?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Automatic segmentation of prostate magnetic resonance (MR) images has great significance for the diagnosis and clinical application of prostate diseases. It faces enormous challenges because of the low contrast of the tissue boundary and the small effective area of the prostate MR images. In order to solve these problems, we propose a novel end-to-end professional network which consists of an Encoder-Decoder structure with dense dilated spatial pyramid pooling (DDSPP) for prostate segmentation based on deep learning. First, the DDSPP module is used to extract the multi-scale convolution features in the prostate MR images, and then the decoder is used to capture the clear boundary of prostate. Competitive results are produced over state of the art on 130 MR images which key metrics Dice similarity coefficient (DSC) and Hausdorff distance (HD) are 0.954 and 1.752 mm respectively. Experimental results show that our method has high accuracy and robustness.
Keywords:
1. Introduction
Prostate cancer is one of the most common cancers in men. Statistics from the National Cancer Institute has shown that 164,690 new prostate cancer patients are expected to be added in 2018, which is the highest proportion of all male cancers [Citation1]. The diagnosis of prostate disease has always been the focus of imaging research. Currently common imaging techniques for prostate imaging include rectal ultrasound (TRUS), computed tomography (CT), and magnetic resonance imaging (MRI). Compared with other imaging methods, the image quality of MRI is clearer for distinguishing prostate anatomical regions and more sensitive to diseased tissue. Therefore, MRI is recognized as the most effective method for diagnosing cancerous prostate, and plays an important role in assessing the nature of prostate lesions [Citation2].
Clinical manual segmentation of the prostate often requires expert manual interaction, which is time-consuming, poorly reproducible, and dependent on specialist experience. The automatic segmentation can improve the repeatability of the results and the clinical efficiency, which has important clinical significance. Many semi-automatic or fully automated methods [Citation3–6] have been proposed for the segmentation of various organs and tissues in medical images, but automatic prostate segmentation is difficult. The main reasons affecting magnetic resonance images segmentation are as follows: (1) Prostate tissue has low contrast with other surrounding tissues and it is difficult to distinguish the boundaries of it. (2) There is less effective information available in a MR image because of the small size of prostate tissue. (3) The shape of prostate is varied, which brings difficulty to segmentation algorithm. Complicated algorithm time consumption may delay clinical diagnosis.
Litjens et al. [Citation7] used the anatomy, gray value and texture features to classify the internal body elements of the prostate that realize the full segmentation of the internal and external contour. The outer contour is segmented manually and used as the initialization of the inner contour, which makes the whole prostate segmentation time-consuming and laborious. Zhang et al. [Citation8] proposed a new prostate MRI two step segmentation method based on the edge distance adjustment level set evolution to realize the full segmentation of the prostate contour. The segmentation effect of this method depends more on the quality of the segmented image, and it takes a long time to train a large number of atlas for the segmentation of an image. Jia et al. [Citation9] proposed a coarse-to-fine prostate segmentation approach based on a probabilistic atlas-based coarse segmentation. Mahapatra et al. [Citation10] proposed a fully automated method for prostate segmentation using Random forests and graph cuts.
In recent years, deep learning has outperformed state of the art in many fields such as computer vision and medical image processing. The availability of large annotated medical imaging data now makes it feasible to use deep convolutional neural networks (DCNNs) for medical image segmentation and classification [Citation11]. Zhu et al. [Citation12] proposed a model named Deeply-Supervised CNN to segment prostate MR images. Karimi et al. [Citation13] put forward a prostate MR image segmentation strategy based on CNN and statistics. Zhan et al. [Citation14] used deconvolution neural network to segment the MR images of prostate. These methods solve the problem of the segmentation of the prostate MR image in some degree. However, the automatic segmentation of the prostate MR image is still a huge challenge, due to the large variability in prostate contour, the interference of the surrounding tissue and the imaging artifact.
In this paper a deep neural network is proposed to solve the above problems. It contains an Encoder-Decoder structure with dense dilated spatial pyramid pooling (DDSPP). Firstly, DDSPP is used to extract multi-scale features of MR images, and then decoder is used for up-sampling to obtain prediction information.
2. Method
2.1. DDSPP
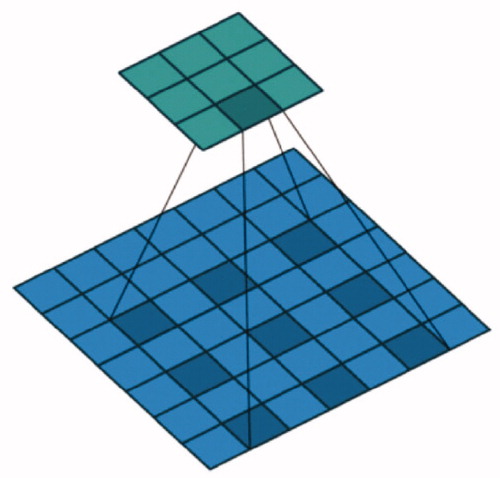
Dilated convolution [Citation15] which named atrous convolution in [Citation16], it can increase the receptive field exponentially without reducing the spatial dimension ().
The dilated convolution can increase the receiving field of the convolution kernel. When the convolution kernel size is the rate is R and the receptive field of the convolution kernel is:
(1)
(1)
When rate = 6, the convolution kernel size is
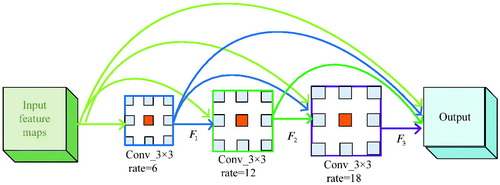
In Deep Labv3 [Citation17], ASPP (Atrous Spatial Pyramid Pooling) is used to obtain multi-scale context information, and the prediction results are obtained by up-sampling directly. The ASPP and cascaded modules with dilated convolution are shown in .
Figure 2. ASPP and cascaded modules with dilated convolution: (a) ASPP. (b) Cascaded modules with dilated convolution.
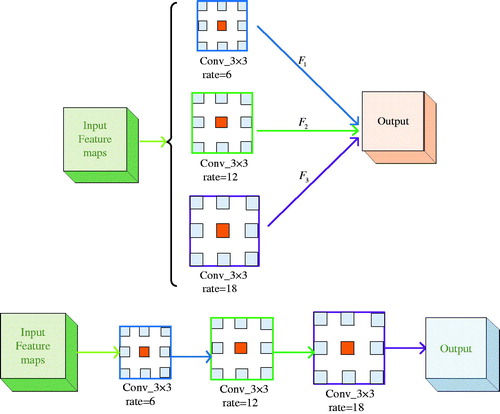
Combining the advantages of the ASPP and cascaded modules with dilated convolution, the DDSPP which can generate more scale features over a wider range is designed. It’s shown in .
Stacking two convolutional layers together can give us a larger receptive field. Suppose we have two convolution layers with the filter size and
respectively, the receptive field is:
(2)
(2)
When rate = 6, 12 will result in a new receptive field of size
shows the contrast of receptive field between DDSPP module and ASPP module. Where
are the receptive field of the dilated convolution of rate = 6,12,18 in the ASPP module and the stacking receptive field as shown in .
Table 1. Comparison of receptive field between ASPP and DDSPP modules in the same rate.
It is obvious that dense connections between stacked dilated layers are able to compose feature pyramid with much denser scale diversity. The receptive fields of DDSPP are larger than the ASPP.
2.2. Encoder-decoder
Encoder-Decoder architecture is successful in many computer vision tasks, such as human pose estimation [Citation18], object detection [Citation19], and semantic segmentation [Citation20–23]. The Encoder-Decoder network includes an encoder module and a decoder module. The encoder module gradually reduces the feature maps and captures higher semantic information and the decoder module gradually recovers space information.
2.3. Network architecture
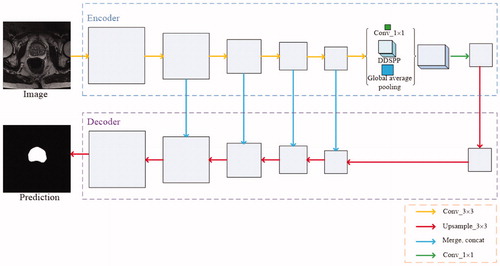
In this paper encoder with DDSPP is used to get information on MR images of the prostate, which can get the edge information of the prostate more clearly. And then gradually recover the details of the prostate through the up- sampling. With the image convolution and pooling, the resolution decreases, deconvolution the feature map will lead to the rough output and loss of many details directly. Therefore, we connect the low-level features and high-level features to produce more accurate results. The network architecture is shown in .
3. Experiments and evaluation
3.1. Experimental results
The experimental platform for this paper is tensorflow1.6, Intel(R) Core(TM) i9-7900X CPU 3.30 GHz and NVIDIA GeForce TITAN XP GPU. MICCAI Grand Challenge: Prostate MR Image Segmentation 2012 (PROMISE12) [Citation24] and NCI-ISBI 2013 Challenge: Automated Segmentation of Prostate Structures (ASPS13) datasets are used in this experiment. Pictures are removed which don’t contain prostate information or unclear in order to reduce the proportion of negative samples as shown in . There are a total 1392 images, the size of the image is 256 × 256. Of the 1392 images in this experiment, 1200 images are used for training and the remaining 192 images are used to test the algorithm.
Figure 5. Pictures which removed because don’t contain prostate such as (a) and (b) or unclear such as (c).
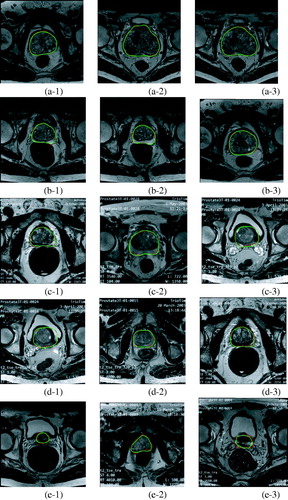
We show the qualitative examples of the segmentation results in . From Figure (a-1–a-3), Figure (b-1–b-3), Figure (c-1–c-3), Figure (d-1–d-3) we can know that our method can accurately segment the prostate MR images and overcome the effects of the around tissues and identify prostate tissue as entire section. Figure (d-1–d-3) show small difference between our results in green and the ground truth in red, because the prostate tissue has so low contrast with other surrounding tissues and the shape of them become so small. Significantly, our method shows a delightful impact in the prostate MR images segmentation.
3.2. Performance evaluation
Evaluation process aims to measure the performance of proposed scheme. In this paper the segmentation precision of prostate MR images can be evaluated from shape distance and area overlap. In this experiment, two parameters are used to quantitatively evaluate segmentation algorithm. The performance is Dice similarity coefficient (DSC), accuracy, Intersection over Union (IoU) and Hausdorff distance (HD).
DSC calculates the degree of similarity between the two contour regions. DSC is computed as
(3)
(3)
Accuracy and IoU are defined as
(4)
(4)
(5)
(5)
where true positive (TP) represents the common area of manual segmentation and algorithm segmentation. True negative (TN) represents the manual segmentation of external and algorithmic segmentation of the external common area. False positive (FP) represented in the algorithm segmentation area, but outside the manual segmentation area and false negative (FN) represents an area that is contained within the manual outline but is missing by the algorithm.
HD reflects the biggest difference between the two contour points set. Suppose there are two sets and
then the HD between the two-point sets is defined as
(6)
(6)
(7)
(7)
(8)
(8)
where A is a combination of manually segmented contour point coordinates, B is a combination of algorithm segmented contour point coordinates.
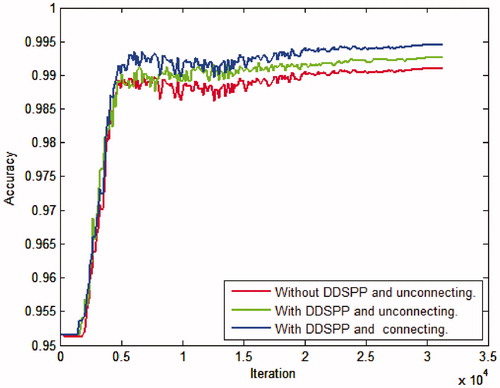
At first, we compare the consequence between original network which without DDSPP and unconnecting the low-level features and high-level features and with DDSPP while connecting and unconnecting the low-level features and high-level features. It is observed from .
From we can get the best results using DDSPP architecture and connecting the low-level features and the high-level features.
We emphatically compared our method with advanced methods [Citation9,Citation14] which using our datasets, and convolution neural networks; including U-net [Citation23], PixelNet [Citation24], and DeepLabV3+ [Citation20] which are the prominent network so far in the domain of semantic segmentation. U-net and PixelNet was based on VGG-16 model, DeepLabV3+ was based on Resnet.
As we all know, the bigger the DSC and the smaller the HD value is, the closer the predict result to the ground truth is. The detailed comparison between different methods is illustrated in from which we can see that the proposed method outperforms other methods. Dice similarity coefficient (DSC) and Hausdorff distance (HD) over other methods are up to 0.954 and 1.752 mm, respectively.
Table 2. Quantitative evaluation of different methods.
4. Discussion
In this study, a deep neural network is proposed to capture the boundary of the prostate. The proposed study leverages on the inherent advantages of dilated convolution, Encoder-Decoder architecture and deep learning. Having successfully trained, tested and validated on the 1392 prostate MR images, we were able to consistently achieve a good qualitative and quantitative results. Thus, we may be able to offer a robust segmentation framework, for the automated segmentation study of the prostate tissues.
Compared with the state-of-the-art methods [Citation12–14], our approach has several advantages. Such as the high rate of accuracy and the bigger DSC is. Most traditional methods [Citation7,Citation8] rely on the shape of prostate to segment. It’s not only time-consuming but also low in accuracy. Though the method used in [Citation23] based on U-Net convolutional neural network also can avoid above steps as ours, it attached much importance on reducing prediction time, which resulted in relatively poor accuracy. We also compare our method with PixelNet [Citation24] and DeepLabV3+ [Citation20]; the results show that our method has higher segmentation performance.
It suggests that the proposed algorithm for the segmentation of the prostate tissues in MR images can be welled in clinical diagnosis research.
5. Conclusion
In this paper, we demonstrate that the inherently difficult problems of MR images can be solved well with deep learning. We have proposed a robust automatic prostate segmentation network jointly utilizing Encoder-Decoder architecture applied with dense dilated spatial pyramid pooling. On the one hand, DDSPP can get more receptive fields shows that resampling features of different scales is effective and can accurately and efficiently classify areas of an arbitrary scale. On the other hand, connecting the low-level features and high-level features enhances the robustness of the algorithm, and produces competitive results in decoder part. The experimental results show that the proposed method has better robustness and accuracy than other methods, which means remarkable performance in prostate segmentation.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30.
- Sharp G, Fritscher KD, Pekar V, et al. Vision 20/20: perspectives on automated image segmentation for radiotherapy. Med Phys. 2014;41:050902
- Martin S , Patil N , Gaede S , et al. A Multiphase Technological Validation of a MRI Prostate Cancer Computer Autosegmentation Software Algorithm. Int J Radiat Oncol. 2011;81(2):S817.
- Yang M, Li X, Turkbey B, et al. Prostate segmentation in mr images using discriminant boundary features. IEEE Trans Biomed Eng. 2013;60:479–488.
- Gao Q, Asthana A, Tong T, et al. Hybrid decision forests for prostate segmentation in multi-channel MR Images. Proceedings of the International Conference on Pattern Recognition; June 22-25. Ghent (Belgium): IEEE Computer Society; 2015. p. 3298–3303.
- Havaei M, Davy A, Warde-Farley D, et al. Brain tumor segmentation with deep neural networks. Med Image Anal. 2017;35:18–31.
- Litjens G, Debats O, Wendy VDV, et al. A pattern recognition approach to zonal segmentation of the prostate on MRI. International Conference on Medical Image Computing & Computer-assisted Intervention. Med Image Comput Comput Assist Interv. 2012;15:413–420.
- Zhang Y, Peng J, Gang L, et al. Research on the segmentation method of prostate magnetic resonance image based on level set. Chin J Sci Instrum. 2017;38:416–424.
- Jia H, Xia Y, Song Y, et al. Atlas registration and ensemble deep convolutional neural network-based prostate segmentation using magnetic resonance imaging. Neurocomputing. 2018;275:1358–1369.
- Mahapatra D, Buhmann JM. Prostate MRI segmentation using learned semantic knowledge and graph cuts. IEEE Trans Biomed Eng. 2014;61:756–764.
- Qayyum A, Anwar SM, Awais M, et al. Medical image retrieval using deep convolutional neural network. Neurocomputing. 2017;266:8.
- Zhu Q, Du B, Turkbey B, et al. Deeply-supervised CNN for prostate segmentation. Proceedings of the International Joint Conference on Neural Networks. Wuhan: IEEE; 2017. p. 178–184.
- Karimi D, Samei G, Kesch C, et al. Prostate segmentation in MRI using a convolutional neural network architecture and training strategy based on statistical shape models. Int J Comput Assist Radiol Surg. 2018;13:1–9.
- Tian Z, Liu L, Fei B. Deep convolutional neural network for prostate MR segmentation. Proceedings of the Society of Photo-Optical Instrumentation Engineers. Orlando, Florida (USA): Society of Photo-Optical Instrumentation Engineers (SPIE) Conference Series; February 11, 2017. 101351L.
- Zhan S. Deconvolutional neural network for prostate MRI segmentation. J Image Graph. 2017;22.4:516–522.
- Yu F, Koltun V. Multi-scale context aggregation by dilated convolutions. Proceedings of the arXiv: Computer Vision and Pattern Recognition; Boston, MA; November 23, 2015. p. 1–13.
- Chen LC, Papandreou G, Kokkinos I, et al. DeepLab: semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected CRFs. IEEE Trans Pattern Anal Mach Intell. 2018;40:834–848.
- Lin TY, Dollar P, Girshick R, et al. Feature pyramid networks for object detection. 2016. p. 936–944.
- Fu CY, Liu W, Ranga A, et al. DSSD: deconvolutional single shot detector. Hawaii (USA): CVPR; 2017.
- Chen LC, Zhu Y, Papandreou G, et al. Encoder-decoder with atrous separable convolution for semantic image segmentation. Wellington (New Zealand): CVPR; 2018. p. 833–851.
- Shelhamer E, Long J, Darrell T. Fully convolutional networks for semantic segmentation. IEEE Trans Pattern Anal Mach Intell. 2017;39(4):640–651.
- Noh H, Hong S, Han B. Learning deconvolution network for semantic segmentation. Proceedings of the IEEE International Conference on Computer Vision. Chile (SA): IEEE Computer Society; 2015. p. 1520–1528.
- Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation. Vol. 9351. Boston (USA): CVPR; 2015. p. 234–241.
- Bansal A, Chen X, Russell B, et al. PixelNet: Representation of the pixels, by the pixels, and for the pixels. Hawaii (USA): CVPR; 2017.