Abstract
The types of kernel function and relevant parameters’ selection in support vector machine (SVM) have a major impact on the performance of the classifier. In order to improve the accuracy and generalization ability of the model, we used mixed kernel function SVM classification algorithm based on the information entropy particle swarm optimization (PSO): on the one hand, the generalization ability of classifier is effectively enhanced by constructing a mixed kernel function with global kernel function and local kernel function; on the other hand, the accuracy of classification is improved through optimization for related kernel parameters based on information entropy PSO. Compared with PSO-RBF kernel and PSO-mixed kernel, the improved PSO-mixed kernel SVM can effectively improve the classification accuracy through the classification experiment on biomedical datasets, which would not only prove the efficiency of this algorithm, but also show that the algorithm has good practical application value in biomedicine prediction.
Introduction
Support vector machine (SVM) is a new machine learning method proposed by Vapnik [Citation1] based on statistical learning theory in the 1990s. It was established on the structural risk minimization principle, and it could obtain very good generalization ability with limited information of samples. SVM, using kernel methods, transforms the non-linear unseparable problem in low-dimensional space into the linearly separable problem in high-dimensional feature space and solves the “dimension disaster” problem when computing in the high-dimensional feature space.
In recent years, there have been many scholars applying SVM classification algorithm in the biomedical field to make a prediction or classification on disease or cancer gene, and significant results have been achieved. For example, Khodor et al. [Citation2] analyzed the autonomic nervous system response during head-up tilt test (HUTT) and extracted relative features of the first 15 min of tilting position. The SVM prediction results showed that these indices might be good predictors of positive response in patients with reflex syncope. Yu et al. [Citation3] used detrended fluctuation analysis (DFA) method to analyze Intron-containing and intronless gene sequences, SVM classifier with Gaussian radial basis kernel function was performed on this feature space which proved that this method significantly improved the accuracy over those existing techniques. Mao et al. [Citation4] used random forest and SVM with radial basis kernel function (RBF) to differentiate retained introns (RIs) and constitutively spliced introns (CSIs) in Arabidopsis. Chen et al. [Citation5] added the information of mutation and chromosome as gene features which improved the prediction performance of five primary sites (large intestine, liver, skin, pancreas, and lung). The results demonstrated that the somatic mutation information was useful for prediction of primary tumor sites with machine learning modeling. Spilka et al. [Citation6] promoted Sparse SVM classification that permitted to select a small number of relevant features and to achieve efficient fetal acidosis detection.
While applied to practical problems, the selection of kernel function and parameters of SVM is critical, it could directly affect the performance of the SVM classifier, the selection of kernel function is the key in the SVM classification. At present, studies in the kernel function theory mainly concentrated in three aspects: the first is the properties of the kernel function [Citation7]; the second is the structure (or improved) method of the kernel function [Citation8,Citation9]; the third is the parameter selection of kernel function.[Citation10]
There are many kinds of kernel functions, commonly used kernel functions are the following: linear kernel function, polynomial kernel function, radial basis kernel function (RBF) and sigmoid kernel function. In 2003, Keerthi et al. [Citation11] studied the RBF kernel and its two involved kernel parameters (penalty parameter c and the kernel width g), and also analyzed the performance of the SVM classifier while kernel parameters took different values; the results showed that if RBF kernel model was used, there was no need to consider the linear support vector machine; Lin et al. [Citation12] explained that the sigmoid kernel and RBF kernel had similar performance in certain parameters. Ricatte et al. [Citation13] compared the advantages and the disadvantages of sigmoid kernel and linear kernel in image classification. In general, the RBF kernel is most widely used owing to its strong learning ability and fewer parameters. Some scholars tried to make a better classification performance by improving the classic kernel function. For example, Remaki et al. [Citation14] thought that the Gaussian kernel had two practical limitations: information loss caused by the unavoidable Gaussian truncation and the prohibitive processing time due to the mask size, so they proposed a new kernel family derived from the Gaussian with compact supports, and an application of extracting handwritten data from noisy document images was presented to evaluate the effectiveness of the improved algorithm.
In 2001, Scholkopf et al. [Citation15] studied the nature of the kernel function in support vector machines, and for the first time proposed that kernel function was divided into global kernel function and local kernel function. Smits et al. [Citation16] pointed out that the extrapolation performance of global kernel function was better; however, the interpolation performance of local kernel function was better. For the first time, they used the linear combination of two kinds of different kernel functions and showed that using mixtures of kernels can result in having both good interpolation and extrapolation abilities, but the study did not analyze the effect of weight coefficient for SVM classification results. In 2004, Lanckriet et al. [Citation17] proposed the multiple kernel-learning frameworks composed of multiple basic kernel and proved that SVM of multiple kernel had a series of advantage that single kernel SVM did not have. In 2009, Jagarlapudi et al. [Citation18] proposed mixed-norm regularization for Multiple Kernel Learning (MKL), in the meanwhile Cortes et al. [Citation19] discussed the possibility of nonlinear combination of basic kernels. Kumar et al. [Citation20] analyzed a linear combination of two common kernel function and compared their classification accuracy rate while given different weight coefficient, the result provided some reference for the assignment of weight coefficient. In 2011, Hovestadt et al. [Citation21] improved the classification accuracy while setting the weight coefficient of mixed kernel function empirically, and achieved effective weight range which could increase the classifier performance. Although the classification accuracy was improved, the method of empirically assigning the mixed weight yet had a certain randomness.
After the selection of kernel function, related kernel parameters should be determined. Marcou et al. [Citation22] used the matrix similarity measurement to select kernel parameters. Chapelle et al. [Citation23] tuned multiple parameters for pattern recognition SVMs automatically by minimizing some estimates of the generalization error of SVM using a gradient descent algorithm over the set of parameters. Usual methods for choosing parameters become intractable as soon as the number of parameters exceeds two. The experimental results assessed the feasibility of the approach for a large number of parameters (more than 100) and demonstrated an improvement of generalization performance. But the quantitative accuracy of the estimate used for the gradient descent is only marginally relevant, which raises the question of how to design good estimates for parameter tuning rather than accurate estimates. In recent years, the emerging intelligent optimization algorithms have been applied to SVM parameter selection problem. Fernandez et al. [Citation24] used genetic algorithm (GA) for parameter optimization, which improved the generalization ability of SVMs, then Subasi et al. [Citation25] used particle swarm optimization (PSO) algorithm to optimize the parameters, and proved that the improvement of the performance of PSO-SVM was more significant compared with other classification methods. Ardjani et al. [Citation26] compared the performance of GA and PSO for parameters optimization in SVM, and thought that PSO was more efficient in coding and optimization strategy, moreover its computation cost is lower than GA. Huang et al. [Citation27] used the discrete PSO-SVM for feature selection and parameters optimization, the experiment proved that this method can effectively select features and improve classification accuracy. In this paper, we use the PSO optimization algorithm based on information entropy to optimize the penalty parameter, the kernel parameter and the weight coefficient of the mixed kernel, which can effectively improve the generalization ability of classifier. The experimental results showed that the improved algorithm significantly increased the classification accuracy compared with the traditional PSO-RBF kernel SVM algorithm.
The structure of the article is as follows: the second section is mixed kernel function SVM classification algorithm based on the particle information entropy PSO optimization. The third section is the results of the experiment, compared with PSO-RBF kernel and PSO-mixed kernel SVM classification, the results demonstrate that the improved PSO-mixed kernel SVM can significantly improve the generalization performance of support vector machine, as with the precision of classification, which demonstrates the effectiveness of the improved algorithm. The fourth section is the discussion of the experimental results. The fifth section is the conclusion of this study.
Proposed algorithm
Kernel function selection
According to Scholkopf’s [Citation15] theory, there are two main types of kernels, namely local and global kernels. In local kernels, only the data that are close or in the proximity of each other have an influence on the kernel values, so it has strong learning ability and weak generalization ability. However, a global kernel allows data points far away from each other to have an influence on the kernel values as well, which have strong generalization ability and weak learning ability. The commonly used kernel functions are as follows: linear kernel K(x, xi) = x ⋅ xi;
polynomial kernel K(x, xi) = ((x*xi) + 1)d;
RBF kernel K (x, xi) = exp (−gamma*ǁx-xiǁ2);
sigmoid kernel K(x, xi) = tanh (k(x, xi)−δ)
gives the mapping characteristics of RBF kernel function, polynomial kernel function, linear kernel function, and sigmoid kernel function, the value of test point a = 0.2 [Citation16]. shows that RBF kernel is local kernel, the input test points of which are influenced only by the data in the proximity of it, the mapping values of the data points far away from the test point are approaching to zero. shows that linear kernel, polynomial kernel, and sigmoid kernel are global kernel whose input test points can be influenced by the data points far away from it.
Figure 1. Mapping characteristics based on four kinds of kernel functions (a) RBF kernel function, (b) polynomial kernel function, (c) linear kernel function, and (d) sigmoid kernel function.
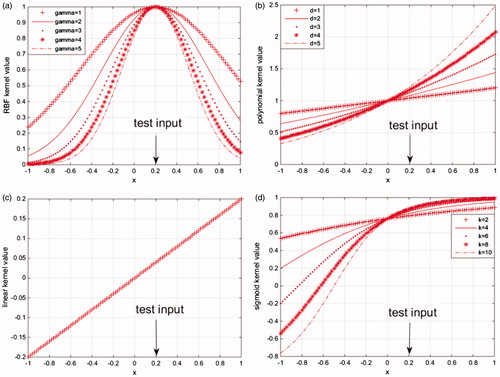
Compare the figures of polynomial kernel function and linear kernel function, we can find out that their mapping properties are the same when d = 1 used in the polynomial kernel function, that is to say linear kernel function is a special case of the polynomial. Compare the figures of polynomial kernel function and sigmoid kernel function, we can find out that the test points of polynomial kernel function can be influenced by the entire set of data points either it is near or far, which could generate good extrapolation capability. But the global performance of the sigmoid kernel function is better on the left side of the test input point, the global performance is relatively poor on the right side of the test input point.
In order to establish a model with both good interpolation capability and good extrapolation capability as well, it is necessary to make full use of the respective advantages of local kernel function and global kernel function. The local kernel function has good learning ability while the global kernel function has good generalization ability. The finite performance of the existing kernel functions can be solved by using a convex combination of these two kernels, thus the mixed kernel function not only has better learning ability but also has better generalization ability. According to the composition conditions of kernel functions,[Citation28] the linear mixture of kernels is still a kernel function. So the mixture of kernels may be defined as
(1)
where u is the mixing weight coefficient. shows the mapping characteristics of which the RBF kernel function combine linearly with polynomial kernel function, linear kernel function, and sigmoid kernel function separately, the value of test point a = 0.2, the mixing weight coefficients u is fixed to 0.2, 0.4, 0.6, and 0.8.
Figure 2. Mapping characteristics based on three kinds of mixed kernel functions (a) RBF kernel mixes with linear function, (b) RBF kernel mixes with polynomial kernel function, and (c) RBF kernel mixes with sigmoid kernel function.
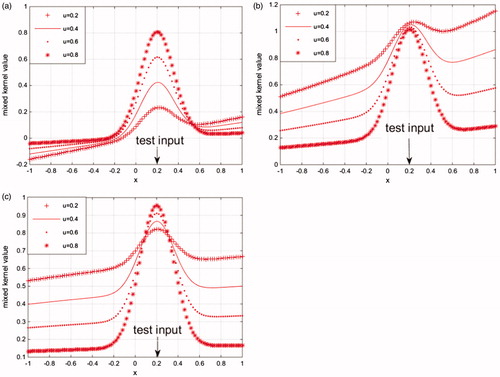
From , we can see that the mixed kernel not only has a local effect but also has a global effect. The global performance of the linear combination of RBF kernel and linear kernel is great, but its local performance declines with the reduction of the weight coefficient u (see ). The global performance of the linear combination of RBF kernel and sigmoid kernel is great, but its global performance declines with the reduction of the weight coefficient u (see ). However, when the weight coefficient of the mixed kernel function of RBF kernel and polynomial kernel changes, its global performance and local performance changed little (see ), thus its stability is better than the two former combination. In conclusion, we choose RBF kernel and polynomial as a mixed kernel function of support vector machine in this paper.
The particle swarm optimization algorithm
The weight coefficient u was fixed subjectively in most studies on the mixed kernel function,[Citation20,Citation21] which makes the research results with great randomness. In order to get a better classification effect, we choose the particle swarm optimization algorithm to optimize the parameters of support vector machine in this paper, which can further enhance the learning ability and generalization ability of the classifier.
In the model of particle swarm optimization algorithm, the feasible solution of every optimization problem is considered as a particle, the state of each particle is described by a group of position vectors and velocity vectors. The particles find their own historical optimal solution and the optimal solution of group by changing its velocity and position,[Citation29] and then update their velocity and position through the velocity and position equations, the velocity updating formula is
(2)
the position updating formula is
(3)
The basic PSO algorithm just needs to determine a few parameters, so it is easy to use. But the main drawback of this algorithm is that the particle is easily trapped in local minima, therefore, the search accuracy is not high enough.
Some scholars put forward the improved algorithm based on fitness feedback,[Citation30] because particles are moving toward the individual and global optimal position, so this improved algorithm has reduced their volatility in a certain extent, and also enhanced their movement trend to the position of the optimal fitness. However, this single index only considers the selection of population but not the variety, once after the particles trapped in local minima it is hard to get out of it, this kind of defect is especially remarkable when solving problem with more than one local extremum.
Information entropy
In 1948, Shannon [Citation31] put forward the concept of entropy, and used it as a measure of information. For a specific system, on the one hand, if its randomness is very great and chaotic, the information entropy of this system is large. On the other hand, if a system is definite with certain rules, the information entropy of the system is very small.
For the particle swarm, its information entropy reflects the distribution of the particles in the solution space, the greater the information entropy is, the sparser the distribution of particles are; the smaller the information entropy is, the denser the distribution of particles are. In order to effectively prevent algorithm falls into local optimum and improve the convergence speed and precision of the algorithm, we used cooperation feedback of information entropy and fitness to improve the diversity of particle population.
The information entropy of particles can effectively evaluate the diversity of particle population. Mutating the particle at a certain probability when the diversity decreases, which can restrain the premature convergence of particle and allows the particles to jump out of local optimum in time to find a better solution; on the contrary, that makes the particles have the ability to explore new areas from the early stages of iteration to the end. The calculation formula of particle information entropy [Citation32] is as follows:
(4)
(5)
where NHδ xi (B) is the information entropy of the ith particle, NHδ(B) is the information entropy of the whole group. The proposed variation conditions in this paper are as follows:
(6)
(7)
(8)
where j is the current step number, i is the index of particle, N is the number of particles. Formula (6) means that the information entropy of the ith particle is less than the information entropy of whole particle swarm, which is to say that the optimization process at this time is turning bad, and the population diversity is reducing gradually. In formula (7), Fg is the global optimal fitness, Fm is the average fitness of all particles at the current iteration; if the absolute value of the difference between these two is less than
the movement trending to the most optimal position of the particles should be enhanced at this point. Formula (8) is the mutation probability of the particle, which gradually reduce with the increase of the number of iterations. Formula (6) is the evaluation of the information entropy of the particle, formula (7) is the fitness evaluation of the particle, formula (8) is the mutation probability, particles which satisfy these three conditions will mutate. The mutation method is to randomly project 20 new particles into the entire solution space, the position formula of the particles is
(9)
where pop(j,:) indicates the position of the jth particle, popmax and popmin indicate the boundary range of the particle. Calculate the fitness according to the location of the new particles, if it is less than the fitness of itself in the history, then update the position of this particle. This process would not only increase the range that particle search in the whole solution space but also increase diversity of the particle population, which can inhibit the local convergence effectively. This improved algorithm is especially suitable for function optimization problems with high dimension and more than one local extremum.
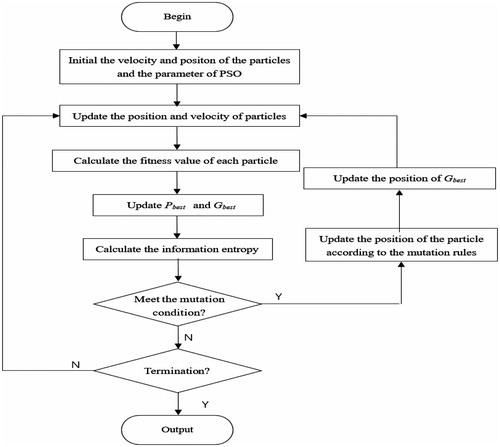
Flowchart of the improved algorithm
The traditional PSO-RBF kernel SVM method need to optimize two parameters (the RBF kernel width g, penalty parameter c), the improved PSO-mixed kernel SVM need to optimize three parameters: the RBF kernel width g, penalty parameter c, and the mixed weight coefficient u. The basic process of the mixed kernel function SVM classification algorithm based on the information entropy particle swarm optimization algorithm is given as follows.
Step 1: Randomly generate the initial velocity and position of the particles.
Step 2: Initialize the parameter of PSO.
Step 3: Update the position and velocity of particles according to formulas (2) and (3).
Step 4: Calculate the fitness value of each particle, the particle's Pbest is set as the current optimal position; Gbest is set as the global optimal position in the initial population.
Step 5: Mutate the particle if it meets the mutation condition according to formulas (4)–(8); if the fitness is better than Pbest’s, update Pbest with the position of the new particle.
Step 6: Turn to Step 7 if termination criterion is satisfied; return to Step 3 otherwise.
Step 7: Output the value of the parameters c, g, u, and the classification accuracy rate of the test set; the flowchart of the algorithm is shown in .
Experimental results
Datasets
In order to verify the effectiveness of the algorithm, we selected five datasets from the UCI repository [Citation33] and the cancer gene repository [Citation34] to conduct classification problems. The basic description information of these datasets is shown in .
Table 1. Main characteristics of datasets.
Parkinsons dataset [Citation35] was extracted from the UCI repository, it includes 195 samples, the number of features is 22, the number of classes is 2, and the training set and test set of the sample are 101 and 94, respectively. Statlog (Heart) [Citation36] dataset was extracted from the UCI repository, it includes 270 samples, the number of features is 13, the number of classes is 2, and the training set and test set of the sample are 150 and 120, respectively. DLBCL-B [Citation37] dataset was extracted from the cancer gene repository, it includes 180 samples, the number of features is 661, the number of classes is 3, and the training set and test set of the sample are 99 and 81, respectively. Diabetes dataset [Citation38] was extracted from the UCI repository, it includes 768 samples, the number of features is 8, the number of classes is 2, and the training set and test set of the sample are 500 and 268, respectively. Lung-A dataset [Citation39] was extracted from the cancer gene repository, it includes 197 samples, the number of features is 1000, the number of classes is 4, the training set and test set of the sample are 131 and 66, respectively.
Results
The classification experiment of training and testing was conducted in Matlab2015b development environment. The parameters of the particle swarm optimization algorithm were set as follows: learning factor c = 1.4945, the number of particles is 40; the number of iterations is 100, the training set used five-fold cross validation. The optimization parameter of PSO-RBF kernel, PSO-mixed kernel and improved PSO-mixed kernel support vector machine and the classification accuracy rate of the test set are shown in .
Table 2. The optimization parameters c, g, u of PSO-RBF kernel, PSO-mixed kernel and improved PSO-mixed kernel SVM and classification accuracy rate of the test set.
From , we can see that the weight coefficient u of the Parkinsons dataset, DLBCL-B dataset and Lung-A dataset are both above 0.5, that is to say RBF kernel function accounted for the dominant position in the classification process among these two datasets, which means that the datasets are relatively dense after mapping to the high dimensional space. However, the weight coefficient u of the Statlog (Heart) dataset, Diabetes dataset are both less than 0.5, indicating that polynomial kernel function accounted for a dominant position in the classification among these datasets, which can reflect that the datasets are relatively sparse after mapping to the high dimensional space. The distribution of the data points of different datasets after mapping to the high-dimensional space is unknown, which demonstrate that the previous method of assigning u value empirically is not reliable.
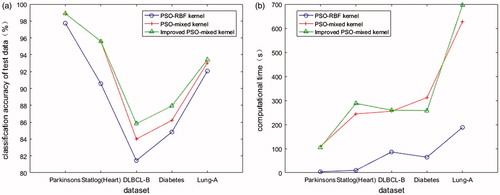
The performance of the classifier is mainly evaluated by the classification precision of the test set, the classification accuracy rate, and computational time line chart of the optimized PSO-RBF kernel, PSO-mixed kernel, and improved PSO-mixed kernel support vector machine is presented in .
As is shown in , compare PSO-mixed kernel with PSO-RBF kernel, five classification accuracy rate all improved, indicating that linear combination of the polynomial kernel and RBF kernel is better than single RBF kernel on the generalization ability; compare improved PSO-mixed kernel with PSO-mixed kernel, the classification accuracy rate of Parkinsons dataset and Statlog (heart) dataset is unchanged, but the classification accuracy rate of DLBCL-B dataset, Diabetes dataset, and Lung-A dataset improved in a certain extent. The features and instances of these three dataset are relatively larger, the distribution of the data points in high-dimensional space may be more complex after using mixed kernel function. And the precision of improved PSO is better in the process of optimization under this condition, thus the final classification accuracy has been further improved. From , we can see that the computational time of improved PSO-mixed kernel and PSO-kernel is longer than PSO-RBF kernel due to the extra parameter optimization and inner product computing (mostly). This paper mainly focused on testifying the effectiveness of the classification algorithm, and we did not process the feature selection. So we had to believe that the computational time will be greatly reduced after a series of feature selection steps which will further improve the classification accuracy.
Discussion
The results showed that the classification accuracy of the algorithm in this paper is higher than the tradition PSO-SVM algorithm. Mostly because the mixed kernel SVM has both local and global characters, it can efficiently extract the mapping characters of datasets and has higher adaptability to complex data. Moreover, the improved PSO based on information entropy can optimize the parameters of SVM especially the weight coefficient which can greatly play the role of the mixed kernel functions. The optimization of different data sets would obtain different weights; it also suggests that the empirically assignment method of the mixed kernel weight is not reliable. From a public data set website, we have found the classification application of the same three datasets used as in this paper. For example, Igor et al. re-implemented Assistant, a system for top down induction of decision trees, using RELIEFF as an estimator of attributes at each selection step. The results were compared with some other well-known machine learning algorithms (like LFC, Assistant-I, Assistant-R, Naive Bayes, k-NN). As a result, the accuracy of Statlog (Heart) and Diabetes datasets used Naive Bayes algorithm is 76.3% and 84.5%, respectively. Besides, Monti et al. presented a new methodology of class discovery and clustering validation tailored to the task of analyzing gene expression data over multiple runs of a clustering algorithm (such as K-means, model-based Bayesian clustering, and SOM), so as to account for its sensitivity to the initial conditions. The accuracy of model-based Bayesian clustering classification on Lung-A dataset used this improved method reached 90.4%. Anyway, compared with the method mentioned above, the accuracy of our method on these three datasets is higher. In general, the training time of the classification model is longer than the conventional kernel SVM due to the extra parameter optimization and inner product computing, but it is acceptable when we consider the computational time of test data after the model has been built. This paper focused on testifying the effectiveness of the classification algorithm, and we did not process the feature selection. So we had believe that the computational time will be greatly reduced after a series of feature selection steps which will further improve the classification accuracy too. We only analyzed the linear combination of several common kernel functions, the future work can focus on using linear combination of other kernel functions, or even discuss the cases of non-linear combination, we can also process different feature selection methods before classification, or the PSO algorithm can be further improved to promote the precision of the parameters, so as to further enhance the performance of the classifier.
Conclusions
The mixed kernel function SVM classification algorithm based on the information entropy particle swarm optimization contains two strategies. On the one hand, the mixed kernel function used in this paper is the linear combination of polynomial kernel and RBF kernel in support vector machine, and its learning ability and generalization ability is enhanced than RBF kernel. On the other hand, the PSO algorithm based on information entropy is used to search the relative kernel parameters, the feedback of information entropy can effectively improve the diversity of particles and improve the precision of parameter optimization, thus further improved the classification accuracy rate. Compared with PSO-RBF kernel and PSO-mixed kernel, the improved PSO-mixed kernel SVM can effectively improve the classification accuracy of the classification experiment of biomedical datasets, and it also shows that the algorithm has good practical application value in biomedicine prediction.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Funding
This work is supported by the National Basic Research Program of China (2014CB744600), the National Natural Science Foundation of China (61602017 and 61420106005), the Beijing Outstanding Talent Training Foundation (2014000020124G039), the Beijing Natural Science Foundation (4164080), the International Science & Technology Cooperation Program of China (2013DFA32180), the Grant-in-Aid for Scientific Research (C) from Japan Society for the Promotion of Science (26350994), the Beijing Municipal Science and Technology Project (D12100005012003), the Beijing Municipal Administration of Hospitals Clinical Medicine Development of Special Funding (ZY201403), and the Beijing Municipal Science and technology achievement transformation and industrialization projects funds (Z121100006112057).
References
- Vapnik V. The nature of statistical learning theory. Berlin, Heidelberg: Springer Science & Business Media; 2013.
- Khodor N, Matelot D, Carrault G, et al. Kernel based support vector machine for the early detection of syncope during head-up tilt test. Physiol Meas. 2014; 35:2119–2134.
- Yu C, Deng M, Zheng L, et al. DFA7, a new method to distinguish between intron-containing and intronless genes. PLoS One. 2014;9:e101363.
- Mao R, Kumar PKR, Guo C, et al. Comparative analyses between retained introns and constitutively spliced introns in Arabidopsis thaliana using random forest and support vector machine. PLoS One. 2014;9:e104049.
- Chen Y, Sun J, Huang LC, et al. Classification of cancer primary sites using machine learning and somatic mutations. Biomed Res Int. 2015;2015:1–9.
- Spilka J, Frecon J, Leonarduzzi R, et al. Sparse support vector machine for intrapartum fetal heart rate classification. IEEE J Biomed Health Inform. 2016;99:1.
- Kung SY. Kernel methods and machine learning. Cambridge: Cambridge University Press; 2014.
- Dioşan L, Rogozan A, Pecuchet JP. Improving classification performance of support vector machine by genetically optimising kernel shape and hyper-parameters. Appl Intell. 2012;36:280–294.
- Yuan L, Chen F, Zhou L, et al. Improve scene classification by using feature and kernel combination. Neurocomputing. 2015;170:213–220.
- Binol H, Bal A, Cukur H. Differential evolution algorithm-based kernel parameter selection for Fukunaga–Koontz transform subspaces construction. Proceedings of the SPIE, High-Performance Computing in Remote Sensing V; October 2015. p. 9646.
- Keerthi SS, Lin CJ. Asymptotic behaviors of support vector machines with Gaussian kernel. Neural Comput. 2003;15:1667–1689.
- Lin HT, Lin CJ. A study on sigmoid kernels for SVM and the training of non-PSD kernels by SMO-type methods. Neural Comput. 2003;1–32.
- Ricatte T, Garriga G, Gilleron R, et al. Learning from multiple graphs using a sigmoid kernel//machine learning and applications (ICMLA). IEEE on the 12th International Conference, Vol. 2; 2013. p. 140–145.
- Remaki L, Cheriet M. KCS-new kernel family with compact support in scale space: formulation and impact. IEEE Trans Image Process. 2000;9:970–981.
- Scholkopf B, Smola AJ. Learning with kernels: support vector machines, regularization, optimization, and beyond. Cambridge: MIT Press; 2001.
- Smits GF, Jordaan EM. Improved SVM regression using mixtures of kernels. IJCNN'02. Proceedings of the 2002 International Joint Conference on IEEE, Vol. 3. 2002. p. 2785–2790.
- Lanckriet GRG, Cristianini N, Bartlett P, et al. Learning the kernel matrix with semidefinite programming. J Mach Learn Res. 2004;5:27–72.
- Jagarlapudi SN, Dinesh G, Raman S, et al. On the algorithmics and applications of a mixed-norm based kernel learning formulation. Adv Neural Inf Process Syst. 2009;844–852.
- Cortes C, Mohri M, Rostamizadeh A. Learning non-linear combinations of kernels. Adv Neural Inf Process Syst. 2009;22:396–404.
- Kumar A, Ghosh SK, Dadhwal VK. Study of mixed kernel effect on classification accuracy using density estimation. Proceedings of the ISPRS Commission VII Symposium 36 (Part 7); 2006.
- Hovestadt T, Binzenhöfer B, Nowicki P, et al. Do all inter‐patch movements represent dispersal? A mixed kernel study of butterfly mobility in fragmented landscapes. J Anim Ecol. 2011;80:1070–1077.
- Marcou GG, Horvath D, Varnek A. Kernel target alignment parameter – a new model ability measure for regression tasks. J Chem Inf Model. 2016;56:6–11.
- Chapelle O, Vapnik V, Bousquet O, et al. Choosing multiple parameters for support vector machines. Mach Learn. 2002;46:131–159.
- Fernandez M, Caballero J, Fernandez L, et al. Genetic algorithm optimization in drug design QSAR: Bayesian-regularized genetic neural networks (BRGNN) and genetic algorithm-optimized support vectors machines (GA-SVM). Mol Divers. 2011;15:269–289.
- Subasi A. Classification of EMG signals using PSO optimized SVM for diagnosis of neuromuscular disorders. Comput Biol Med. 2013;43:576–586.
- Ardjani F, Sadouni K, Benyettou M. Optimization of SVM multiclass by particle swarm (PSO-SVM)//Database Technology and Applications (DBTA). 2010 2nd International Workshop on IEEE; 2010. p. 1–4.
- Huang CL, Dun JF. A distributed PSO–SVM hybrid system with feature selection and parameter optimization. Appl Soft Comput. 2008;8:1381–1391.
- Mercer J. Functions of positive and negative type, and their connection with the theory of integral equations. Philos Trans R Soc Lond. Ser A: Containing Papers of a Mathematical or Physical Character. 1909;209:415–446.
- Kennedy J. Particle swarm optimization. In: Encyclopedia of machine learning. US: Springer. 2010. p. 760–766.
- Tang Y, Wang Z, Fang J. Feedback learning particle swarm optimization. Appl Soft Comput. 2011;11:4713–4725.
- Shannon CE. A mathematical theory of communication. ACM SIGMOBILE Mobile Comput Commun Rev. 2001;5:3–55.
- Hu Q, Pan W, An S, et al. An efficient gene selection technique for cancer recognition based on neighborhood mutual information. Int J Mach Learn Cybern. 2010;1:63–74.
- Machine Learning Repository; 2016. Available from: http://archive.ics.uci.edu/ml/.
- Cancer Program Datasets; 2016. Available from: http://www.broadinstitute.org/cgi-bin/cancer/datasets.cgi.
- Little MA, McSharry PE, Roberts SJ, et al. Exploiting nonlinear recurrence and fractal scaling properties for voice disorder detection. BioMed Eng Online. 2007;6:23.
- Kononenko I, Simec E, Robnik-Sikonja M. Overcoming the myopia of inductive learning algorithms with RELIEFF. Appl Intell. 1997;7:39–55.
- Hoshida Y, Brunet JP, Tamayo P, et al. Subclass mapping: identifying common subtypes in independent disease data sets. PLoS One. 2007;2:e1195.
- Michalski RS, Mozetic I, Hong J, et al. The multi-purpose incremental learning system AQ15 and its testing application to three medical domains. Proceedings of AAAI; 1986. p. 1041–1045.
- Monti S, Tamayo P, Mesirov J, et al. Consensus clustering: a resampling-based method for class discovery and visualization of gene expression microarray data. Mach Learn. 2003;52:91–118.