Abstract
Purpose: Mutual information (MI) has been a preferred choice of similarity measure for multi-modal image registration, but the accuracy and robustness of MI are not satisfactory as MI only considers the global intensity correlation while ignoring local and structural information. To address this problem, we combine MI with local and structural information.
Method: We bring structural information extracted by a modified Accelerated Segment Test (FAST) algorithm into MI. Traditional FAST is transferred into 3 D for the first time, and the 3 D-FAST based structural information is added into MI as another channel, thereby incorporating spatial and geometric information with intensity information in the registration.
Result: The robustness and accuracy of the proposed method were demonstrated in three experiments. The average registration errors of our method were 1.17, 1.33 and 1.20 compared to 1.47, 1.63 and 1.40 of LMI in T1-T2, T1-PD and T2-PD registration respectively.
Discussion: In this paper, we use the structural similarity computed by 3 D-FAST as the conditional information to encode spatial and geometric cues into LMI. In all of these three experiments, our method shows to be more robust and accurate than common registration methods based on information theory.
Introduction
Multi-modal medical image registration is a challenging problem commonly faced in image-guided interventions and image fusion [Citation1]. The main difficulty of the multi-modal image registration comes from the great variability of tissue or organ appearance when imaged with different physical principles, which results in the lack of a general rule to establish structure correspondence. Therefore, efforts to tackle this problem focus mainly on the design of similarity measures. Existing similarity measures in multi-modal image registration can be roughly divided into three categories: (1) similarity measures by matching sparse structural points, (2) transferring different modalities into the same modality and then registering by simple unimodal similarity measures, and (3) similarity measures based on information theory.
In the first category, structural points, such as corners and edges, are extracted from the two images to be registered, then point set registration methods like the Iterative Closest Point (ICP) [Citation2] method, Gaussian Mixture Model (GMM) [Citation3] based point set registration method and Thin Plate Spline-Robust Point Matching (TPS-RPM) [Citation4] can align these structures and register images at the same time. However, it is difficult to extract a sufficient amount of corresponding structural points in some scenarios of the deformable registration of medical images, and the errors between the extracted corresponding points may influence the accuracy of registration.
In the second category, multi-modal images are transferred into the same modality, then a simple similarity measure such as Sum of Absolute Difference (SAD) can register the unimodal images. This category can be further divided into two sub-categories. In the first sub-category, one image is transformed into the modality of the other image. For example, Ultrasound (US) images are simulated from MR/CT images according to the US imaging physics, and then registered to real US images in [Citation5,Citation6]. In the second sub-category, the multi-modal images are transformed into images of an intermediate modality. For example, Wachinger et al. [Citation7] transformed multi-modal images into entropy or Laplacian images. Heinrich et al. [Citation8,Citation9] and Jiang et al. [Citation10,Citation11] transferred multi-modal images to unimodal images with local structure descriptor named ‘Modality Independent Neighborhood Descriptor’ (MIND), ‘Self-Similarity Context’ (SSC), ‘discriminative Local Derivative Pattern’ (dLDP) and ‘modality independent Local Binary Pattern’ (miLBP). Although methods in this category have higher registration speed, they may not be applicable for some tissues or organs with less structural information and the modality transformation may lose a large amount of image information.
The third category explores the statistical dependency between the contents (usually intensity) of the two images. Mutual Information (MI), Cross Correlation (CR) and their variants fall into this category [Citation1] and they are usually more accurate and more generally applicable than voxel-wise matching in the categories above [Citation12]. Traditionally, information theory-based methods only consider the statistical relationship of intensity and ignore the spatial and geometric information. Therefore, they are unable to drive small local deformation and often have some problems with spatially-varying intensity inhomogeneity. Several variants have been proposed to overcome these problems. For example, Klein et al. [Citation13] proposed Local Mutual Information (LMI). Rivas et al. [Citation14] proposed a Robust PaTch-based cOrrelation Ratio (RaPTOR) by blocking the intensity values of the fixed image on the joint probability density distribution graph and computing CR locally to resist spatial intensity inhomogeneity. Staring et al. [Citation15] proposed a graph-based implementation of α-Mutual Information (α-MI) using more features. Rivas et al. [Citation16,Citation17] presented a self-similarity conditioned LMI named Contextual Conditioned Mutual Information (CoCoMI) and a self-similarity conditioned α-MI (SeSaMI). Although α-MI, RaPTOR, SeSaMI and CoCoMI are of higher accuracy and robustness, they all need complex pre-processing and are time-consuming (performing self-similarity estimations on a volume of size 1003 voxels takes about 5 h on one core of a 3 GHz processor before down-sample strategies in [Citation17]). Therefore, how to improve the accuracy while keeping low time complexity is still an open question in the field of multimodal deformable registration.
In this paper, we propose a structure conditioned mutual information using a novel local structure descriptor. Three aspects are addressed to make our method more feasible for 3 D deformable registration. First, we extend the traditional 2 D corner detector FAST to a 3 D structural descriptor using a fast clustering method, which makes the structure description fairly fast. Second, 3 D-FAST can decompose the image into two parts: one part with more structures and the other region with less structures. Then our similarity measure is calculated on the region with more structures instead of the whole volume in traditional MI or the randomly selected region in LMI. Third, the similarity of structure is calculated by comparing 3 D-FAST which acts as the conditional information of MI (MI on selected regions in our method). Thereby, the proposed method can incorporate spatial and geometric information with intensity information in the registration.
Background
Local mutual information
Mutual information of a fixed image If and a moving image Im can be defined by EquationEquation (1)(1) . The two images are geometrically aligned when MI is maximal.
(1)
Where Ω is the image domain, ϕ is a collection of deformation parameters, T is the transformation modeled by ϕ, pf and pm are the marginal intensity probabilities of If and Im, and pfm is the joint intensity probability of If and Im. As MI only considers the global intensity distribution, it is easy to be disturbed by local deformation or inhomogeneity. Klein et al. [Citation13] proposed LMI by adding spatial information in EquationEquation (2)(2) .
(2)
Where Ωj ⊂ Ω are spatial neighborhoods, and N is the number of these neighborhoods which are randomly selected in [Citation13].
Fast
FAST is a corner detector [Citation18] proposed by Rosten et al, the advantage of FAST is the higher speed than other well established corner detection algorithms such as Harris, SURF, SUSAN.
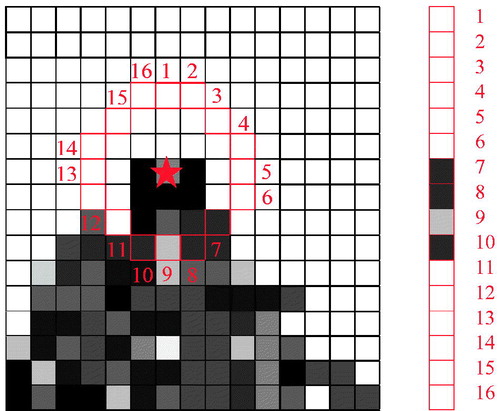
Select a pixel P (indicated by a red star in ) with grey value of IP which is to be identified as an interest point or not. Taking the neighbor circle of R = 3 as example, FAST is calculated as showed in :
Circle of 16 pixels with R = 3 surrounding P are numbered from 1 to 16
Set a threshold intensity value T. If there are “N” contiguous pixels with intensities being either above IP+T or below IP-T, point P is considered as an interest point, where N is a parameter set by users. Machine learning method [Citation19] can be adapted to check the number of contiguous pixels.
Up to now, FAST can only be used on 2 D images. Even though there are algorithms [Citation20,Citation21] that extended FAST to 3 D spatio-temporal domain, they can’t be used on 3 D image. For example, Yu et al. [Citation20] considered three circles in three dimensions (X, Y and Time (T)) in the XY plane, XT plane and YT plane, an interest point is detected when the FAST criterion is satisfied in the XY plane and in one of XT and YT planes. Koelstra et al. [Citation21] considered the proportion of the surrounding pixels as criterion while ignoring their consistency.
Proposed method
In this section, we will elaborate the proposed method denoted as FastMI. FastMI takes structural similarity as the conditional information of MI. The similarity measure (structure conditioned MI) of the registration in our method is described in section 3.1, followed by the modified 3 D-FAST in section 3.2, which is used to measure the similarity of structures. Section 3.3 gives the final cost function. Section 3.4 details the optimization of the cost function. The framework of the registration using FastMI is summarized in .
Table 1. The framework of the registration using FastMI.
Structure conditioned MI
When considering the structural similarity as an extra ‘channel’ of intensity-based mutual information between images under registration, information theory can be used to describe the relationship between the three types of information [Citation22]. The three channel mutual information is given by [Citation23] in EquationEquation (3)(3) .
(3)
Where p(if, im, ufm(x)) is the joint probability of intensities of the fixed image If and the moving image Im co-occurring with the similarity of structure (defined by ufm(x) and will be explained in section 3.2), if and im are the bins of intensity distribution, and x is the voxel of the overlap region.
As ufm(x) is only related to the corresponding voxels in the two images, we can assume that the marginal probability p(ufm(x)) is constant at each voxel x. The joint probability p(if, im, ufm(x)) can be replaced by EquationEquation (4)(4) in the form of conditioned probability.
(4)
Simplify EquationEquation (3)(3) with EquationEquation (4)
(4) , the three channel mutual information is given by EquationEquation (5)
(5) .
(5)
Where pufm(if,im) is the structure conditioned probability, and p(ufm(x)) is taken off as a constant. Quantitative-qualitative measure [Citation24] of information is used in the calculation of pufm(if,im). This measure consists of two aspects of an event: the quantitative one and the qualitative one. The quantitative one is related to the probability of occurrence, and the qualitative one is related to its utility for the fulfillment of the goal. In image registration, images are better aligned when their local structure is more similar. Therefore, structure similarity ufm(x) acts as the qualitative one of the probability of joint intensity. The contribution of voxel x to point (if, im) of the structured conditioned joint intensity histogram is defined by EquationEquation (6)(6) .
(6)
Where χ is the Gaussian Parsons window, and ρp is the width of the window. The similarity measure (the structure conditioned MI) of FastMI is given by EquationEquation (7)(7) where pufm(if,im) is calculated in EquationEquation (8)
(8) .
(7)
(8)
Modified 3 D-FAST
For the measurement of structural similarity, we proposed the modified 3 D-FAST algorithm which extends FAST on 2 D images to that on 3 D volumes. A fast clustering method [Citation25] was adapted to detect contiguous regions on the spherical surface fulfilling FAST criterions instead of contiguous lines on a circle in traditional 2 D FAST.
As 3 D-FAST can split the image into a set of non-overlapping regions with more or less structural information, we perform the registration on spatially meaningful regions with more structural information instead of the randomly selected region in EquationEquation (2)(2) .This modified 3 D-FAST algorithm has two goals: (1) calculate the structural similarity of two voxels, and (2) find local neighborhoods with more structural information for the computation of FastMI. 3 D-FAST achieves these two goals by the following steps:
For each voxel x in overlap regions of the two images, we select 90 blocks weighted by Gaussian kernel with center points on the spherical surface surrounding x. Set a threshold intensity value M, and then x can be encoded as a vector b composed of 90 elements in EquationEquation (9)
(9) . bn (n = 1, 2, …, 90) in EquationEquation (9)
(9) is calculated by EquationEquation (10)
(10) , where an is the absolute difference between the nth patch’s Gaussian weighted intensity and the intensity of the central voxel x.
(9)
(10)
Select six elements of vector b on the sagittal, coronary and vertical axis of voxel x and form a new vector c in EquationEquation (11)
(11) . Voxel x is detected to be with more structural information when satisfying EquationEquation (12)
(12) where K is an integer from one to six set by users.
(11)
(12)
Build a lookup table Q for finding the spatial distance between these blocks according to their indexes in vector b. Q is independent of images and can be calculated before registration.
Find the indexes of nonzero elements in b and form a distance matrix D of these indexes using the lookup table Q. The matrix D is then used to get the maximal contiguous region of the spherical surface surrounding x with a fast clustering method on Science 2014 [Citation25].
Count the number of blocks in the maximal contiguous region as the structural value of x. The structural similarity ufm(x) of Iƒ and Im at voxel x is defined by the Gaussian distance of their structural value in EquationEquation (13)
(13) , Where Ff(x) and Fm(x) are the structural value of Iƒ and Im at voxel x, α is the weight of structural information.
(13)
Structure conditioned MI using modified 3 D-FAST (FastMI)
The objective function for the registration is given by EquationEquation (14)(14) . The first term FastMI is the similarity measure. The second term ω is the regularization penalty item with ∇ being the gradient operator of transformation field T.
(14)
In this paper, cubic B-spline functions are used to model the transformation in EquationEquation (15)(15) where i, j and k are the indexes of the control nodes, and B represents the B-spline basis functions (see [Citation26] for more details).
(15)
Image blocks Ωj ⊂ Ω are regions with more structural information detected by 3 D-FAST are the neighborhoods for FastMI calculation as in EquationEquation (16)(16) where N is the number of this kind of neighborhoods.
(16)
Optimization
Gradient descent method [Citation27] is adopted in this paper for the optimization of the objective function. The update equation of the gradient descent method is shown in EquationEquation (17)(17) where at is the step size and ∇ΔϕC is the gradient of the objective function w.r.t. ϕ at the current position ϕt.
(17)
For the gradient of the objective function, we follow work in [Citation28] and derive a Taylor expansion of FastMI as in EquationEquation (18)(18) where N is the number of parameters.
(18)
Then we simplify EquationEquation (18)(18) by ignoring all terms above the second-order and found the gradient of FastMI as:
(19)
A multi-resolution scheme is used to represent coarse-to-fine details of both volumes for fast and robust registration during the optimization.
Results
Three experiments were performed to demonstrate the accuracy and robustness of the proposed method compared with other information theory based methods. The algorithm was implemented using MATLAB with mex file compiled from C. All experiments were performed on a workstation with Intel Xeon CPU (clock speed 3.30 GHz) and 32 G memory. As medical images are usually noisy and not normalized, an intensity normalization is applied mostly before registration. Therefore, intensity normalization is applied in all experiments below.
Accuracy experiment with noise and intensity inhomogeneity
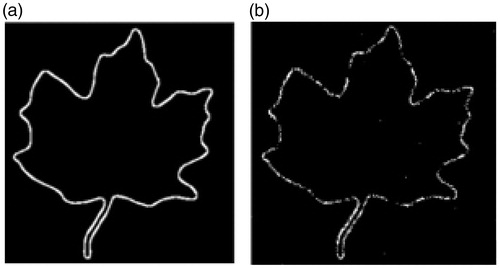
In this experiment, we compared the performance of FastMI to LMI in the presence of noise and intensity inhomogeneity. Images under registration are shown in . For each image pair under registration ( and , and ), we moved the right one along the horizontal and the vertical axis and calculated the similarity value between the image pairs using LMI and FastMI. The results are plotted in . It can be observed from that FastMI can still work when there are severe noise and intensity inhomogeneity while LMI fails, and FastMI has higher convergence speed than LMI. The edge of the leaf in and detected by 3 D-FAST (can be used in 2 D images) are shown in which proved that 3 D-FAST was robust to noise and intensity inhomogeneity.
Figure 2. (a) and (b) are images with opposite intensity. (c) is (a) with Gaussian noise, and (d) is (b) with Gaussian noise and intensity inhomogeneity.
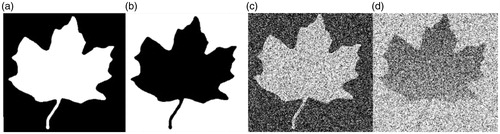
Figure 3. comparison between LMI (top row) and FastMI (bottom row). (a) and (c) are the LMI and FastMI between and with horizontal and vertical shifts, respectively. (b) and (d) are the LMI and FastMI between and .
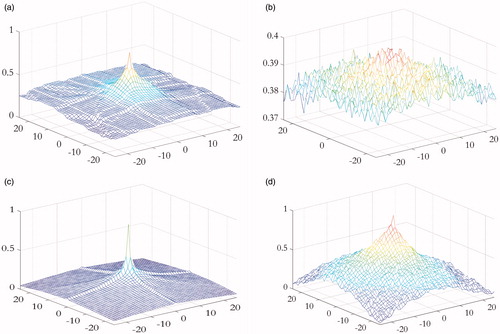
In this experiment, 3 D-FAST was performed using 16 neighborhoods with a 3 × 3 Gaussian filter mask and a threshold of M = 0.5 in EquationEquation (10)(10) . The window’s width of structural distance ρu in EquationEquation (13)
(13) is set to be 4. The parameters were chosen and tuned empirically to achieve the best performance.
Robustness experiment with local deformation
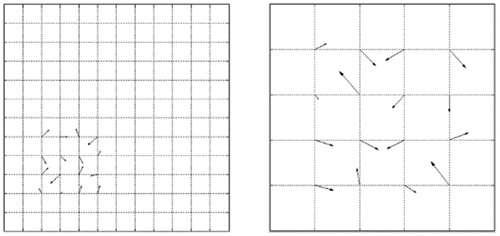
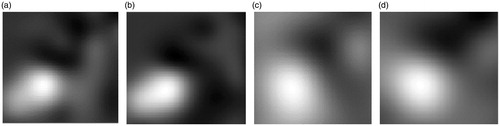
In this experiment, we compared the performance of FastMI and MI in the presence of local deformation. We deformed a T1-MR image with dimension of 100 × 100 on the B-spline nodes in a local region as shown in . A random displacement in the range of -5 to 5 in each dimension was added to each node of the local region. The images before and after deformation were used as the moving and reference images respectively (see in ). shows the normalized horizontal and vertical displacement of pixels in the moving image after registration with MI and FastMI. It can be seen that the deformation field obtained from FastMI is better confined in a local region corresponding to the true deformation.
Figure 7. Displacement field from MI and FastMI based registrations. (a) and (b) are the horizontal and vertical displacement of FastMI, (c) and (d) are the horizontal and vertical displacement of MI.
In this experiment, 3 D-FAST was performed using 16 neighborhoods with a 3 × 3 Gaussian filter mask and a threshold of M = 0.2 in EquationEquation (10)(10) . The window’s width of structural distance ρu in EquationEquation (13)
(13) is set to be 4. The parameters were chosen and tuned empirically to achieve the best performance.
Accuracy experiment on 3 D images
In this experiment, we compared the accuracy of FastMI with LMI and Normalized Cross Correlation (NCR) in three groups (eight registrations in each group) of pairwise registration between T1-T2, T1-PD and T2-PD images from the BrainWeb database. All images used in this experiment were with 3% noise and 20% intensity inhomogeneity. The moving images in each group were obtained by deforming the original image with randomly generated deformation fields while the images of the other modalities were registered to them. In each registration, the mean Target Registration Error (TRE) is calculated at 10000 points detected to be structural using the 3 D-FAST detector in our work as shown in EquationEquation (20)(20) where ‖‖⋅‖‖ is the square operator, n is the number of these structural points.
(20)
In the FastMI registration, 3 D-FAST was performed using 90 neighborhoods with a 3 × 3 Gaussian filter mask and a threshold of M = 0.1 in EquationEquation (10)(10) . The window’s width of structural distance ρu in EquationEquation (13)
(13) is set to be 20. The parameters were chosen and tuned empirically to achieve the best performance.
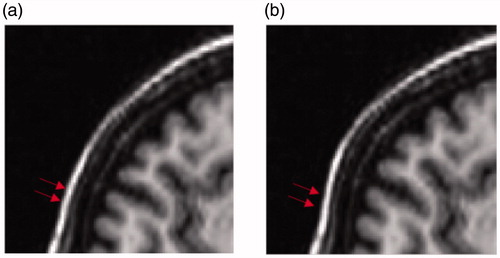
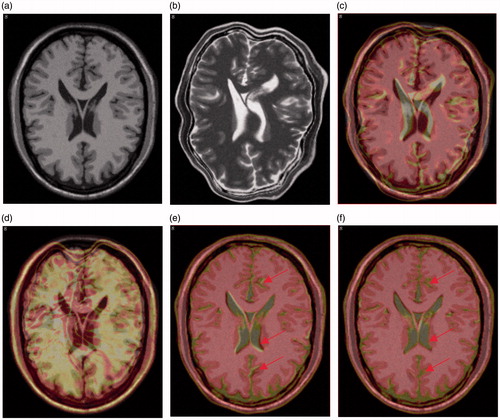
lists the TRE of each groups (eight registrations in each group) by performing registration with CC, LMI and our method, where FastMI shows higher accuracy than other methods with the same terminate conditions. shows the results of overlaying the 90th slice of the moving image on that of the fixed image before and after registration with different methods in the first registration of T1-T2 image pairs. Some clearly improved alignments at the ventricles and the gyri can be observed with our method (indicated by arrows).
Figure 8. The mTRE of pairwise registrations between T1-T2, T2-PD and T1-PD using CC, LMI and the proposed method. The results of CC are not shown in T1-T2 and T1-PD as their mTRE are too large.
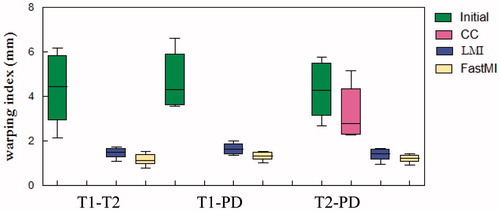
Figure 9. Visual comparison of registration accuracy in the first case of T1-T2 registration. (a) and (b) are the original 90th T1 slice and the artificially deformed T2 slice in registration, respectively. (c) is the result of overlaying the 90th slice of the moving image in (b) on the 90th slice of fixed image in (a) before registration. (d), (e) and (f) are the overlaying after registration with CC, LMI and the proposed method, respectively.
Discussion
In the context of deformable multi-modal medical image registration, MI may lead to undesired results. In this paper, we propose a new algorithm named FastMI, which can combine the structural, spacial and intensity information for image registration. In our work, the structural similarity calculated by 3 D-FAST was added into MI as the conditional information of intensity correlation. It is also possible to condition other correlation such as entropy and gradients of intensity by this structural similarity.
In our experiments, FastMI achieved better performance than common multi-modal registration when there were noise, intensity inhomogeneity and local deformation. As we can see from and in the first experiment, FastMI is robust to noise and intensity inhomogeneity, and has higher convergence speed. The reason is that 3 D-FAST can detect structural voxels and is not easily influenced by noise and inhomogeneity as showed in . In addition, the structural similarity ufm is more sensitive to the local misalignment than the intensity correlation similarity.
There are two reasons for the better performance of FastMI on local deformation in the second experiment. Firstly, FastMI is calculated on local regions with more structural information. So it performs better than MI which is calculated on the whole image and LMI which is calculated on random selected local regions. Secondly, LMI only considers the statistical intensity relationships between both images and may have the same intensity relationships when the voxels in the images have different intensity, while FastMI are more robust by considering both intensity and structural relationships.
Compared with other variants of MI using structural information, such as SeSaMI and CoCoMI, FastMI is faster in structural detection as it doesn’t need time-consuming preprocessing steps, and 3 D-FAST can analyze images in high speed by taking 3 0 ∼ 45 sec on a volume with dimension of 217 × 181 × 181 on one core of a 3.3 GHz processor (self-similarity estimations on a volume of size 1003 takes about 5 h on one core of a 3 GHz processor before down-sample strategies in SeSaMI and CoCoMI).
Additionally, the parameters can influence the performance of registration. How to choose proper parameters is a crucial problem. For example, we may ignore some detailed structure of image if threshold T is too large while the registration might be influenced by noise if T is too small. The window’s width of structural distance ρu cannot be too large because the structural information might be over-smoothed and it cannot be too small because the structural information might be influenced by noise. And so on.
One limitation of our method is the speed. FastMI cannot be real-time. It is possible to speed it up by GPU, parallel computing or more sophisticated optimization schemes which is our future works.
Conclusion
In this paper, we extend traditional 2 D-FAST corner detector to a 3 D structure descriptor using a fast clustering method, which is the first FAST method used on 3 D images to our best knowledge. And then we used the structural similarity computed by 3 D-FAST as the conditional information to encode spatial and geometric cues into LMI. The method we proposed is demonstrated to be more robust and accurate than the common registration methods based on information theory.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Sotiras A, Davatzikos C, Paragios N. Deformable medical image registration: a survey. IEEE Trans Med Imaging. 2013;32:1153–1190.
- Yang J, Li H, Campbell D, et al. Go-ICP: a globally optimal solution to 3D ICP point-set registration. IEEE Trans Pattern Anal Mach Intell. 2016;38:2241–2254.
- Jian B, Vemuri BC. Robust point set registration using gaussian mixture models. IEEE Trans Pattern Anal Mach Intell. 2011;33:1633–1645.
- Cosse A. Diffeomorphic surface-based registration for MR-US fusion in prostate brachytherapy. 2012; p 903–907.
- Wein W, Brunke S, Khamene A, et al. Automatic CT-ultrasound registration for diagnostic imaging and image-guided intervention. Medical Image Analysis. 2008;12:577.
- Fuerst B, Wein W, Müller M, et al. Automatic ultrasound-MRI registration for neurosurgery using the 2D and 3D LC(2) Metric. Med Image Anal. 2014;18:1312–1319.
- Wachinger C, Navab N. Entropy and Laplacian images: structural representations for multi-modal registration. Medical Image Analysis. 2012;16:1–17.
- Heinrich MP, Jenkinson M, Bhushan M, et al. MIND: Modality independent neighbourhood descriptor for multi-modal deformable registration. Med Image Anal. 2012;16:1423–1435.
- Heinrich MP, Jenkinson M, Papiez BW, et al. Towards realtime multimodal fusion for image-guided interventions using self-similarities. 2013; p 187.
- Jiang D, Shi Y, Chen X, et al. Fast and robust multimodal image registration using a local derivative pattern. Med Phys. 2017;44:497.
- Jiang D, Shi Y, Yao D, et al. miLBP: a robust and fast modality-independent 3D LBP for multimodal deformable registration. Int J Cars. 2016;11:997–1005.
- PWP J, BAM J, AV M. Mutual-information-based registration of medical images: a survey. IEEE Trans Med Imaging. 2003;22:986–1004.
- Klein S, Van-Der-Heide U, Lips I, et al. Automatic segmentation of the prostate in 3D MR images by atlas matching using localized mutual information. Med Phys. 2008;35:1407–1417.
- Rivaz H, Chen JS, Collins DL. Automatic Deformable MR-Ultrasound Registration for Image-Guided Neurosurgery. IEEE Trans Med Imaging. 2015;34:366–380.
- Staring M, VDH U, Klein S, et al. Registration of cervical MRI using multifeature mutual information. IEEE Trans Med Imaging. 2009;28:1412–1421.
- Rivaz H, Karimaghaloo Z, Fonov VS, et al. Nonrigid registration of ultrasound and MRI using contextual conditioned mutual information. IEEE Trans Med Imaging. 2014;33:708–725.
- Rivaz H, Collins DL. Self-similarity weighted mutual information: a new nonrigid image registration metric. Berlin Heidelberg: Springer; 2012.
- Rosten E, Drummond T. Machine learning for high-speed corner detection. 2006; p 430–443.
- Rosten E, Porter R, Drummond T. Faster and better: a machine learning approach to corner detection. IEEE Trans Pattern Anal Mach Intell. 2008;32:105–119.
- Yu TH, Kim TK, Cipolla R. Real-time action recognition by spatiotemporal semantic and structural forests. 2010; p 1–12.
- Koelstra S, Patras I. The fast-3D spatio-temporal interest region detector. 2009; p 242–245.
- Reza FM. An introduction to information theory. New York, USA: McGraw-Hill Book Company, Inc; 1961.
- Wachinger C, Navab N. Simultaneous Registration of Multiple Images: Similarity Metrics and Efficient Optimization. IEEE Trans Pattern Anal Mach Intell. 2013;35:1221–1233.
- Luan H, Qi F, Xue Z, et al. Multimodality image registration by maximization of quantitative-qualitative measure of mutual information. Pattern Recogn. 2008;41:285–298.
- Rodriguez A, Laio A. Machine learning. Clustering by fast search and find of density peaks. Science. 2014;344:1492.
- Lee S, Wolberg G, Shin SY. Scattered data interpolation with multilevel B-splines. IEEE Trans Visual Comput Graphics. 1997;3:228–244.
- Klein S, Staring M, Pluim JP. Evaluation of optimization methods for nonrigid medical image registration using mutual information and B-splines. IEEE Trans Image Process. 2007;16:2879.
- Studholme C, Drapaca C, Iordanova B, et al. Deformation-based mapping of volume change from serial brain MRI in the presence of local tissue contrast change. IEEE Trans Med Imaging. 2006;25:626–639.