M. James. 2006. Grasses of South Australia: An Illustrated Guide to the Native and Naturalised Species. Kent Town, Australia: Wakefield Press.
- Katoh, K., and D. M. Standley. 2013. “MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability.” Molecular Biology and Evolution 30 (4): 772–780.10.1093/molbev/mst010
- Keir, A. F., and W. D. Vogler. 2006. “A Review of Current Knowledge of the Weedy Species Themeda quadrivalvis (Grader Grass).” Tropical Grasslands 40 (4): 193.
- Kellogg, E. A., and L. Watson. 1993. “Phylogenetic Studies of a Large Data Set. I. Bambusoideae, Andropogonodae, and Pooideae (Gramineae).” The Botanical Review 59 (4): 273–343.10.1007/BF02857419
- Langmead, B., and S. L. Salzberg. 2012. “Fast Gapped-Read Alignment with Bowtie 2. 2012.” Nature methods 9 (4): 357–359.
- Langmead, B., C. Trapnell, M. Pop, and S. L. Salzberg. 2009. “Ultrafast and Memory-Efficient Alignment of Short DNA Sequences to the Human Genome.” Genome Biology 10 (3): R25.10.1186/gb-2009-10-3-r25
- Lehmann, C. E. R., and C. L. Parr. 2016. “Tropical Grassy Biomes: Linking Ecology, Human Use and Conservation.” Philosophical Transactions of the Royal Society B: Biological Sciences 371 (1703): 20160329.10.1098/rstb.2016.0329
- Lehmann, C. E. R., T. M. Anderson, M. Sankaran, S. I. Higgins, S. Archibald, W. A. Hoffmann, N. P. Hanan, et al. 2014. “Savanna Vegetation-Fire-Climate Relationships Differ Among Continents.” Science 343 (6170): 548–552.10.1126/science.1247355
- Li, H., B. Handsaker, A. Wysoker, T. Fennell, J. Ruan, N. Homer, G. Marth, G. Abecasis, and R. Durbin. 2009. “The Sequence Alignment/Map format and SAMtools.” Bioinformatics 25 (16): 2078–2079.10.1093/bioinformatics/btp352
- Linder, P. H., and P. J. Rudall. 2005. “Evolutionary History of Poales.” Annual Review of Ecology, Evolution, and Systematics 36 (1): 107–124.10.1146/annurev.ecolsys.36.102403.135635
- Lundgren, M. R., G. Besnard, B. S. Ripley, C. E. R. Lehmann, D. S. Chatelet, R. G. Kynast, M. Namaganda, et al. 2015. “Photosynthetic Innovation Broadens the Niche within a Single Species.” Ecology Letters 18 (10): 1021–1029.10.1111/ele.12484
- Mathews, S., R. E. Spangler, R. J. Mason-Gamer, and E. A. Kellogg. 2002. “Phylogeny of Andropogoneae Inferred from Phytochrome B, GBSSI, and ndhF.” International Journal of Plant Sciences 163 (3): 441–450.10.1086/339155
- Maurin, O., T. J. Davies, J. E. Burrows, B. H. Daru, K. Yessoufou, A. M. Muasya, M. Bank, and W. J. Bond. 2014. “Savanna Fire and the Origins of the ‘Underground Forests’ of Africa.” New Phytologist 204 (1): 201–214.10.1111/nph.12936
- McNaughton, S. J. 1985. “Ecology of a Grazing Ecosystem: The Serengeti.” Ecological Monographs 55 (3): 259–294.10.2307/1942578
- Mills, A. J., T. G. O’Connor, J. S. Donaldson, M. V. Fey, A. L. Skowno, A. M. Sigwela, R. G. Lechmere-Oertel, and J. D. Bosenberg. 2005. “Ecosystem Carbon Storage under Different Land Uses in Three Semi-Arid Shrublands and A Mesic Grassland in South Africa.” South African Journal of Plant and Soil 22 (3): 183–190.10.1080/02571862.2005.10634705
- Milton, S. J. 1993. “Studies of Herbivory and Vegetation Change in Karoo Shrublands.” PhD Thesis, University of Cape Town, South Africa.
- Morales, V. F. 2014. “Taxonomy, Distribution and Functional Traits in Themeda Forssk. (Poaceae).” Masters Thesis, University of Edinburgh, United Kingdom.
- Morgan, J. W., and I. D. Lunt. 1999. “Effects of Time-Since-Fire on the Tussock Dynamics of a Dominant Grass (Themeda triandra) in a Temperate Australian Grassland.” Biological Conservation 88 (3): 379–386.10.1016/S0006-3207(98)00112-8
- Olofsson, J. K., M. Bianconi, G. Besnard, L. T. Dunning, M. R. Lundgren, H. Holota, M. S. Vorontsova, et al. 2016. “Genome Biogeography Reveals the Intraspecific Spread of Adaptive Mutations for a Complex Trait.” Molecular Ecology 25 (24): 6107–6123.10.1111/mec.2016.25.issue-24
- Osborne, Colin P. 2008. “Atmosphere, Ecology and Evolution: What Drove the Miocene Expansion of C4 Grasslands?” Journal of Ecology 96 (1): 35–45.
- Patel, R. K., and M. Jain. 2012. “NGS QC Toolkit: A Toolkit for Quality Control of Next Generation Sequencing Data.” PLoS One 7 (2): e30619.10.1371/journal.pone.0030619
- Paterson, A. H., J. E. Bowers, R. Bruggmann, J. Grimwood, H. Gundlach, G. Haberer, U. Hellsten, et al. 2009. “The Sorghum bicolor Genome and the Diversification of Grasses.” Nature 457: 551–556.
- Pennington, R. T., and M. Lavin. 2016. “The Contrasting Nature of Woody Plant Species in different Neotropical Forest Biomes Reflects differences in Ecological Stability.” New Phytologist 210 (1): 25–37.10.1111/nph.13724
- Prasad, V., C. A. E. Strömberg, A. D. Leaché, B. Samant, R. Patnaik, L. Tang, D. M. Mohabey, S. Ge, and A. Sahni. 2011. “Late Cretaceous Origin of the Rice Tribe Provides Evidence for Early Diversification in Poaceae.” Nature Communications 2: 480.10.1038/ncomms1482
- du Preez, C. C., and H. A. Snyman. 1993. “Research Note: Organic Matter Content of a Soil in A Semi-Arid Climate with Three Long-Standing Veld Conditions.” African Journal of Range & Forage Science 10 (2): 108–110.10.1080/10220119.1993.9638333
- Reynolds, C., and G. S. Cumming. 2016. “Seed Dispersal by Waterbirds in Southern Africa: Comparing the Roles of Ectozoochory and Endozoochory.” Freshwater Biology 61 (4): 349–361.10.1111/fwb.2016.61.issue-4
- Sage, R. F., and M. Stata. 2015. “Photosynthetic Diversity Meets Biodiversity: The C4 Plant Example.” Journal of Plant Physiology 172: 104–119.10.1016/j.jplph.2014.07.024
- Sánchez-Ken, J. Gabriel, and L. G. Clark. 2010. “Phylogeny and A New Tribal Classification of the Panicoideae sl (Poaceae) based on Plastid and Nuclear Sequence Data and Structural Data.” American Journal of Botany 97 (10): 1732–1748.10.3732/ajb.1000024
- Scheiter, S., S. I. Higgins, C. P. Osborne, C. Bradshaw, D. Lunt, B. S. Ripley, L. L. Taylor, and D. J. Beerling. 2012. “Fire and Fire‐Adapted Vegetation Promoted C4 Expansion in the Late Miocene.” New Phytologist 195 (3): 653–666.
- Simon, M. F., R. Grether, L. P. de Queiroz, C. Skema, R. T. Pennington, and C. E. Hughes. 2009. “Recent Assembly of the Cerrado, A Neotropical Plant Diversity Hotspot, by in situ Evolution of Adaptations to Fire.” Proceedings of the National Academy of Sciences 106 (48): 20359–20364.10.1073/pnas.0903410106
- Simpson, K. J., B. S. Ripley, P.-A. Christin, C. M. Belcher, C. E. R. Lehmann, G. H. Thomas, and C. P. Osborne. 2016. “Determinants of Flammability in Savanna Grass Species.” Journal of Ecology 104 (1): 138–148.10.1111/1365-2745.12503
- Snyman, H. A., L. J. Ingram, and K. P. Kirkman. 2013. “Themeda triandra: A Keystone Grass Species.” African Journal of Range & Forage Science 30 (3): 99–125.10.2989/10220119.2013.831375
- Spriggs, E. L., P.-A. Christin, and E. J. Edwards. 2014. “C4 Photosynthesis Promoted Species Diversification during the Miocene Grassland Expansion.” PLoS One 9 (5): e97722.10.1371/journal.pone.0097722
- Veldkamp, J. F. 2016. “A Revision of Themeda (Gramineae) in Malesia with a New Species from Laos.” Blumea-Biodiversity, Evolution and Biogeography of Plants 61 (1): 29–40.10.3767/000651916X691349
- Vuorio, V., A. Muchiru, R. S. Reid, and J. O. Ogutu. 2014. “How Pastoralism Changes Savanna Vegetation: Impact of Old Pastoral Settlements on Plant Diversity and Abundance in South-Western Kenya.” Biodiversity and Conservation 23 (13): 3219–3240.10.1007/s10531-014-0777-4
- Zerbino, D. R., and Ewan Birney. 2008. “Velvet: Algorithms for de novo Short Read assembly using de Bruijn Graphs.” Genome Research 18 (5): 821–829.10.1101/gr.074492.107