Abstract
Background: Compared with the traditional point-based registration in the image-guided neurosurgery system, surface-based registration is preferable because it does not use fiducial markers before image scanning and does not require image acquisition dedicated for navigation purposes. However, most existing surface-based registration methods must include a manual step for coarse registration, which increases the registration time and elicits some inconvenience and uncertainty.
Methods: A new automatic surface-based registration method is proposed, which applies 3D surface feature description and matching algorithm to obtain point correspondences for coarse registration and uses the iterative closest point (ICP) algorithm in the last step to obtain an image-to-patient registration.
Results: Both phantom and clinical data were used to execute automatic registrations and target registration error (TRE) calculated to verify the practicality and robustness of the proposed method. In phantom experiments, the registration accuracy was stable across different downsampling resolutions (18–26 mm) and different support radii (2–6 mm). In clinical experiments, the mean TREs of two patients by registering full head surfaces were 1.30 mm and 1.85 mm.
Conclusion: This study introduced a new robust automatic surface-based registration method based on 3D feature matching. The method achieved sufficient registration accuracy with different real-world surface regions in phantom and clinical experiments.
Introduction
Image-to-patient registration is the process of establishing the spatial transformation between the coordinate systems of the real-patient space (the position of calibrated tracking system) and the digital image space (the volumetric reconstruction of imaging data). It is a crucial step in image-guided neurosurgery systems (IGNSs) [Citation1–5]. The accuracy, robustness, and convenience of the image-to-patient registration directly determine the quality of the application of IGNS [Citation5]. However, most current technologies that are widely used for image-to-patient registration have their limitations.
Two types of noninvasive image-to-patient registration methods are available in clinical application: point-based registration [Citation5] and surface-based registration [Citation1–4]. After the successful use of a point-based registration for decades, surface-based registration methods emerged to avoid some inherent disadvantages of the former in terms of cost and convenience [Citation1–4]. In surface-based registration, a point cloud is collected from the surface of the patient’s head via a laser range scanner [Citation1–4] or a tracked probe [Citation6] in the real-patient space, and the spatial transformation is obtained by matching the real-patient point cloud with the head surface point cloud extracted from the image space. The accuracy of surface-based registration is closely correlated with the scanning areas. As previously reported [Citation2], the registration accuracy decreases away from the center of the point clouds that are used for matching. Recently, a portable 3D scanner has been used to acquire a point cloud from the skin surface of the entire head to achieve sufficient registration accuracy [Citation1]. However, most existing commercial surface-based registration methods for IGNS must include a manual step for coarse registration that offers a good initial pose for the following fine registration. The manual intervention for coarse registration increases the registration time and elicits some inconvenience and human error.
This study introduced a new robust surface-based automatic registration method based on 3D feature matching for point cloud registration [Citation7–12]. The proposed method is more robust and convenient in IGNS than existing surface-based registration methods.
Materials and methods
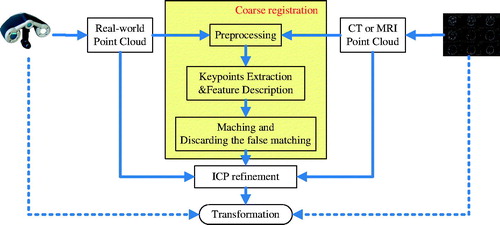
The proposed registration method is illustrated in . First, the point cloud of a patient’s head surface was constructed in the image space from computed tomographic (CT) or magnetic resonance (MR) image of the patient, and the real-patient point cloud of the patient’s head surface was acquired using a 3D laser range scanner. Next, the two point clouds were downsampled to similar spatial resolution by preprocessing. Then, keypoints were extracted and the local feature descriptor of each keypoint was calculated for matching. Keypoint correspondence was obtained by matching the features, and false matchings were discarded by considering geometric constraints like the one in the study by Buch et al. [Citation11]. Once the correspondence of the keypoints between the two point clouds was obtained, a coarse transformation between them was calculated. The last step was refining the transformation via the iterative closest point (ICP) algorithm [Citation13], which was known for its simple concept, good accuracy, and shorter runtime.
Acquisition and preprocessing of the 3D point cloud
A portable 3D scanner was used to obtain the raw real-patient point cloud, and the raw CT or MR image point cloud was efficiently obtained by the threshold method [Citation4]. The raw data usually included some outliers. Therefore, first the outliers were removed with a StatisticalOutlierRemoval filter [Citation9], which used point neighborhood statistics to filter outliers. After that, a subsequent downsampling step was performed to prepare the two point clouds to similar mesh resolutions. This was done as follows: dividing the point cloud into multiple cube-shaped regions with the desired resolution and keeping only one point from every region. The downsampling process was for two purposes: (1) it reduced the number of points in the two point clouds and increased the speed of subsequent steps; (2) similar mesh resolutions improved the robustness of feature matching because the 3D local feature descriptor was more repeatable in the similar mesh resolution of the two point clouds [Citation7]. It was worth noting that despite the downsampling, the accuracy of the image-to-patient registration did not decrease because the ICP algorithm was used to refine the registration results in the last step with the point clouds at their original mesh resolutions, as in .
Keypoint extraction and feature description
Before calculating the local feature descriptors, it was necessary to decide which points from the point cloud should be used, and this process was called keypoint detection. Several methods are available to detect keypoints in the literature [Citation8,Citation12]; the intrinsic shape signatures (ISS) method was used in this study. ISS scanned a surface and chose only points with large variations in the principal direction, which were ideal for being keypoints. The keypoints detected in a head phantom are depicted in . The next step was to describe the features of the keypoints with the Fast Point Feature Histogram (FPFH) [Citation10], and this step was one of the most important steps in the proposed registration method. Many different feature descriptors have been developed for different application scenarios [Citation7]. The FPFH algorithm encoded information about the geometry surrounding a keypoint by analyzing the differences between the directions of the normals in its vicinity using a multidimensional histogram. The readers are referred to the study by Rusu et al. [Citation10] for the details of FPFH descriptor. According to Guo’s conclusion [Citation7], the support radius, which indicated how many points participated in calculating the feature in the keypoint’s neighborhood, affected the time and robustness of registration. Therefore, 18–26 mm was tested for support radius in the present study. The proposed method showed high robustness while the support radius changed in this range. No larger range was tested because the application scenario remained fairly consistent in terms of the scale of the point clouds.
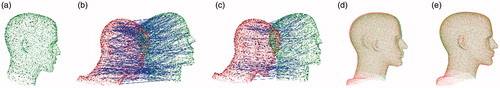
Figure 2. Illustration of the proposed registration process with a head phantom. Real-patient point cloud is green; CT point cloud is red. (a) Keypoint detection. The black dots indicate keypoints. (b) Keypoint correspondence. The blue lines indicate the correspondences. (c) Correspondences after discarding false matchings. (d) Coarse registration. (e) ICP refinement. (color online).
Matching and discarding false correspondences
Once the features were computed in the two point clouds, features from different point clouds were matched to obtain point-to-point correspondences using the Euclidean distances between feature vectors. A k-d tree [Citation14] was used to perform a nearest neighbor search to speed up the matching process. Furthermore, a threshold was set to discard the long-distance feature correspondences.
After matching, a list of correspondences were achieved between the keypoints in the image point cloud and those in the real-patient point cloud, but accurate registration still could not be obtained due to a substantial number of false matchings. A correspondence grouping method was implemented, as described in the study by Buch et al. [Citation11], to discard false correspondences based on geometric consistency. show the results of keypoint matching before and after discarding false correspondences in the registration of a head phantom.
ICP refinement
Although most of the false correspondences could be discarded, it could not be guaranteed that all remaining correspondences were consistent with the correct registration. For this reason, a robust and outlier-tolerant method was used, that is, the random sample consensus (RANSAC) method [Citation15], which was a common method for solving this issue in feature correspondences, to perform coarse registration. RANSAC could yield a coarse registration that was not accurate enough for the clinical application of IGNS, but it was a good start point for further refinement. In the final step, the ICP algorithm was used to obtain an accurate registration, as depicted in . Because of the coarse registration, the ICP algorithm could begin from a good initial position and converge at an accurate registration.
Results
The method was tested in the image-to-patient registration of one head phantom and two clinical patients to verify the robustness and practicability of the proposed method in IGNS. In all experiments, the real-patient 3D point clouds were obtained with a portable 3D scanner (Go!SCAN; Creaform, Levis, Quebec, Canada). The point cloud of the entire head surface was scanned using a previously proposed method [Citation1]. Furthermore, the facial region from the whole head point cloud was clipped to test the method with only facial surfaces because sometimes the whole head could not be scanned for various reasons, such as hair, in a clinical setting. The two clinical 3D point clouds were reconstructed from MR images (T1 sequences), which were scanned with fiducial markers on the head. These markers were used to perform point-based registration in actual image-guided neurosurgery. However, the plastic phantom with no fiducial markers for simulating practical application was scanned using a high-resolution 64-slice CT scanner with a slice thickness of 0.625 mm. As in previous studies [Citation1–4], the target registration error (TRE) was calculated as the metric of registration accuracy using EquationEquation (1)(1) :
(1)
where T is a rigid transformation that is the output of image-to-patient registration, and p and q denote the corresponding points from the image space and the real-patient space, respectively. Because of no fiducial markers, three distinct anatomic target points on the phantom were selected: the tip of the nose and the top of two helixes (outside edges of the ears). In the clinical experiments, four skin markers were selected as the target points to calculate TRE. All experiments were conducted on a computer with 3.4-GHz Intel Core i7-6700 CPU and 64 GB of RAM. The method was written in unoptimized C++ and used the PCL library [Citation9].
Robustness
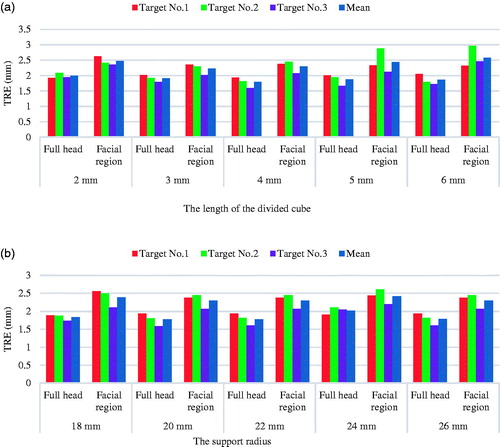
According to Guo’s conclusion [Citation7], the support radius, noise, and mesh resolution could affect the robustness of FPFH, but for the proposed method in IGNS, the noise introduced in the acquisition step could be reduced by proper operation and using a high-precision surface scanner. Therefore, experiments were conducted on the influence of varying support radius and mesh resolution. In addition, the method was tested using different real-patient surface regions to validate its robustness. and show the visual registration results using different real-patient surface regions (full head region and facial region). The registration results with different support radii and mesh resolution are summarized in . As expected, the average TRE of the facial region registration was slightly larger than that of the registration with full head, but it was still sufficient for IGNS. As shown in , the registration accuracy was stable across different downsampling resolutions and different support radii.
Figure 3. Facial region registration. The real-patient point cloud is green; the CT point cloud is red. (a) Only the facial region was scanned in the real-patient space. (b) Point cloud of the CT data. (c) Coarse registration. (d) ICP refinement. (e) An example of failed registration using a previously described method [Citation3]. The real-patient point cloud is golden; the CT point cloud is gray. (color online).
![Figure 3. Facial region registration. The real-patient point cloud is green; the CT point cloud is red. (a) Only the facial region was scanned in the real-patient space. (b) Point cloud of the CT data. (c) Coarse registration. (d) ICP refinement. (e) An example of failed registration using a previously described method [Citation3]. The real-patient point cloud is golden; the CT point cloud is gray. (color online).](/cms/asset/e83b12fb-0f84-4923-bf97-6ee8c96b19ef/icsu_a_1389411_f0003_c.jpg)
Figure 4. (a) TREs (mm) of each selected target point at different downsampling resolutions (the support radius was 18 mm). (b) TREs (mm) of each selected target point at different support radii for calculating feature descriptor (the edge length of the downsampling cube was 6 mm). (color online).
Furthermore, the proposed method was compared with another coarse registration method: 4-point congruent sets (4PCS) [Citation16] used in [Citation3] for spatial registration in IGNS, which was a heuristic method with certain contingency. More concretely, the method was compared with the original 4PCS algorithm [Citation16], whose source code was publicly available. The 4PCS method could not always return acceptable coarse registrations between a facial region and the whole head. Six failed facial registrations were found in 25 tests using 4PCS, and a typical failed case is illustrated in . In contrast, the proposed method consistently returned acceptable coarse registrations in all tests.
Run time
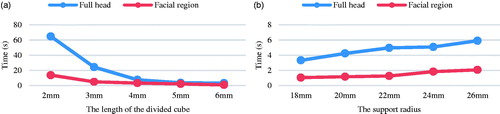
The time of the whole process of image-to-patient registration in IGNS was divided into two parts: point cloud acquisition time and registration time. First, the point cloud from the image space could be prepared before operation. However, the time for acquiring real-patient point cloud depended on the scanned region. The facial region could be easily scanned instantaneously (<1 s), but the time would be longer for scanning the whole head (10–20 s). Second, the registration time depended on the support radius and the mesh resolution of the downsampled points indicated by the length of divided cube for downsampling. As expected, the registration time increased as the support radius increased and decreased as the length of divided cube increased, as shown in . The reason was that when the length of divided cube for downsampling increased, the number of points in the downsampled point cloud would decrease and when the support radius increased, the number of points that participated in calculating features would increase. The last ICP refinement took 1–10 s depending on the number of points in the two original point clouds.
Clinical feasibility
Data from two patients were used to verify the feasibility of the proposed method in a clinical setting. As in the phantom experiments, both facial region and full head registrations were executed. depicts the visual results of the registration using full head surface and a facial region, and the TREs are listed in . The TREs show that the method was accurate enough for IGNS. Again, two examples () show that the previously described 4PCS method [Citation3,Citation16] was not robust for facial registration.
Figure 6. Clinical registration using the proposed method and a previously described 4PCS method [Citation3,Citation16]. I and II indicate patients one and two, respectively. (a) Point cloud from the real-patient surface. (b) Point cloud from MRI. (c) Full head registration. (d) Facial registration. (e) Failed facial registration using the 4PCS method. The real-patient point cloud is golden; MR image point cloud is gray. (color online).
![Figure 6. Clinical registration using the proposed method and a previously described 4PCS method [Citation3,Citation16]. I and II indicate patients one and two, respectively. (a) Point cloud from the real-patient surface. (b) Point cloud from MRI. (c) Full head registration. (d) Facial registration. (e) Failed facial registration using the 4PCS method. The real-patient point cloud is golden; MR image point cloud is gray. (color online).](/cms/asset/82e7d81a-560f-44cf-9b31-3748365ae34f/icsu_a_1389411_f0006_c.jpg)
Table 1. TREs (mm) of the clinical experiments. Numbers 1, 2, 3, and 4 are the numbers of four target points, and I and II indicate patients one and two, respectively.
Discussion
This study introduced a new markerless robust automatic image-to-patient registration method for IGNS that was an easy approach for improving the convenience of surface-based registration in IGNS. The method applied 3D surface feature description and matching algorithms to obtain point correspondences for coarse registration and used ICP refinement in the last step to obtain an accurate image-to-patient registration. To verify the practicality and robustness of the proposed method, both phantom and clinical data were used to execute automatic registrations, and the TRE was calculated. The method achieved sufficient accuracy registration with different real-world surface regions and different parameters in experiments, indicating that it could gain robust registration accuracy. In addition, the method was compared with another automatic surface-based registration method [Citation3] for IGNS using phantom and clinical data. The experimental results also revealed that the proposed method, which was markerless and could provide sufficient accuracy, was more robust for IGNSs.
The main limitation of the proposed method was that the accuracy of the real-world point cloud must be satisfied. A low-precision 3D scanner always leads to a low-precision surface registration in IGNS. Therefore, a good 3D scanner is essential. Furthermore, the method should be tested on a larger number of real patients in the future.
Conclusions
This study proposed a new robust automatic surface-based registration framework for IGNSs based on 3D feature matching. The experiments showed that the proposed method was feasible in neuronavigation and could provide sufficient registration accuracy.
Acknowledgements
This study was supported by National Natural Science Foundation of China (project 81471758) and Shanghai Natural Science Foundation (project 17ZR1401500). This study was partially funded under the Program of Shanghai Academic/Technology Research Leader project 16XD1424900 and the National Science and Technology Support Program (No.2015BAK31B01).
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Fan Y, Jiang D, Wang M, et al. A new markerless patient-to-image registration method using a portable 3D scanner. Med Phys. 2014;41:101910
- Wang MN, Song ZJ. Properties of the target registration error for surface matching in neuronavigation. Comput Aided Surg. 2011;16:161–169.
- Chen X, Song Z, Wang M. Automated global optimization surface-matching registration method for image-to-patient spatial registration in an image-guided neurosurgery system. J Med Imaging Health Inform. 2014; 4:942–947.
- Potter M, Yaniv Z. Cost-effective surgical registration using consumer depth cameras. In: Webster RJ, Yaniv ZR, editors. Proceedings of SPIE 2016.
- Hong J, Hashizume M. An effective point-based registration tool for surgical navigation. Surg Endosc. 2010;24:944–948.
- Simpson AL, Burgner J, Glisson CL, et al. Comparison study of intraoperative surface acquisition methods for surgical navigation. IEEE Trans Biomed Eng. 2013;60:1090–1099.
- Guo Y, Bennamoun M, Sohel F, et al. A comprehensive performance evaluation of 3D local feature descriptors. Int J Comput Vis. 2016;116:66–89.
- Tombari F, Salti S, Di Stefano L. Performance evaluation of 3D keypoint detectors. Int J Comput Vis. 2013;102:198–220.
- Rusu RB, Cousins S. 3D is here: Point Cloud Library (PCL). IEEE International Conference on Robotics and Automation ICRA, 2011.
- Rusu RB, Blodow N, Beetz M. Fast Point Feature Histograms (FPFH) for 3D Registration. IEEE International Conference on Robotics and Automation-ICRA, 2009. p. 1848–1853.
- Buch AG, Kraft D, Kamarainen J, et al. Pose estimation using local structure-specific shape and appearance context. IEEE International Conference on Robotics and Automation ICRA 2013. p. 2080–2087.
- Zhong Y. Intrinsic shape signatures: a shape descriptor for 3D object recognition. International conference on computer vision 2009. p. 689–696.
- Besl PJ, Mckay ND. A method for registration of 3-D shapes. IEEE Trans Pattern Anal Mach Intell. 1992;14:239–256.
- Bentley JL. Multidimensional binary search trees used for associative searching. Commun Acm. 1975;18:509–517.
- Fischler MA, Bolles RC. Random sample consensus - a paradigm for model-fitting with applications to image-analysis and automated cartography. Commun Acm. 1981;24:381–395.
- Aiger D, Mitra NJ, Cohen-Or D. 4-points congruentsets for robust pairwise surface registration. ACM Trans Graph. 2008;27. DOI:10.1145/1399504.1360684