 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
The omicron infection rate led many countries to lock down. The variant risk assessment and new ones have been a high uncertainty. In this study, the model training is used across all data. By this point in the modeling process, the optimal hyperparameters that gave the best performance on the outer loop validation dataset have already been identified. Specific validation data given for our measurements are taken daily and set some arguments to skip between validation data. The results showed 58 days for UK research at 9 areas in two months displayed that the Northwest Enn. It confirmed most infection cases: values in November 2021 were 11,305,9811 for n (confirmed cases), 86.8% of infection rate, confidence low was 86.14% and 87.40 % for high confidence high. Values for November 22, 2021, were 16,985, and 15,415 for high confirmed cases, and 94.4% of infection rate, confidence low was 93.99, and 94.83 for high, respectively.
Highlights
The study is to make a model to epidemic modeling EpiNow to estimate the effective reproduction and related statistics.
The study is to analyze spread characteristics of S-gene positivity of the omicron.
This study is to estimate and predict the omicron data from the UK to obtain the trend in epidemiology.
The results of this study suggest that the omicron variants have elusiveness, transmissibility, and changeability.
1. Introduction
Omicron has been outbreaking in the world. The UK Health Security Agency (UKHSA) identified all variants and amended its variant classification system to give a clearer indication and observe potentially significant changes in biological properties compared to the current dominant variants. The S gene belongs to BA. 2. XE that has been confirmed community transmission within the UK. BA.2 was found it rarely has spike gene deletion at position 69/70 and S-gene target positive (SGTP) on diagnostic assays with targets in the area. In England there was 97.6% compared to 93.7% on 20 March 2022 [Citation1]. Two scientists confirmed that Omicron has been transmitted and evaded vaccines and course reinfections to more than 20 countries [Citation2]. Some researchers observed the seroprevalence impact on 7010 participants. The results found that a decoupling of hospitalizations and deaths from the infections while omicron was circulating [Citation3]. Some researchers explained that if the infection of SAR-CoV-2, the risk of severe outcomes is substantially lower for omicron than for delta, with higher reductions for more severe endpoints and significant variation with age [Citation4]. A report has confirmed that after the first detection of omicron, the outbreak threshold within 5–10 days, and other omicron variants came to 14 days and up to 35 days [Citation5].
Time-series analysis of COVID-19 cases and forecasting future trends have been made in fewer professional journals. A researcher tried to use a Bayesian structure time-series model to forecast the COVID-19 cases in the following time and made some progressive results [Citation6]. A study from a researcher used the Arima method to analyze the omicron data and obtained satisfactory effects [Citation7]. Some scientists applied regression models to analyze the association between symptomatic infection with the Omicron or Delta variants and vaccination by comparing the odds of prior 3 dose vaccination vs unvaccinated obtained good results [Citation8]. Thus, we think that using time-series analysis of the omicron and its variant needs to be a strengthening area of the weakness.
The study of this article aims to estimate the omicron properties, its irregular nature of changeability, and susceptibility as well as the impact of omicron on communities from the density of population.
2. Methods
2.1. Data
We obtained the data from the UK Health Security Agency which is responsible for pre-testing every member of every community from the impact of infectious infectious diseases. It includes 9 areas in the UK. The variables contain UKHSA region, specimen date, n, per cent, sgtf, total, low conference interval, and high conference interval between 1 November and 28 December 2021. The total number of observations is 943, including adults and children. The UKHSA region represents 9 areas in the UK Health Security Agency: Landon, East Midlands, East of England, Northeast, Northwest, Southeast, Southwest, West Midlands, Yorkshire, and Humber. ‘Specimen date’ indicates the date that collected cases. ‘total’ represents the total number of cases daily that suspected Omicron with the symptoms. ‘n’ in the dataset indicates the number of people who were confirmed as Omicron infection daily. ‘percent’ is the value that equals n/total. ‘sgtf’ means the Omicron S-gene target failure as a percentage of cases. Typically, England data reported are up to 6 pm on 30 December, Northern Ireland data collected are up to 4 pm on 30 December; Scotland and Wales’ data are up to 5 pm on 30 December. Hospitalization and vaccination data are up to 30 December and might be lagged up to 48 hours [Citation9].
2.2. Omicron variant characteristics
The Omicron variant classification was based on the UKHSA amendment for the variant classification system to give a clear indication that variants have significantly generated change in biological features that differentiate from the current dominant variants. Current variants were classified as Omicron sub-lineage BA.1 VOC-21NOV-01, Omicron sub-lineage BA.1 VOC-22 NOV-01, V-22APR-02(XE), V-22APR-03(Omicron sub-lineage BA.4), V-22APR-04(Omicron sub-lineage BA.5), V-22OCT-01(AY.4.2), V-20DEC-01(Alpha, B.1.1.7), V-21APR-02(Delta, and all sub-lineages). Also, XD is the lineage that has the Omicron S gene and combines a Delta genome which is mainly found in the France and UK. XE is a BA.1/BA.2 recombinant that includes the S gene belonging to BA.2. It could be transmitted within England. The overall proportion of SGTP caused by the relevant assay in England on 3 April 2022 for 97.6%. The assay that detects S gene target failure processed the number of samples and it has fallen which will affect the reliability of estimation.
2.3. Modeling for this data
2.3.1. The modeling epidemic dynamic characteristics
In the epidemic modeling there are three basic models: SIS endemic, SIR epidemic, and SIR endemic as the spread of infectious diseases. This study considers the susceptible exposes-infectious-recovered (SEIRD) epidemic model. If we assume the total population of the areas as the analyzed subject is T. Also, we assume that only some infected persons at the onset of groups in this area had been transmitted. So, susceptible person P would be exposed to E with the force of infection I. Then, the transmissive model could come into I. We supposed that unobserved (N) and observed (A) infectious persons spread the infectious groups as follows:
where
is the infectious rate, and the effective population sizes should be
, where
is the total population size, and the number
is isolated persons who were able to be infected by infectious patients within their contracted groups.
included those whose persons separated infected ones for social contacts and self-isolating persons. Also, we assumed that the contact proportion for observed infectious is a factor (
). Then, if a fraction
of exposed persons was introduced to the observed infected class (
) with the rate
. If those persons come from the infectious class, that is
which is a faction 1-d to regaining and would have deleting class (m), and the remainder(d). on the other hand, for unobserved ones, individuals come to the deleting state by rate
[Citation10].
Since the reproduction number R is a measure of the transmissibility for the epidemic disorder, sometimes it represents basic reproduction values such as . If there exists susceptible individuals for a population change, we often use
to express value that reflects various response measures. Thus, EpiNow2 can provide some important advantages over other packages. For example, it could delay reporting and estimate
, especially as data are not complete. Also, we can estimate
for dates of infection in that it represents a change in
In this article, the omicron infection over space and time has happened in the world. The UK omicron incidence characteristics have some differences from other regions in the world. The author used epidemic modeling EpiNow to estimate the effective reproduction and related statistics such as the doubling time, and future incidence. Figure presents the omicron incidence numbers, report cases on different dates, epidemic curve, estimate R, and explored Sl distribution, respectively. After that, the ggplot function displays 9-panel observation values in the UK in Figures and . The last modeling is to forecast omicron trends and optimal hyperparameters in the UK as time-series modeling professes [Citation11] We generate a model of the expected reproduction number at the level of upper-tier local authorities l during a single day m starting on November 1, 2021, which includes local restrictions, mobility indicators, residual temporal variation, and proportion of positive test S-gene positive. The function is as follows:
where
is an intercept in the level of upper-tier local authorities that corresponds to
while the UK omicron infection in November and December 2021,
is 1 when there is intervention
is in place and 0 otherwise.
refers to the relative mobility in context (such as home, job, and public place) at time
in the level of upper-tier local authorities in which
as measured via Google, and
is a time-varying component, modeled either as a region-specific thin-plate regression spline, and there is the sum of a static regional parameter and the UK spline. In this paper, we suppose the baseline model in a static regional parameter, when this model got the most explicable parameter estimates, and we can assume reproducibility of numbers with the relaxation steps and spread of S-gene positivity. Also, the spline version is referred to as capture confounding with unmeasured covariates over time, and it was as lower bounds on effect estimates. At last, we reached a fitted model in each
separately. On the other hand, we want to point out that the important parameter is
, which is the relative change in reproduction number in the presence of positive s-gene;
is the proportion out of all positive tests for COVID-19 where the S-gene was tested positive, and the reproduction number in any given level of upper-tier local authorities.
2.3.2. Statistical analysis
We estimated the daily suspected and confirmed cases. And then, we analyzed and forecasted these variables using time-series regression with a forecast machine learning model. This technique included train-test split, which created the models on data train and evaluated their out-of-sample performance on data test; data preparation, which set up a list of datasets for model training, and there is one forecast horizon; windows establishing that partitioned the training data in the outer loop of nested cross-validation. The validation was in contiguous blocks of window length so that we forecast over multi-step ahead forecast horizons; historical data forecasting that took some numbers from trained models of train model and a list of used defined forecast functions. Model selection to estimate the LASSO model more stable and accurate and retrain the model by the entire training dataset to get our final models-1 for forecast horizon. To overview the cases in the UK, we used a pie chart that was based on the data frame entries.
3. Results
UK’s population is 68.4 million of which 1.77 persons day daily [Citation12]. Omicron is a variant of the changeable and contagious epidemiology. In many countries and areas, the omicron outbreak presents suddenly increasing numbers daily. This paper analyzed detailed variant surveillance that made the variant risk assessments and designation of new SARS-CoV-2 variants such as Omicron variants S-gene and another confirmed SGFT. We observed 943 cases in 9 UK omicron infections in November and December the suspected and confirmed cases in Northwest reached our highest values (Table ) in two single days.
Table 1. The most Omicron suspected and confirmed cases in each area in the UK were in November and December 2021.
In addition, we found that the Omicron variant’s S-gene and another confirmed SGFT existed in 9 UK areas and cases rapidly increased to a higher value in a couple of days. Northwest and London were the most significant areas. In addition, the author analyzed UK omicron by using epidemic modeling. In Figure , we can see that the results displayed the cases of omicron would be increased trends: the new confirmed cases by infection increased (5806 cases). The rate of growth was 0. 032. These suggested that omicron variants are strongly contagious and abruptly increased.
Figure 1. COVID-19 Epidemic modeling of the datasets. The new confirmed cases by infection date look like 5806 (between 1536 and 18,234). The effective reproduction numbers are 1.1 for 0.64 and 1.7. The rate of growth is 0.032 between – 0.032 and 0.17. Doubling or halving time (days) is 21 days. The unsure value is 0.32.
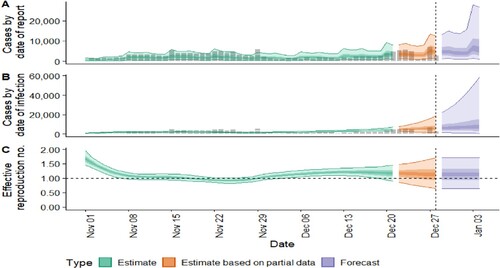
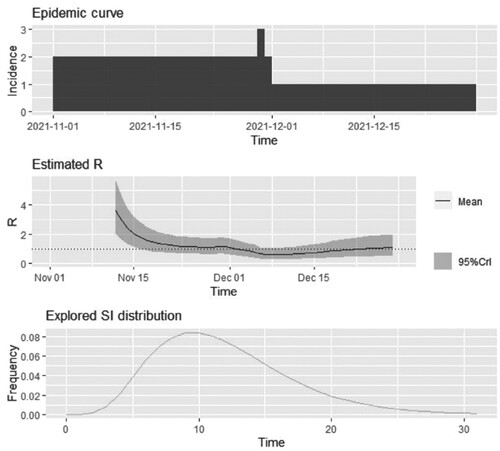
Next, the authors analyzed the dataset by estimating from the Epidemic curve, Estimated R, and Explored SI distribution in Figure . In the Epidemic curve, the incidence on December 1 2021, reached 3; in Estimated R, the trend appeared from higher to lower; the explored SI distribution seemed wavy. These changes reflected the observed values that were influenced by the epidemic.
Figure 2. The Serial internal estimates from the data. It shows that it looks like to be highest incidence on December 1, 2021. The values of the mean are kpt under 4. The frequencies of the explored Sl distribution look like to be shaped waves. The highest value of frequency was 0.08.
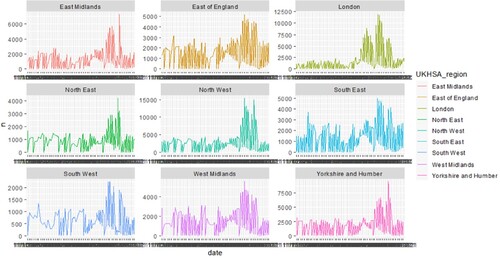
For forecasting case trends, we used the ggplot function to estimate the values of confirmed and suspected cases no matter what the symptoms happened. In London, there were from lower, higher, and last lower. For this point, it was particularly prominent. Rather, the Omicron infection cases in other UK regions kept the values under 5000 dailies. We also found that the southwest region least increase. It reflected that more populations and more density could become an epidemiological environment.
Like suspected cases, the most increase of confirmed cases daily was in Northwest and London, and the least one was the southwest region. These showed that more populations and multiple cultural areas were susceptible to epidemic diseases (Figure ). We should emphasize preventive measurement and health policies in these areas.
Figure 3. Overview of confirmed cases in the UK. 9 panels show the observation values, in each area in the UK. The model is a ggplot function that included specimen date (x-axis) and suspected cases of Omicron symptoms during November and December 2021 (y-axis). It looks like significant pandemic mitigation strategies. The colors are mainly used to represent 9 different research areas.
The authors also estimated and predicted the suspected cases to increase daily. It showed that, after December 28, 2021, suspected and confirmed increasing cases daily should continue to up to a higher peak (Figure ).
Figure 4. Forecast of datasets. We allow the model training to across all data. By this point in the modeling process, the optimal hyperparameters that gave the best performance on the outer loop validation datasets have already been identified.
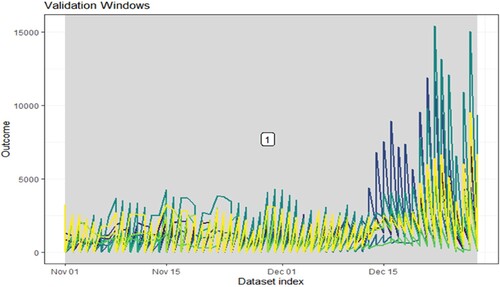
Incorporating the optimal hyperparameters in a final model would occur in a newly defined modeling wrapper function. The model with ‘1’ validation datasets that are given for our measurements are taken daily and set 460 arguments to skip between validation datasets. Also, custom validation windows were defined with vectors of start and stop dates. The blue panel represents feature, the green one indicates forecast, and the yellow one expresses no feature. The outside research dates, after December 28 2021, seem to have more infections and suspected cases in the UK.
4. Discussion and limitations
The UK Health Security Agency conducted an amendment for its variant classification system to give a clearer indication. This study is to explore some methods to find some epidemiological characteristics in the UK. The results suggest that the regions Omicron spread rapidly out may occur in higher population density, or big cities, people’s activity frequency. Our results show that Omicron infection cases were climbing up in the same UK area in a single day. Also, our result findings included the highest values of suspected and confirmed cases had most Omicron infection frequency came to 4000–4800, the median value was about 4500; the median value of confirmed cases/suspected cases was 70%; moreover, there was a narrow difference between confidence low and confidence high; the out of research time, after December 28 2021, forecasting values may be increasing. These findings have epidemiological significance. The study for Omicron analysis is currently a few reported in professional journals. Poland researchers reported that the Omicron variant (B.1.1.529) could be the exponential growth over four-week periods in the two most populated of South Africa’s provinces [Citation13]. An England researcher reported a Friday’s daily data was 12,395 patients [Citation14]. Some researchers analyzed 5 impacted countries including the USA, India, France, the UK, and Italy and found rapid growth in these countries in 2.5–4 days [Citation15]. A couple of researchers used fit Cox proportional hazard models to compare time to any hospital admission and the association with new-onset respiratory symptoms, etc. The results found that 16,982 cases were infected as non-SGFT (Delta [B.1.617.2]) in southern California in the United States [Citation16].
However, due to the limitation of these research data, we cannot further explore the impact of different environments, different age groups, different religions, and different policies of preventive measurement on Omicron variants. For example, how to analyze the prevalence of symptoms for an omicron infection from those of the delta [Citation17]. How to figure out the problem of prevention if omicron infects children [Citation18]? Currently, based on the data available, our study did not further estimate the impact of Omicron and its variants on communities. In addition, how to decrease the odds of hospitalization GGTF versus non-SGTF infection [Citation19]. Second, how to identify omicron variant mutation in clinical samples? When there are several variants with the original wild type of strain [Citation20]. Third, in our study of 943 observations with 58-day omicron infection cases, but time series was not long enough. The nonlinear association between the confidence interval and SGTF was unknown.
Further analysis should have more detailed information such as the presence of a relationship between the Omicron variants and different age groups whether they have been vaccinated [Citation21], or they have obtained chronic diseases. Unfortunately, data in this study lack age groups (Please see the Appendix), we cannot analyze that age groups affect the omicron variants. Also, we hope that future research will include longer and newer time series.
In summary, in terms of the current study of time series of omicron infection cases daily in the UK, we obtained the results including that the population living in density regions existed at high risk of omicron infections, even more, possible to multiple time infections; the existence of a significant relationship between narrow confidence interval and high total numbers infected and confirmed with Omicron variants; the association of sharp increasing cases daily with strong contagious omicron variant mutation; in 58-day time-series data the suspected and confirmed cases in the future could be continued to have increasing trends in the UK. We predict the data will have a trend of increasing status in the future (Figure ).
5. Conclusion
In this study, we analyzed the Omicron in THE UK November and December outbreaks present suddenly increasing numbers daily and estimated omicron variants S gene and SGFT. In forecasting case trends, we used the ggplot function to estimate the values of confirmed and suspected cases no matter what symptoms happened. We think that in London, there was from lower, higher, and last lower. For this point, it was particularly prominent. Rather, the Omicron infection cases in other UK regions kept the values under 5000 dailies. We also found that the southwest region least increases. These results remind us that public health in the areas where there is a population should be improved and strengthened to prevent epidemics in the future.
Data availability and materials statement
https://www.gov.uk/government/publications/covid-19-omicron-daily-overview.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Notes on contributors
Liming Xie
Liming Xie is a PhD student in the department of statistics at North Dakota State University. 1340 Administration Ave, Fargo, ND 58108, USA.
Xiyu Deng
Xiyu Deng is an MD and professor in the department of internal medicine, The First Hospital of Wuchang, Wuhan, Hubei 430060, China.
Xiaoyan Xie
Xiaoyan Xie is a Charge Nurse and RN in the department of preventive Disease, Wuchang Center of Disease Control and Prevention, Wuhan, Hubei 430061, China.
Shanshan Hu
Shanshan Hu receiced a master's in data Science program at Rutgers University School of Arts and Sciences, New Brunswick, NJ 08901. Her favorite research topics cover the statistical modeling of epidemiological and genomic data.
References
- UK Health Security Agency. SARS-CoV-2 variants of concern and variants under investigation in England Technical briefing 40, 8 April 2022.
- Ewen C, Heidi L. How bad is omicron? what scientists know so far. Nat News. 2021;600, 2021: 197–199.
- Shabir MA, Gaurav K, et al. Population immunity and COVID-19 severity with omicron variant in South Africa. N Engl J Med 2022 Apr 7; 386 (14): 1314–1326.
- Tommy N, Neil FM, et al. Comparative analysis of the risks of hospitalisation and death associated with SARS-CoV-2 omicron (B.1.1.529) and delta (B.1.617.2) variants in England: a cohort study. Lancet. 2022;399:1303–1312. doi:10.1016/S0140-6736(22)00462-7
- Alexander LL, Ramon L-R, et al. Has omicron changed the evolution of the pandemic? JMIR Public Health Surveill. 2022;8(1):e35763, doi:10.2196/35763
- Liming X. The analysis and forecasting COVID-19 cases in the United States using Bayesian structural time series models. Biostat Epidemiol. 2021;6(1):1–15. doi:10.1080/24709360.2021.1948380.
- Debmalya R. New omicron variant- time series analysis. Dev Genius 2022, Apr 3.
- Emma AK, Katherine F-DE, et al. Association between 3 doses of mRNA COVID-19 vaccine and symptomatic infection caused by the SARS-CoV-2 omicron and delta variants. JAMA. 2022;327(7):639–651. doi:10.1001/jama.2022.0470
- Champredon D, Dushoff J, Earn David JD. Equivalence of the erlang-distributed SEIR epidemic model and the renewal equation. SIAM J Appl Math. 2018;78(6):3258–3278. doi:10.1137/18M1186411
- UK Health Security Agency. COVID-19: Omicron daily overview, Details, 2021. https://www.gov.uk/government/publications/covid-19-omicron-daily-overview.
- World population review. United Kingdom population. 2022 (Live). 2022.
- Frederic G, Marek K, et al. The spread of SARS-CoV-2 variant omicron with a doubling time of 2.0–3.3 days can be explained by immune evasion. MDPI Viruses. 2022;14:294, doi:10.3390/v14020294
- Reuters. Omicron hospitalization risks around one third of Delta, UK. Healthcare and Pharmaceuticals. 2021, December 31.
- Ranjan R. Omicron impact in India: analysis of the ongoing COVID-19 third wave based on global data. medRxiv. 2022. doi: 10.1101/2022.01.09.22268969
- Lewnard A, Hong V, et al. Clinical outcomes among patients infected with omicron (B.1.1.529) SARS-CoV-2 variant in southern California. medRxiv. 2022;28(9):1933–1943.
- Cristina M, Anna VM, et al. Symptom prevalence, duration, and risk of hospital admission in individuals infected with SARS-CoV-2 during periods of omicron and delta variant dominance: a prospective observational study from the ZOE COVID study. Lancet. 2021;399(10335): 1618–1624. doi: 10.1016/S0140-6736(22)00327-0.
- Max K. Does omicron hit kids harder? scientists are trying to find out. Nat News. 2022. doi:10.1038/d41586-022-00309-x.
- Nicole W, Waasila J, et al. Early assessment of the clinical severity of the SARS-CoV-2 omicron variant in South Africa: a data linkage study. Lancet. 2022;399:437–446.
- Yamin S, Wenchao L, et al. Origin and evolutionary analysis of the SARS-CoV-2 omicron variant. J Biosaf Biosecurity. 2022;4(1):33–37.
- William HS, Elizabeth M, et al. Generation time of the alpha and delta SAR-CoV-2 variants: an epidemiological analysis. Lancet Infect Dis. 2022;22(5):603–610. doi:10.1016/S1473-3099(22)00001-9
- Nick A, Stowe J, et al. COVID-19 vaccine effectiveness against the omicron (B.1.1.529) variant. N Engl J Med. 2022;386(16):1532–1546. doi:10.1056/NEJMoa2119451
