Testing for tuberculosis (TB), using chest radiography for screening and microbiology for confirmation, is indicated in everyone considered to be at high risk of TB disease or with signs and symptoms of TB.
Chest radiography is an integral part of the TB diagnostic algorithm but is not specific for the diagnosis of pulmonary TB (PTB) and cannot provide a conclusive diagnosis on its own.
Every effort should be made to obtain a microbiological diagnosis, which requires demonstration of acid-fast bacilli (AFB) on smear microscopy and/or culture or requires detection of Mycobacterium tuberculosis (M. tuberculosis) nucleic acids using nucleic acid amplification tests (NAATs).
Phenotypic drug susceptibility testing (DST) should be routinely performed for first positive culture isolates obtained from each new TB patient.
Presence of AFB on smear microscopy, positive TB culture or NAAT in patients not previously diagnosed with TB represent laboratory critical values and should immediately be reported to the submitting clinician.
All patients newly diagnosed with active TB should be reported to public health authorities.
The use of tuberculin skin test (TST) or interferon gamma release assays (IGRA) for the diagnosis of TB disease in adults is not recommended.
NAAT testing is not recommended for monitoring treatment response or determining contagiousness after treatment has been initiated.
Rapid molecular tests should be performed to predict drug resistant-TB (DR-TB) on new positive cultures and/or samples with a new positive NAAT. The use of these tests does not eliminate the need for conventional phenotypic DST.
KEY POINTS
1. Introduction
This chapter will cover radiologic testing for suspected pulmonary TB disease and microbiologic investigations for samples from all sites (pulmonary and extra-pulmonary), from microscopy to DST, including molecular prediction of drug resistance.
2. Radiology
2.1. Chest radiography
The lungs are the most commonly involved organ in TB and, from a public health perspective, the most important. In Canada, in 2017, 69.4% of all notified cases were classified as PTB.Citation1 Despite the increased availability of more-detailed imaging techniques, such as computed tomography (CT), chest radiography remains the mainstay of chest imaging for PTB. This overview is focused on chest imaging in adults.
2.1.1. Chest radiograph
Chest radiographs are important to both the diagnosis and management of PTB. They are accessible and inexpensive in most settings, they can be easily acquired and interpreted at point of care and they are safe: a single chest radiograph exposes the patient to an amount of radiation that is roughly equivalent to what they are exposed to naturally over the course of about 10 days. Serial chest radiographs provide additional information in that they allow one to detect change and therefore help to identify and chart the progress of TB disease.
PTB can manifest on chest radiograph in a variety of ways.Citation2 Key patterns important to the diagnosis of TB disease are summarized in the following sections.
2.1.2. Key diagnostic patterns
The classic radiographic presentation of PTB in immunocompetent adults is upper lung zone disease (ie, involving the apical or posterior segments of the upper lobes or the superior segments of the lower lobes), with or without cavitation but with no discernable adenopathy. This was the pattern observed in 56.2% of culture-positive PTB patients in a recent population-based cohort study in Canada; in 67.0% of those patients with this pattern, the sputum microscopy was positive.Citation8 The presence of cavitation in such patients increases the probability of a higher semi-quantitative smear grade (from 1-2+ to 3-4+),Citation3 which may explain in part why smear-positivity and cavitation are independent risk factors for transmission.Citation4–6 More details are provided in Chapter 2: Transmission and Pathogenesis of Tuberculosis.
Intrathoracic adenopathy, typically involving the left or right hilar and/or right paratracheal lymph nodes, is a common feature of primary PTB in children. This pattern of progressive primary PTB may also present in adolescents or adults.
A calcified pulmonary nodule, usually in the lower part of upper lobes or the upper part of lower lobes and close to the pleura, is consistent with a granuloma and generally indicates remote TB infection.
Unusual or atypical patterns include solitary pleural effusion, lower lobe TB, a diffuse micronodular (miliary) pattern and, occasionally, a normal chest radiograph.
2.1.3. Key factors that can influence the chest radiographic presentation of PTB
Older age and immunosuppressing conditions that are known to increase the risk of TB (eg, human immunodeficiency virus/acquired immunodeficiency syndrome (HIV/AIDS), transplant immunosuppressionCitation7,Citation8 and renal failureCitation9–11 also increase the likelihood of an atypical radiographic presentation of PTB. Another high-risk medical condition, silicosis, can, itself, alter the appearance of the chest radiograph in such a way as to make it difficult to discern concomitant PTB. Patients with past PTB treated with collapse therapy, such as thoracoplasty, can relapse years later with atypical radiographic abnormalities, most of which are unrelated to the current episode.
2.1.4. Chest radiographs during pregnancy
As a general rule, the risk to the fetus of undiagnosed PTB far outweighs any risk from radiation exposure. Some simple steps, however, can minimize the risk to the fetus of radiation exposure. First, avoid chest radiographs during the first trimester if possible; second, limit the exposure to a single posterior-anterior (PA) view; and third, double-shield the abdomen, both front and back.
2.1.5. Accuracy and limitations of chest radiography
Sensitivity: If any abnormality is considered, the chest radiograph has more than 95% sensitivity;Citation12 if only those key patterns listed above are included, the sensitivity is reduced substantially. A normal radiograph may sometimes occur in someone who is sputum culture-positive and living with HIV, especially those with advanced immunosuppression; close contacts of sputum smear-positive PTB; and patients with extra-pulmonary TB. Such patients may or may not have symptoms referable to the respiratory tract.
Specificity: Specificity is greater when only chest radiographic abnormalities suggestive of PTB are considered.Citation12 If sensitivity is improved, by considering any abnormality as possible TB, then specificity is reduced from 89% to 75%.
Inter-reader variability: One of the greatest problems associated with chest radiograph reading is that interpretation is highly variable. Even with experienced chest radiologists, there is poor agreement between and within readers regarding the presence of cavitation, hilar adenopathy or the likelihood of TB disease.
In summary, chest radiograph is an imperfect tool. The sensitivity in people with symptoms is high, therefore a negative chest radiograph can be a helpful, albeit imperfect, rule-out test. However, it cannot be used as a stand-alone test to rule in PTB.
2.2. Other radiologic methods
2.2.1. Computed tomography
The major advantage of computed tomography CT is increasing the specificity of the diagnosis of TB; therefore, CT is often not necessary in the acute setting, particularly when the disease is already suspected, with appropriate precautions implemented and microbiologic testing underway. CT may be able to better show distinct findings, such as cavitation or endobronchial spread with tree-in-bud nodules, and may be helpful in cases in which the chest radiograph does not show “classic” findings of PTB.Citation13 Although CT has twice the sensitivity to detect cavities,Citation14 it is the presence or absence of cavitation on chest radiograph that is entered on Canada’s case report form and that is used in treatment-duration decisions. CT findings can also better correlate with AFB smear microscopy positivity.Citation15–17 Even in AFB smear-negative patients, CT may suggest the risk that a patient will be TB-culture positive when findings consistent with PTB are present.Citation18 CT may be of value in the severely immunocompromised patient with a normal or near-normal radiograph by revealing abnormal lymph nodes or subtle parenchymal disease.
2.2.2. Magnetic resonance imaging
The accuracy of magnetic resonance imaging (MRI) is similar to CT in describing findings related to culture-positive PTB.Citation19 MRI’s greatest utility, however, is in the diagnosis and management of extra-pulmonary TB (see Chapter 7: Extra-pulmonary Tuberculosis). MRI is a reasonable consideration for use in select patients for whom there is a desire to avoid ionizing radiation.
2.2.3. FDG-PET/CT
The noninvasive imaging tool 18F-fluoro-2-deoxyglucose-positron emission tomography/computed tomography (FDG-PET/CT) is used primarily for cancer diagnosis and staging. It identifies areas of active inflammation by mapping where cells with high metabolic demand take up the radioactively labeled glucose analogue. The radiotracer accumulates within inflammatory cells, such as macrophages and neutrophils, and can be quantified as a standardized uptake value (SUV). FDG-PET/CT cannot reliably differentiate PTB lesions from malignant lesions or other infections/inflammatory conditions.Citation20 Radiotracer uptake magnitude is also unrelated to TB disease activity, thus limiting the role of FDG-PET/CT in the diagnosis of PTB.Citation21 Conceivably, FDG-PET/CT may be helpful in identifying other sites of disease in patients with an already confirmed tracer-positive site of involvement.
2.2.4. Future developments in imaging for TB
Deep learning artificial intelligence software for chest radiography detection of PTB have achieved sensitivity and specificity similar to human readers,Citation22,Citation23 exceeding thresholds for triage test criteria by the World Health Organization (WHO).Citation24 This may be valuable in closing diagnostic gaps in resource-limited and remote settings.
In the future, electronic medical records may be able to link clinical and computer-detected radiographic features to aid diagnosis.Citation25 The intensity and duration of treatment may eventually be tailored to the clinical and radiographic presentation of PTB.
Recommendation
We strongly recommend that posterior-anterior and lateral chest radiography should be an integral part of TB diagnosis but should be accompanied by confirmatory microbiological tests for TB disease because of its low specificity (good evidence).
Good practice statements
Chest radiography findings suggestive of pulmonary TB should be immediately reported to the ordering physician.
In pregnant women suspected of having TB, a posterior-anterior chest radiograph should be performed, as the risk to the fetus of undiagnosed pulmonary TB far outweighs any risk from radiation exposure.
3. Microbiology
The role of the mycobacteriology TB laboratory is to detect, isolate, identify and perform DST on clinically significant mycobacteria from clinical specimens. Mycobacterial culture, using liquid broth and/or solid culture media, is considered the gold standard for diagnosis, and the use of broth-based culture methods for DST is the standard of practice in North America.Citation26–28 The most widely used rapid tests are 1) staining and microscopic examination for acid-fast organisms (AFB smear), and 2) NAATs. Appendix 1 provides more detail on TB laboratory methodologies.
3.1. Clinical samples
Given the critical importance of microbiology for TB diagnosis, it is important to ensure that specimens are correctly collected and processed to achieve valid results. All specimens should be collected in sterile, leak-proof, laboratory-approved containers and accompanied by a completed requisition form providing the patient identifier, the ordering physician’s name, the date and time of collection and the specimen type and collection site. As much as possible, specimens collected for initial diagnosis should be obtained before the initiation of anti-TB therapy.Citation26,Citation29
Once collected, specimens should be transported to the laboratory promptly. If transport or processing within 1 hour is not possible, samples should be kept at 2-8 °C (not frozen) and protected from light until transport.
3.1.1. Sputum
At least 3 sputum specimens, optimally 5-10mL each, should be collected and tested with microscopy as well as culture. While available evidence shows that the incremental yield of the third sputum smear is only an additional 2-5%,Citation30,Citation31 the incremental yield of the third culture may be as high as 5-10%, especially in HIV-infected people.Citation31,Citation32
While it is conventional to collect three separate morning sputum specimens, it is well known that this scheme is inconvenient to patients, making drop-outs during diagnosis common. Published research has demonstrated the feasibility of “frontloaded” diagnosis of TB using specimens collected on the same day and shown that the diagnostic yield is undiminished.Citation33
3.1.2. Induced sputum
A systematic review of 23 studies reported that the overall success rate of sputum induction was high, ranging from 76.4 to 100%, while adverse events associated with sputum induction were infrequent and mild.Citation34 The sensitivity of microscopy is variable, presumably because the bacteria are diluted by the inhaled saline. Yield of induced sputum is generally higher than nasopharyngeal aspiration and gastric lavage,Citation34 or stool samples.
It is important that sputum induction be performed with large volumes of 3% hypertonic saline. For best results, an ultrasonic nebulizer should be used that can administer 5 to 6 mL per minute over 15 minutes.Citation35 With this use, virtually all patients will produce sputum, and a single sputum induction will have equivalent or better yield than fiberoptic bronchoscopy.Citation36 Sputum induction has also been performed successfully in very young childrenCitation37 (see Chapter 9: Pediatric Tuberculosis). Although the specimen often appears watery, it can be handled in the laboratory in the same way as spontaneously expectorated sputum.
3.1.3. Bronchoscopy
Bronchoscopy may be used to facilitate the diagnosis of TB when spontaneous and induced sputum are unavailable, or if another disease, such as lung cancer, is suspected.Citation38 Used solely for the diagnosis of active TB, however, it entails risk and discomfort for the patient, is expensive and can contribute to nosocomial spread of TB if not performed in an appropriate environment with protection of staff. In addition, the overall yield of bronchoscopy in prospective series of patients is only 77%.Citation39–42 If bronchoscopy is done, post-bronchoscopy sputum should be sent for AFB testing, as this has a yield similar to that of bronchial washings and lavage.
3.1.4. Gastric aspirate
This technique was introduced in the early 20th century and is still used in some centers.Citation43 The primary indications are investigation of possible TB in children who cannot expectorate sputum or, for the same reason, elderly patients with dementia. The technique is relatively simple and is described in Chapter 9: Pediatric Tuberculosis.
3.1.5. Stool
As young children swallow their sputum, recovery of M. tuberculosis from stool samples may be a way to diagnose TB disease. Collection of stool samples is noninvasive and doesn’t require specialized equipment/expertise. However, there are no standardized recommendations for stool processing for culture and NAAT testing and various studies have demonstrated relatively lower sensitivity of stool specimens vs sputa or gastric aspirates, as well as higher potential for culture contamination.Citation44 As such, stool cultures are currently not recommended in Canada for the purposes of PTB diagnosis.
3.1.6. Other specimen types
A variety of specimen types may be collected for diagnosing extra-pulmonary TB. In general, tissue has a higher yield than liquid/biologic fluids. The general best practices for collection and transport of these specimens are the same as for those routinely used for diagnosis of pulmonary TB. Handling of these specimens in the laboratory may differ with respect to the ability to perform AFB smears and/or molecular testing, as well as full culture set-up. Given that extra-pulmonary samples can have a smaller number of bacteria and there is often a small volume of sample, priority should be given to culture over other assays, such as microscopy or NAAT testing. Additional considerations specific to individual specimen types are described in Appendix 1 of this chapter and in Chapter 7: Extra-pulmonary Tuberculosis.
Recommendations
We strongly recommend that in all persons with suspected pulmonary TB, at least three (either spontaneous or induced) sputum specimens should be collected and tested with microscopy and culture (good evidence).
We strongly recommend that three sputum specimens (either spontaneous or induced) should be collected on the same day, at least 1 hour apart (good evidence).
We conditionally recommend that sputum samples should be collected in sterile containers without any transport medium and transported to the mycobacteriology laboratory within 1 day or stored at 2-8 °C until transport (poor evidence).
3.2. Laboratory testing for the diagnosis of TB
3.2.1. Smear microscopy
Sputum smear microscopy is the most widely used test for TB disease.Citation26 Two stains are widely used: 1) the traditional Ziehl-Neelsen or Kinyoun staining, which requires a light or bright field microscopy and 2) the auramine-rhodamine stain, which requires fluorescence microscopy (see Appendix 1). In most high-income countries (including Canada), fluorescence microscopy is standard practice because it can be read at a lower magnification than the classic Ziehl-Neelsen or Kinyoun stain, thus allowing slides to be read more quickly.Citation28 The sensitivity of all staining methods, however, is inferior to that of culture. The threshold of detection of AFB in concentrated specimens using a fluorochrome stain is 5,000-10,000Citation45,Citation46 bacteria/mL of sputum and is 100,000 bacteria/mL using the Ziehl-Neelsen stain. The threshold of detection in unconcentrated smears is 10-fold higher, resulting in much lower sensitivity. This is important to remember, since often “stat” smears are unconcentrated. In contrast, as few as 10 viable bacteria can be detected by culture.
The specificity of the AFB smear is high for mycobacteria, but it is important to remember that all nontuberculous mycobacteria (NTM) will be AFB-positive. Other organisms, such as Nocardia and other actinomycetes, can be weakly acid-fast, but these are less common. Therefore, a positive AFB smear almost always indicates the presence of mycobacteria, but not necessarily M. tuberculosis.Citation28
When acid-fast organisms are seen, the number of bacteria is reported semi-quantitatively, from 0 to 4+ as detailed in Appendix 1.
Smear microscopy is both rapid and inexpensive and identifies the most infectious TB patients (see Chapter 11: Tuberculosis Contact Investigation and Outbreak Management).Citation47 Although it has long been used to help assess contagiousness and manage isolation, the test has well-known limitations:
Sensitivity is modest and variable (20-80%) depending upon the type of specimen, patient population, stain used, time used to examine and the experience of the microscopist. Thus, multiple sputum smears are recommended to increase the overall sensitivity. Sensitivity is higher for respiratory than for non-respiratory specimens, particularly body fluids.
In low TB-incidence settings, smear microscopy has lower specificity — a positive smear could be due to NTM.
Smear microscopy has lower sensitivity in childhood TB and extra-pulmonary disease, especially in HIV-infected people.
Smear microscopy cannot be used to determine drug resistance.
Recommendations
We strongly recommend that smear microscopy should be performed on concentrated samples where technically feasible (good evidence).
We strongly recommend fluorescent microscopy to maximize the sensitivity of smear microscopy (good evidence).
3.2.2. Mycobacterial culture
Mycobacterial culture can be performed on all specimen types so long as it is received in appropriate condition.Citation26 Culture for M. tuberculosis is considered the gold standard in diagnosis, as it is more sensitive than microscopy or currently available NAAT tests. Culture allows for the identification of the pathogen, serves as the basis for DST and can provide isolates for molecular epidemiology using deoxyribonucleic acid (DNA) fingerprinting or genome sequencing. Standards and technical details for mycobacterial culture are described in Appendix 1.
A single positive culture for M. tuberculosis, in general, is considered definitive for active disease. However, it is important to remember that cultures occasionally can be falsely positive due to cross-contamination within the laboratory. When clinical suspicion is low, a report of a single positive culture, especially with a negative smear and a long detection time, should raise the possibility of a false-positive result. If this is a clinical possibility, the lab should be contacted to do further investigations.
Culture results typically take 2-to-8 weeks, depending on the culture method used and the number of bacteria in the inoculum. Once there is evidence of growth, labs will assign a presumptive identity of M. tuberculosis complex or non-tuberculous mycobacterium before further testing is done to provide a species name (see Appendix 1 for details).
Recommendations
We strongly recommend that every specimen of sufficient volume from patients with suspected TB undergo testing with both smear microscopy and culture; for very low volumes (< 2 mL),Citation48 culture should take precedence over smear microscopy (good evidence).
We strongly recommend that mycobacteria culture testing should include liquid medium culture at a minimum and both liquid and solid culture whenever possible (good evidence).
3.2.3. Nucleic acid amplification tests
The amplification of nucleic acids for the diagnosis of TB from specimens produces a faster result than conventional culture methods.Citation49,Citation50 Some of these assays also provide predictions about drug resistance, discussed further in the following section on drug-resistance testing.
On the one hand, the sensitivity of NAATs to detect TB is high (>95%) in sputum smear-positive samples and they are, therefore, used to provide a rapid presumptive diagnosis of TB while awaiting culture results.Citation49,Citation51 On the other hand, the sensitivity of NAATs is lower (50-70%) when smear-negative/culture-positive specimens are tested.Citation51 The difference in sensitivity is because the analytic sensitivity of NAATs is around 100 bacteria per mL – this is lower than the analytic sensitivity of AFB smear (5000-10,000 bacteria per mL) but higher than the analytic sensitivity of culture (fewer than 10 bacteria per mL).Citation49–52 The sensitivity of NAATs is also lower in extra-pulmonary specimens.Citation52 Negative NAAT results alone should not be used to rule out TB and considerations for patient management should be informed by other factors, such as the epidemiologic risk factors, clinical suspicion and results of culture.
Because certain cartridge-based NAAT technologies (eg, the Xpert, by Cepheid) are simple and can be implemented in peripheral laboratories, NAATs may be potentially useful in remote settings, where there is no on-site capacity for routine smear microscopy and cultures. In such settings, NAAT results can be available within hours and could potentially help reduce diagnostic delays. However, it is important to note that the use of Xpert in these settings should not replace conventional smears and cultures.
NAAT assays are continuously evolving. Front-line healthcare providers should be aware of when a test is replaced, as this could affect the negative or positive predictive values of the results. For example, the widely useful GeneXpert platform has recently introduced the Xpert MTB/RIF Ultra, which has greater sensitivity but reduced specificity compared to the first-generation assay.Citation52 The greater sensitivity of the Ultra (91%) might guide a clinician to consider another diagnosis if the test is negative in a patient with a low pretest probability. In high-burden countries the Ultra had a lower specificity of 96%, whereas retrospective studies demonstrated that in low-burden settings where there is limited TB transmission, the specificity of Ultra is high (99.3%, 95%CI 96-99).Citation53 In the absence of other corroborating information supporting a diagnosis of TB, one might consider the possibility of a false-positive. Further details on these assays are provided in Appendix 1. If in doubt, providers should contact the laboratory to ask more details on the NAAT test in use and its limitations.
Because NAATs can amplify nonviable AFB, they are not recommended for use in monitoring TB treatment response or to assess patients’ contagiousness after treatment initiation.
Recommendations
We strongly recommend that in all new smear-positive patients, at least one acid-fast bacilli positive respiratory sample should be tested with a Health Canada-approved or -validated laboratory-developed nucleic acid amplification test (good evidence).
We conditionally recommend that in smear-negative patients suspected of having TB, a nucleic acid amplification test may be performed on one acid-fast bacilli negative sample upon request by the physician or public health (poor evidence).
We strongly recommend against using nucleic acid amplification test results for monitoring TB treatment response or patient contagiousness after start of therapy (good evidence).
We conditionally recommend that, in remote settings where there is currently no on-site capacity for routine smear microscopy and culture, an automated cartridge-based nucleic acid amplification test can be used to make rapid decisions on TB treatment and isolation. However, specimens should also be sent to clinical/reference laboratories for smear microscopy and culture in such contexts (poor evidence).
3.3. Diagnosis of drug resistance
The diagnosis of drug-resistant TB can be made in 2 ways: 1) phenotypic (culture) and 2) molecular (or genomic) methods. Phenotypic DST should be routinely performed for all first-positive-culture isolates obtained from each new TB case. Molecular methods can be applied to samples (such as the Xpert MTB/RIF, previously detailed) or to positive cultures. For molecular methods, we deliberately use the term “resistance prediction” rather than “resistance testing” because a) these assays do not provide direct evidence for resistance, and b) for reasons detailed in the following section, they can give false-positive results that are refuted by the phenotypic DST done on the same sample.
For phenotypic testing, details are provided in Appendix 1. A phenotypic DST is done from a positive culture and is expected to provide results within 14 days from receipt of the positive culture. If the isolate is resistant to first-line drugs, the complete DST can take longer, as a second set of drugs is then tested. Unlike molecular tests, phenotypic DST provides direct evidence that an antibiotic does or does not work (growth of the organism is inhibited by the antibiotic).
Molecular testing involves two conceptually different approaches: targeted assays and whole-genome sequencing (WGS). In targeted testing, a known mutation or set of mutations is sought, to predict resistance. These assays lack perfect sensitivity because they only look at pre-specific parts of selected genes and can overlook mutations at other sites. As an example, ∼10% of isoniazid resistance is due to mutations not tested by line probe assays.Citation54 These assays also suffer from imperfect specificity because they report on whether there is a mutation in a part of the gene, without characterizing whether it is a resistance-conferring mutation.
In contrast, WGS interrogates the entire bacterial chromosome. It is more sensitive because it can detect resistance-associated mutations outside the regions examined in targeted assays. It is more specific because it can discriminate between mutations that are present but do not confer resistance and those that do. WGS is being routinely used in some settings (see Appendix 1) to predict drug sensitivity; in these settings, isolates predicted to be pan-susceptible to first-line drugs do not have any DST done. As a consequence, when WGS is validated in Canadian public health laboratories, providers can expect to receive a report of pan-susceptibility sooner than the current norm (2 weeks). If, on the other hand, there are mutations associated with resistance to any of the front-line drugs, second-line DST testing can be started immediately, leading to several weeks faster reporting for these drugs. The impact on patient care, however, remains to be determined. More information on resistance testing is provided in Appendix 1.
Recommendations
We strongly recommend that phenotypic drug susceptibility testing should be routinely performed for all first-positive-culture isolates obtained from each new TB case (good evidence).
We conditionally recommend that rapid molecular tests for prediction of drug resistance should be performed to rapidly predict drug resistant-TB on new positive cultures. The use of these tests does not eliminate the need for conventional culture-based phenotypic drug susceptibility testing (poor evidence).
We conditionally recommend that while awaiting a positive culture, rapid molecular testing on samples with a new positive nucleic acid amplification test should be performed. The use of these tests does not eliminate the need for a culture and an ensuing culture-based phenotypic drug susceptibility testing (poor evidence).
3.4. Future developments in the laboratory diagnosis of TB
3.4.1. Blood-based assays (serology, gene-expression signatures)
For decades, researchers and the industry had pinned their hopes on blood-based assays for the detection of TB. Unfortunately, TB serologic tests are inaccurate,Citation55,Citation56 prompting the WHO to issue a strong recommendation against their use.Citation57 A newer generation of blood-based tests, looking at gene expression signatures, is under development and evaluation. At the time of writing, none has achieved high enough sensitivity to rule out active TB, nor high enough specificity to be used as a stand-alone test for the diagnosis of TB.Citation58
Another blood-based assay is the IGRA. As described in Chapter 4: Diagnosis of Tuberculosis Infection, the IGRAs cannot distinguish infection from disease; for this reason, a recent WHO policy on IGRAs has discouraged their use for the diagnosis of TB disease. In children, TST and/or IGRA are used as an indicator of recent infection and can be used to support a diagnosis of TB disease, along with clinical data and radiologic and microbiological investigations (see Chapter 9: Pediatric Tuberculosis).
3.4.2. Fluid-based assays (adenosine deaminase)
Laboratories occasionally receive requests to send pleural, pericardial or peritoneal fluid for adenosine deaminase (ADA), which is a marker of serosal inflammation. A systematic review reported a sensitivity and specific of 90% for these tests in high-burden countries. Given that the test can give false-positive results with empyema and certain types of malignancies, it is not recommended in Canada.Citation59 Furthermore, the ADA result can support a diagnosis of TB but cannot inform antibiotic treatment, as there is no DST associated with this result.
3.4.3. Urine-based assays (urinary LAM detection)
Urine-based testing has received increased attention, with studies of patients with advanced HIV/AIDS reporting a more rapid diagnosis of TB and faster initiation of therapy.Citation60 These tests are not offered in Canadian TB labs, owing to the availability of other diagnostic modalities and the low burden of HIV-associated TB (see Chapter 10: Treatment of Active Tuberculosis in Special Populations).
Recommendations
We strongly recommend against the use of serologic TB tests, adenosine deaminase and urine lipoarabinomannan testing for the diagnosis of TB (good evidence).
We strongly recommend against the use of a tuberculin skin test or interferon-gamma release assay for the diagnosis of active TB in adults (good evidence).
Disclosure statement
The CTS TB Standards editors and authors declared potential conflicts of interest at the time of appointment and these were updated throughout the process in accordance with the CTS Conflict of Interest Disclosure Policy. Individual member conflict of interest statements are posted on the CTS website.
Funding
The 8th edition Canadian Tuberculosis Standards are jointly funded by the Canadian Thoracic Society (CTS) and the Public Health Agency of Canada, edited by the CTS and published by the CTS in collaboration with AMMI Canada. However, it is important to note that the clinical recommendations in the Standards are those of the CTS. The CTS TB Standards editors and authors are accountable to the CTS CRGC and the CTS Board of Directors. The CTS TB Standards editors and authors are functionally and editorially independent from any funding sources and did not receive any direct funding from external sources. The CTS receives unrestricted grants which are combined into a central operating account to facilitate the knowledge translation activities of the CTS Assemblies and its guideline and standards panels. No corporate funders played any role in the collection, review, analysis or interpretation of the scientific literature or in any decisions regarding the recommendations presented in this document.
References
- LaFreniere M, Hussain H, He N, McGuire M. Tuberculosis in Canada: 2017. Can Commun Dis Rep. 2019;45(2-3):67–74. doi:10.14745/ccdr.v45i23a04.
- Daley CL, Gotway MB, Jasmer RM. Radiographic Manifestations of Tuberculosis: A Primer for Clinicians. Francis J. Curry National Tuberculosis Center San Francisco; 2003.
- Benito N, Garcia-Vazquez E, Horcajada JP, et al. Clinical features and outcomes of tuberculosis in transplant recipients as compared with the general population: a retrospective matched cohort study. Clin Microbiol Infect. 2015;21(7):651–658. doi:10.1016/j.cmi.2015.03.010.
- Heffernan C, Barrie J, Doroshenko A, et al. Prompt recognition of infectious pulmonary tuberculosis is critical to achieving elimination goals: a retrospective cohort study. BMJ Open Resp Res. 2020;7(1):e000521. doi:10.1136/bmjresp-2019-000521.
- Bailey WC, Gerald LB, Kimerling ME, et al. Predictive model to identify positive tuberculosis skin test results during contact investigations. JAMA. 2002;287(8):996–1002. doi:10.1001/jama.287.8.996.
- Catanzaro A. Nosocomial tuberculosis. Am Rev Respir Dis. 1982;125(5):559–562. doi:10.1164/arrd.1982.125.5.559.
- Golub JE, Bur S, Cronin WA, et al. Delayed tuberculosis diagnosis and tuberculosis transmission. Int J Tuberc Lung Dis. 2006;10(1):24–30.
- Bumbacea D, Arend SM, Eyuboglu F, et al. The risk of tuberculosis in transplant candidates and recipients: a TBNET consensus statement. Eur Respir J. 2012;40(4):990–1013. doi:10.1183/09031936.00000712.
- Fang HC, Lee PT, Chen CL, Wu MJ, Chou KJ, Chung HM. Tuberculosis in patients with end-stage renal disease. Int J Tuberc Lung Dis. 2004;8(1):92–97.
- Andrew OT, Schoenfeld PY, Hopewell PC, Humphreys MH. Tuberculosis in patients with end-stage renal disease. Am J Med. 1980;68(1):59–65. doi:10.1016/0002-9343(80)90166-7.
- Iclal I, Bilge BF, Fusun K, Galip G. Pleuropulmonary tuberculosis in patients with end-stage renal disease: findings on chest radiographs. Transplant Proc. 1999;31(3):1719–1720. doi:10.1016/S0041-1345(99)00076-7.
- Toman K. Tuberculosis Case-Finding and Chemotherapy. Geneva, Switzerland: World Health Organization; 1979.
- Yeh JJ, Yu JK, Teng WB, et al. High-resolution CT for identify patients with smear-positive, active pulmonary tuberculosis. Eur J Radiol. 2012;81(1):195–201. doi:10.1016/j.ejrad.2010.09.040.
- Im JG, Itoh H, Shim YS, et al. Pulmonary tuberculosis: CT findings–early active disease and sequential change with antituberculous therapy. Radiology. 1993;186(3):653–660. doi:10.1148/radiology.186.3.8430169.
- Matsuoka S, Uchiyama K, Shima H, et al. Relationship between CT findings of pulmonary tuberculosis and the number of acid-fast bacilli on sputum smears. Clin Imaging. 2004;28(2):119–123. doi:10.1016/S0899-7071(03)00148-7.
- Ors F, Deniz O, Bozlar U, et al. High-resolution CT findings in patients with pulmonary tuberculosis: correlation with the degree of smear positivity. J Thorac Imaging. 2007;22(2):154–159. doi:10.1097/01.rti.0000213590.29472.ce.
- Yeh JJ, Neoh CA, Chen CR, Chou CY, Wu MT. A high resolution computer tomography scoring system to predict culture-positive pulmonary tuberculosis in the emergency department. PLoS One. 2014;9(4):e93847. doi:10.1371/journal.pone.0093847.
- Nakanishi M, Demura Y, Ameshima S, et al. Utility of high-resolution computed tomography for predicting risk of sputum smear-negative pulmonary tuberculosis. Eur J Radiol. 2010;73(3):545–550. doi:10.1016/j.ejrad.2008.12.009.
- Rizzi EB, Schinina V, Cristofaro M, et al. Detection of Pulmonary tuberculosis: comparing MR imaging with HRCT. BMC Infect Dis. 2011;11:243. doi:10.1186/1471-2334-11-243.
- Yu WY, Lu PX, Assadi M, et al. Updates on (18)F-FDG-PET/CT as a clinical tool for tuberculosis evaluation and therapeutic monitoring. Quant Imaging Med Surg. 2019;9(6):1132–1146. doi:10.21037/qims.2019.05.24.
- Geadas C, Acuna-Villaorduna C, Mercier G, et al. FDG-PET/CT activity leads to the diagnosis of unsuspected TB: a retrospective study. BMC Res Notes. 2018;11(1):464. doi:10.1186/s13104-018-3564-6.
- Murphy K, Habib SS, Zaidi SMA, et al. Computer aided detection of tuberculosis on chest radiographs: An evaluation of the CAD4TB v6 system. Sci Rep. 2020;10(1):5492. doi:10.1038/s41598-020-62148-y.
- World Health Organization. Rapid Communication on Systematic Screening for Tuberculosis. Geneva. 2020. Licence: CC BY-NC-SA 3.0 IGO. Web site: https://www.who.int/publications/i/item/rapid-communication-on-the-systematic-screening-for-tuberculosis. Accessed February 26, 2022.
- Khan FA, Majidulla A, Tavaziva G, et al. Chest x-ray analysis with deep learning-based software as a triage test for pulmonary tuberculosis: a prospective study of diagnostic accuracy for culture-confirmed disease. Lancet Digit Health. 2020;2(11):e573–e581. doi:10.1016/S2589-7500(20)30221-1.
- Heffernan C, Rowe BH, Long R. Engaging frontline providers: an important key to eliminating tuberculosis in Canada, and other high-income countries. Can J Public Health. 2021;112(5):872–876. doi:10.17269/s41997-021-00556-x.
- Lewinsohn DM, Leonard MK, LoBue PA, et al. Official American Thoracic Society/Infectious Diseases Society of America/Centers for Disease Control and Prevention Clinical Practice Guidelines: Diagnosis of Tuberculosis in Adults and Children. Clin Infect Dis. 2017;64(2):111–115. doi:10.1093/cid/ciw778.
- Woods GL, Brown-Elliott BA, Conville PS, et al. Susceptibility Testing of Mycobacteria, Nocardiae, and Other Aerobic Actinomycetes. 2nd ed. https://clsi.org/media/1463/m24a2_sample.pdf. Wayne (PA); 2011. Clinical and Laboratory Standards Institute. Accessed February 26, 2022.
- Association of Public Health Labratories. Mycobacterium tuberculosis:Assessing Your Laboratory. 2019.
- Menzies D, Gonzalo G, Khan K, Public Health Agency of Canada. In: ed. Canadian Tuberculosis Standard, chap 6: Treatment of Latent Tuberculosis Infection, 7th ed. Can Respir J. 2013;20(Suppl A).
- Mase SR, Ramsay A, Ng V, et al. Yield of serial sputum specimen examinations in the diagnosis of pulmonary tuberculosis: a systematic review. Int J Tuberc Lung Dis. 2007;11(5):485–495.
- Monkongdee P, McCarthy KD, Cain KP, et al. Yield of acid-fast smear and mycobacterial culture for tuberculosis diagnosis in people with human immunodeficiency virus. Am J Respir Crit Care Med. 2009;180(9):903–908. doi:10.1164/rccm.200905-0692OC.
- Harvell JD, Hadley WK, Ng VL. Increased sensitivity of the BACTEC 460 mycobacterial radiometric broth culture system does not decrease the number of respiratory specimens required for a definitive diagnosis of pulmonary tuberculosis. J Clin Microbiol. 2000;38(10):3608–3611. doi:10.1128/JCM.38.10.3608-3611.2000.
- Davis JL, Cattamanchi A, Cuevas LE, Hopewell PC, Steingart KR. Diagnostic accuracy of same-day microscopy versus standard microscopy for pulmonary tuberculosis: a systematic review and meta-analysis. Lancet Infect Dis. 2013;13(2):147–154. doi:10.1016/S1473-3099(12)70232-3.
- Hepple P, Ford N, McNerney R. Microscopy compared to culture for the diagnosis of tuberculosis in induced sputum samples: a systematic review. Int j Tuberc Lung Dis. 2012;16(5):579–588. doi:10.5588/ijtld.11.0617.
- Menzies D, Khan K. Canadian Tuberculosis Standards, 6th ed. Ottawa: Canadian Lung Association, Public Health Agency of Canada, Tuberculosis Prevention and Control; 2007.
- Anderson C, Inhaber N, Menzies D. Comparison of sputum induction with fiber-optic bronchoscopy in the diagnosis of tuberculosis. Am J Respir Crit Care Med. 1995;152(5):1570–1574. doi:10.1164/ajrccm.152.5.7582296.
- Zar HJ, Tannenbaum E, Apolles P, Roux P, Hanslo D, Hussey G. Sputum induction for the diagnosis of pulmonary tuberculosis in infants and young children in an urban setting in South Africa. Arch Dis Child. 2000;82(4):305–308. doi:10.1136/adc.82.4.305.
- American Thoracic Society. Diagnostic standards and classification of tuberculosis. Am Rev Respir Dis. 1990;142(3):725–735.
- Chawla R, Pant K, Jaggi OP, Chandrashekhar S, Thukral SS. Fibreoptic bronchoscopy in smear-negative pulmonary tuberculosis. Eur Respir J. 1988;1(9):804–806.
- So SY, Lam WK, Yu DY. Rapid diagnosis of suspected pulmonary tuberculosis by fiberoptic bronchoscopy. Tubercle. 1982;63(3):195–200. doi:10.1016/S0041-3879(82)80030-5.
- Wallace JM, Deutsch AL, Harrell JH, Moser KM. Bronchoscopy and transbronchial biopsy in evaluation of patients with suspected active tuberculosis. Am J Med. 1981;70(6):1189–1194. doi:10.1016/0002-9343(81)90826-3.
- Chan HS, Sun AJ, Hoheisel GB. Bronchoscopic aspiration and bronchoalveolar lavage in the diagnosis of sputum smear-negative pulmonary tuberculosis. Lung. 1990;168(1):215–220. doi:10.1007/BF02719695.
- Bahammam A, Choudhri S, Long R. Utility of gastric aspirates in screening for pulmonary tuberculosis in high risk subjects:The Manitoba experience. Can J Infect Dis. 1999;10(1):69–73.
- Walters E, Demers AM, van der Zalm MM, et al. Stool Culture for Diagnosis of Pulmonary Tuberculosis in Children. J Clin Microbiol. 2017;55(12):3355–3365. doi:10.1128/JCM.00801-17.
- Yeager H, Lacy J, Smith LR, LeMaistre CA. Quantitative studies of mycobacterial populations in sputum and saliva. Am Rev Respir Dis. 1967;95(6):998–1004. doi:10.1164/arrd.1967.95.6.998.
- Hobby GL, Holman AP, Iseman MD, Jones JM. Enumeration of tubercle bacilli in sputum of patients with pulmonary tuberculosis. Antimicrob Agents Chemother. 1973;4(2):94–104. doi:10.1128/AAC.4.2.94.
- Behr MA, Warren SA, Salamon H, et al. Transmission of Mycobacterium tuberculosis from patients smear-negative for acid-fast bacilli. Lancet (London, England). 1999;353(9151):444–449. doi:10.1016/S0140-6736(98)03406-0.
- Global Laboratory Initiative WHO Stop TB Partnership. Mycobacteriology laboratory manual. 2014.
- Ling DI, Flores LL, Riley LW, Pai M. Commercial nucleic-acid amplification tests for diagnosis of pulmonary tuberculosis in respiratory specimens: meta-analysis and meta-regression. PLoS One. 2008;3(2):e1536. doi:10.1371/journal.pone.0001536.
- Flores LL, Pai M, Colford JM, Jr., Riley LW. In-house nucleic acid amplification tests for the detection of Mycobacterium tuberculosis in sputum specimens: meta-analysis and meta-regression. BMC Microbiol. 2005;5:55. doi:10.1186/1471-2180-5-55.
- Sarmiento OL, Weigle KA, Alexander J, Weber DJ, Miller WC. Assessment by meta-analysis of PCR for diagnosis of smear-negative pulmonary tuberculosis. J Clin Microbiol. 2003;41(7):3233–3240. doi:10.1128/JCM.41.7.3233-3240.2003.
- Zifodya JS, Kreniske JS, Schiller I, et al. Xpert Ultra versus Xpert MTB/RIF for pulmonary tuberculosis and rifampicin resistance in adults with presumptive pulmonary tuberculosis. Cochrane Database Syst Rev. 2021;2:CD009593. doi:10.1002/14651858.CD009593.pub5.
- World Health Organization. WHO meeting report of a technical expert consultation: non-inferiority analysis of Xpert MTB/RIF Ultra compared to Xpert MTB/RIF. 2017.
- Nathavitharana RR, Cudahy PG, Schumacher SG, Steingart KR, Pai M, Denkinger CM. Accuracy of line probe assays for the diagnosis of pulmonary and multidrug-resistant tuberculosis: a systematic review and meta-analysis. Eur Respir J. 2017;49(1):1601075. doi:10.1183/13993003.01075-2016.
- Steingart KR, Flores LL, Dendukuri N, et al. Commercial serological tests for the diagnosis of active pulmonary and extrapulmonary tuberculosis: an updated systematic review and meta-analysis. PLoS Med. 2011;8(8):e1001062. doi:10.1371/journal.pmed.1001062.
- Dowdy DW, Steingart KR, Pai M. Serological testing versus other strategies for diagnosis of active tuberculosis in India: a cost-effectiveness analysis. PLoS Med. 2011;8(8):e1001074. doi:10.1371/journal.pmed.1001074.
- Morris K. WHO recommends against inaccurate tuberculosis tests. Lancet. 2011;377(9760):113–114. doi:10.1016/S0140-6736(11)60005-6.
- World Health Organization. WHO Operational Handbook on Tuberculosis. Module 3: diagnosis—Rapid Diagnostics for Tuberculosis Detection 2021 Update. https://www.who.int/publications/i/item/9789240030589. 2021. Licence: CC BY-NC-SA 3.0 IGO. Accessed February 26, 2022.
- Aggarwal AN, Agarwal R, Sehgal IS, Dhooria S. Adenosine deaminase for diagnosis of tuberculous pleural effusion: A systematic review and meta-analysis. PLoS One. 2019;14(3):e0213728. doi:10.1371/journal.pone.0213728.
- Bulterys MA, Wagner B, Redard-Jacot M, et al. Point-Of-Care Urine LAM Tests for Tuberculosis Diagnosis: A Status Update. J Clin Med. 2019;9(1):111.
- Forbes BA, Hall GS, Miller MB, et al. Practical Guidance for Clinical Microbiology Laboratories: Mycobacteria. Clin Microbiol Rev. 2018;31(2):e00038-17. doi:10.1128/CMR.00038-17.
- Government of Canada. Human Pathogens and Toxins Regulations. 2015.
- Government of Canada. Canadian Biosafety Standard (CBS), 2nd ed. 2015. https://www.canada.ca/en/public-health/services/canadian-biosafety-standards-guidelines/second-edition.html. Accessed on February 26, 2022.
- Public Health Agency of Canada. Biosafety Directive for Mycobacterium tuberculosis Complex (MTBC). 2017.
- Transport Canada. Transportation of dangerous goods in Canada. AEAD 17002250 Web site. https://tc.canada.ca/en/dangerous-goods/transportation-dangerous-goods-canada. Published 2021. Accessed February 26, 2022.
- Steingart KR, Henry M, Ng V, et al. Fluorescence versus conventional sputum smear microscopy for tuberculosis: a systematic review. Lancet Infect Dis. 2006;6(9):570–581. doi:10.1016/S1473-3099(06)70578-3.
- Steingart KR, Ramsay A, Pai M. Optimizing sputum smear microscopy for the diagnosis of pulmonary tuberculosis. Expert Rev anti Infect Ther. 2007;5(3):327–331. doi:10.1586/14787210.5.3.327.
- Steingart KR, Ng V, Henry M, et al. Sputum processing methods to improve the sensitivity of smear microscopy for tuberculosis: a systematic review. Lancet Infect Dis. 2006;6(10):664–674. doi:10.1016/S1473-3099(06)70602-8.
- Salfinger M, Pfyffer GE. The new diagnostic mycobacteriology laboratory. Eur J Clin Microbiol Infect Dis. 1994;13(11):961–979. doi:10.1007/BF02111498.
- Clinical and Laboratory Standards Institute. Laboratory Detection and Identification of Mycobacteria. Clsi Guideline M48 2018. M48.
- Association of Public Health Laboratories. AFB Smear Microscopy. https://www.aphl.org/programs/infectious_disease/tuberculosis/TBCore/TB_AFB_Smear_Microscopy_TrainerNotes.pdf. Accessed February 26, 2022.
- Health Canada. Medical Devices Active Licence Listing. https://health-products.canada.ca/mdall-limh/prepareSearch-preparerRecherche.do?type=archived_archive. Published 2012. Updated 2021. Accessed.
- Steingart KR, Sohn H, Schiller I, et al. Xpert(R) MTB/RIF assay for pulmonary tuberculosis and rifampicin resistance in adults. Cochrane Database Syst Rev. 2014;2014(1):CD009593.
- Boehme CC, Nicol MP, Nabeta P, et al. Feasibility, diagnostic accuracy, and effectiveness of decentralised use of the Xpert MTB/RIF test for diagnosis of tuberculosis and multidrug resistance: a multicentre implementation study. Lancet. 2011;377(9776):1495–1505. doi:10.1016/S0140-6736(11)60438-8.
- Tortoli E, Cichero P, Piersimoni C, Simonetti MT, Gesu G, Nista D. Use of BACTEC MGIT 960 for recovery of mycobacteria from clinical specimens: multicenter study. J Clin Microbiol. 1999;37(11):3578–3582. doi:10.1128/JCM.37.11.3578-3582.1999.
- Clinical and Laboratory Standards Institute. Molecular Diagnostic Methods for Infectious Diseases. 3rd Edition. 2015. https://clsi.org/standards/products/molecular-diagnostics/documents/mm03/. Accessed August 8, 2021.
- Clinical and Laboratory Standards Institute. MM09-A2: Nucleic Acid Sequencing Methods in Diagnostic Labratory Medicine; Approved Guideline, 2nd ed.; 2004. https://clsi.org/standards/products/molecular-diagnostics/documents/mm09/. Accessed August 8, 2021.
- Al Zahrani K, Al Jahdali H, Poirier L, Rene P, Menzies D. Yield of smear, culture and amplification tests from repeated sputum induction for the diagnosis of pulmonary tuberculosis. Int J Tuberc Lung Dis. 2001;5(9):855–860.
- Heifets LB, Desmond EP. Clinical Mycobacteriology (Tuberculosis) Laboratory: Services and Methods. In: Cole S, Eisenach K, McMurray D, Jacobs W, eds. Tuberculosis and the Tubercle Bacillus. Washington DC: ASM press; 2005. 49–69.
- Burman WJ, Stone BL, Reves RR, et al. The incidence of false-positive cultures for Mycobacterium tuberculosis. Am J Respir Crit Care Med. 1997;155(1):321–326. doi:10.1164/ajrccm.155.1.9001331.
- Ruddy M, McHugh TD, Dale JW, et al. Estimation of the rate of unrecognized cross-contamination with mycobacterium tuberculosis in London microbiology laboratories. J Clin Microbiol. 2002;40(11):4100–4104. doi:10.1128/JCM.40.11.4100-4104.2002.
- Small PM, McClenny NB, Singh SP, Schoolnik GK, Tompkins LS, Mickelsen PA. Molecular strain typing of Mycobacterium tuberculosis to confirm cross-contamination in the mycobacteriology laboratory and modification of procedures to minimize occurrence of false-positive cultures. J Clin Microbiol. 1993;31(7):1677–1682. doi:10.1128/jcm.31.7.1677-1682.1993.
- Diagnostic Standards and Classification of Tuberculosis in Adults and Children. This official statement of the American Thoracic Society and the Centers for Disease Control and Prevention was adopted by the ATS Board of Directors, July 1999. This statement was endorsed by the Council of the Infectious Disease Society of America, September 1999. Am J Respir Crit Care Med. 2000;161(4 Pt 1):1376–1395.
- Clinical and Laboratory Standards Institute. Susceptibility Testing of Mycobacteria, Nocardia Spp., and Other Aerobic Actinomycetes. 3rd ed.; 2018. https://clsi.org/media/2620/m24ed3_sample.pdf. Accessed February 26, 2022.
- Danchuk SN, McIntosh F, Jamieson FB, May K, Behr MA. Bacillus Calmette-Guerin strains with defined resistance mutations: a new tool for tuberculosis laboratory quality control. Clin Microbiol Infect. 2020;26(3):384 e385–384 e388.
- Penn-Nicholson A, Georghiou SB, Ciobanu N, et al. Detection of isoniazid, fluoroquinolone, ethionamide, amikacin, kanamycin, and capreomycin resistance by the Xpert MTB/XDR assay: a cross-sectional multicentre diagnostic accuracy study. Lancet Infect Dis. 2021; 22(2):242–249.
- Ling DI, Zwerling AA, Pai M. GenoType MTBDR assays for the diagnosis of multidrug-resistant tuberculosis: a meta-analysis. Eur Respir J. 2008;32(5):1165–1174. doi:10.1183/09031936.00061808.
- Meehan CJ, Goig GA, Kohl TA, et al. Whole genome sequencing of Mycobacterium tuberculosis: current standards and open issues. Nat Rev Microbiol. 2019;17(9):533–545. doi:10.1038/s41579-019-0214-5.
- CRyPTIC Consortium and the 100 GP. Prediction of susceptibility to first-line tuberculosis drugs by DNA sequencing. New Eng J Med. 2018;379(15):1403–1415.
- Jajou R, van der Laan T, de Zwaan R, et al. WGS more accurately predicts susceptibility of Mycobacterium tuberculosis to first-line drugs than phenotypic testing. J Antimicrob Chemother. 2019;74(9):2605–2616. doi:10.1093/jac/dkz215.
- Shea J, Halse TA, Lapierre P, et al. Comprehensive Whole-Genome Sequencing and Reporting of Drug Resistance Profiles on Clinical Cases of Mycobacterium tuberculosis in New York State. J Clin Microbiol. 2017;55(6):1871–1882. doi:10.1128/JCM.00298-17.
- Brown AC, Bryant JM, Einer-Jensen K, et al. Rapid Whole-Genome Sequencing of Mycobacterium tuberculosis Isolates Directly from Clinical Samples. J Clin Microbiol. 2015;53(7):2230–2237. doi:10.1128/JCM.00486-15.
- Votintseva AA, Bradley P, Pankhurst L, et al. Same-Day Diagnostic and Surveillance Data for Tuberculosis via Whole-Genome Sequencing of Direct Respiratory Samples. J Clin Microbiol. 2017;55(5):1285–1298. doi:10.1128/JCM.02483-16.
- Doberne D, Gaur RL, Banaei N. Preanalytical delay reduces sensitivity of QuantiFERON-TB gold in-tube assay for detection of latent tuberculosis infection. J Clin Microbiol. 2011;49(8):3061–3064. doi:10.1128/JCM.01136-11.
- Herrera V, Yeh E, Murphy K, Parsonnet J, Banaei N. Immediate incubation reduces indeterminate results for QuantiFERON-TB Gold in-tube assay. J Clin Microbiol. 2010;48(8):2672–2676. doi:10.1128/JCM.00482-10.
- Oxford Immunotec. T-SPOT. TB package insert PI- TB8-IVD- UK-V4. 2012.
- Oxford Immunotec. T-SPOT. TB training guide, TH- TB- UK-V3 300407. http://www.oxfordimmunotec.com/north-america/wp-content/uploads/sites/2/TG-TB-US-V5.pdf. Published 2012. Accessed February 26, 2022.
- Oxford Immunotec. TB T-Cell Xtend package insert, PI-TT.610- US-V4 https://www.tspot.com/resources/resource/t-cell-xtend-package-insert/. Published 2012. Accessed February 26, 2021.
Appendix 1.
Tuberculosis and mycobacteriology laboratory standards: Services and policies
A.1. Introduction
The diagnosis of tuberculosis (TB) is a collaborative process involving physicians and other health care providers, including clinical and reference laboratories. Before offering mycobacteriology services, each laboratory should assess the level of testing required at that site and the capacity and capability to provide these tests, in a safe and reliable manner.Citation26,Citation61 A complete questionnaire for the assessment of a laboratory’s capacity for handling Mycobacterium tuberculosis complex (MTBC) organisms can be found in the publication Mycobacterium tuberculosis: Assessing Your Laboratory, 2019 edition, produced by the Association of Public Health Laboratories.Citation28 This appendix addresses specific standards for a Canadian mycobacteriology laboratory.
A.2. Laboratory Requirements
A.2.1. Biosafety requirements
Microbiologic diagnosis of TB is obtained by the detection of MTBC organisms in clinical samples. The diagnostic approach varies from province to province and can include a combination of front-line labs in remote settings, more specialized hospital-based labs and provincial reference labs. Laboratories that perform culture amplification of human pathogens such as Mycobacterium tuberculosis (M. tuberculosis) must comply with the Human Pathogens and Toxins ActCitation62 and the corresponding operational and physical biosafety requirements outlined in the Government of Canada’s Canadian Biosafety Standard (CBS) Guideline, Second Edition.Citation63 In these guidelines, M. tuberculosis is classified as a Risk Group 3 pathogen, for which biosafety Containment Level (CL) 3 is required for propagative activities. However, certain diagnostic activities can be conducted safely at CL2, as specified in the new MTBC Biosafety Directive.Citation64 This directive is to be used in conjunction with the CBS Guideline to ensure safety of laboratory personnel conducting M. tuberculosis diagnostic activities
A.2.2. Receiving and transporting specimens
Most specimens submitted for mycobacterial culture originate from the respiratory tract, but tissue, sterile body fluids, urine and gastric aspirates are also commonly submitted (see Chapter 3: Diagnosis of Tuberculosis Disease and Drug-resistant Tuberculosis and Chapter 7: Extra-pulmonary Tuberculosis). If a laboratory does not have processing facilities, specimens should be referred to a laboratory that does. Processing should be done within 24 hours of specimen collection to avoid overgrowth by other microorganisms or deterioration of the sample.Citation28 Specimens should be kept refrigerated at 4 °C (except blood culture and cerebrospinal fluid specimens) if not transported immediately.
All types of clinical specimens are potentially contagious and therefore should be handled with the same procedures. Laboratories are required to adhere to the Transportation of Dangerous Goods Act (Canada) and the International Air Transport Association’s Dangerous Goods Regulations (for transport by air) when transporting clinical specimens or cultures to another facility. The accepting facility is required to accept and process the incoming specimens according to the relevant acts and regulations. The most current information on Transport of Dangerous Goods is available through the Government of Canada website.Citation65
A.2.3. Quality assurance and proficiency testing
All laboratories should be accredited by a recognized national/international accrediting organization and should participate in internal and external quality assurance/quality control activities in conjunction with a reference laboratory. These programs will assess the reproducibility and the inter-laboratory variability of the methods used and adherence to standardized testing procedures. For peripheral labs performing nucleic acid amplification tests (NAAT) in remote Canadian settings, quality assurance can be achieved under the oversight of an accredited clinical mycobacteriology laboratory, to ensure expected results with control samples and to cross-validate NAAT results with other results obtained from the same specimens.
A.3. Detecting and identifying mycobacterium tuberculosis complex species
A.3.1. Digestion, decontamination and concentration of specimens
Upon receipt in the laboratory, respiratory samples submitted for TB diagnosis are decontaminated and concentrated. Decontamination ensures that other microorganisms present in samples, such as respiratory bacterial flora, do not outgrow the slow-growing TB in solid and liquid media, rendering the test uninterpretable and hence limiting the sensitivity of culture. Concentration separates mucus and debris from mycobacterial cells, which increases the sensitivity of smear microscopy. Decontamination and concentration processes also improve the analytical performance of molecular assays performed on primary clinical samples by increasing the available TB molecular template and eliminating potential PCR inhibitor substances. To verify the success of the decontamination process, monitoring of contamination rates is recommended.
Primary sample decontamination and concentration are processes which are not automated. They require laboratory personnel time and need to be quality controlled. Notably, sterile samples, such as cerebrospinal fluid and tissue biopsies, do not need to be subjected to decontamination nor concentration procedures and can be processed immediately for microscopy, NAAT and/or culture.
A.3.2. Acid-Fast smear and microscopy
The early and rapid diagnosis of TB still relies on the traditional acid-fast bacilli (AFB) smear. Overall, smears have a reported sensitivity of 20-80%, depending on many factors, including the type of specimen, stain used and the experience of the technologist.Citation66–69 In addition, fluorescence microscopy can be performed by conventional mercury vapor fluorescence microscopes or newer, light-emitting diode microscopes, which have many practical advantages and have been endorsed by the World Health Organization (WHO). The sensitivity of all staining methods is inferior to that of culture. The threshold of detection of AFB in concentrated specimens using a fluorochrome stain is 5,000-10,000 bacteria/mL of sputum and is 100,000 bacteria/mL using a carbol fuchsin stain (eg, Ziehl-Neelsen). Although they are more rapid, “direct smears” without digestion, decontamination and concentration steps are discouraged because of the inherent lack of sensitivity.Citation68 The threshold of detection in unconcentrated smears is 10-fold higher, resulting in much lower sensitivity. Culture is more sensitive than smear and can detect a bacillary load as low as 10 bacteria/mL.
The following guidelines should be observed:
Slides should be individually prepared and fixed to prevent cross-contamination.
Control slides that contain known acid-fast (positive control) and non-acid-fast (negative control, to assess for possibility of contamination) organisms should be run with each batch of smears prepared.
Primary specimen smears should be stained and reviewed using the fluorochrome method. Laboratories should confirm new AFB-positive smears by a second reader. Smears that are questionable should be repeated or can be stained using a carbol fuchsin method for review.
Fluorochrome and carbol fuchsin stain performance should be confirmed with each new lot of reagents by reviewing AFB-positive and AFB-negative control slides prior to reading patient smears.
Smears should be reported following an established grading system, such as that recommended by the WHO,Citation48 Clinical & Laboratory Standards Institute (CLSI)Citation70 or Association of Public Health Laboratories (APHL)Citation71 (see ).
Laboratories should participate in an approved proficiency program that includes acid-fast smears.
The American Thoracic Society, U.S. Centers for Disease Control and Prevention (CDC) and the Canadian Thoracic Society recommend that laboratories not performing a minimum of 15 AFB smears/week should refer specimens to another laboratory or reference laboratory.
A.3.3. Molecular detection of mycobacteria directly from clinical samples
Molecular detection of M. tuberculosis deoxyribonucleic acid (DNA) from clinical samples uses NAATs, of which polymerase chain reaction (PCR) is the most common method. For this reason, clinicians often use the terms NAAT and PCR interchangeably. In addition to commercial assays, there are many protocols for laboratory-developed molecular assays. Unlike standardized, commercial NAATs, in-house NAATs have been historically associated with unreliable results.Citation50 Validation studies should, therefore, be conducted before implementation and the tests used only in accredited laboratories with quality-assurance systems in place.
Previously, performance of NAATs required sophisticated laboratory infrastructure and highly skilled technicians. This changed when partially or fully automated systems such as the Xpert MTB/RIF© test (Cepheid Inc, Sunnyvale, CA) were progressively introduced in Canadian clinical laboratories. Xpert MTB/RIF© test is a cartridge-based, automated, nested, real-time PCR test utilizing the GeneXpert© platform, which can simultaneously detect M. tuberculosis for diagnosis and mutations conferring resistance to rifampicin, a marker of multidrug-resistant TB (MDR-TB), in less than two hours with minimal hands-on technical time. Even with such automated and cartridge-based molecular assays, the risk of contaminating the test site with amplified DNA demands following stringent standardized laboratory processes and implementation of quality-control procedures. At the time of writing, the following assays are commercially available and Health Canada approved: Roche (COBAS® Taqman® MTB; real-time-PCR); Becton Dickson (BD ProbeTec®, strand displacement amplification); Gen-Probe (Amplified Mycobacterium tuberculosis Direct, [AMTD], transcription mediated amplification); Hain Lifescience (GenoType® Mycobacteria Direct, PCR); and Cepheid (Xpert MTB/RIF®, automated cartridge-based nested PCR). The COBAS® Taqman® MTB, AMTD, and Xpert MTB/RIF tests are approved for direct testing on sputum specimens. A living registry of assays can be found by searching the Medical Devices Active License Listing online query website.Citation72
A Cochrane systematic review on the accuracy of Xpert MTB/RIF identified 18 published studies.Citation73 The majority were performed in low- and middle-income countries. Although the test was originally presented as a point-of-care assay, in 17 of the 18 studies, Xpert was performed by trained technicians in reference laboratories. In the meta-analyses for M. tuberculosis detection (15 studies, 7,517 participants), pooled median sensitivity and specificity were 88% (83%, 92%) and 98% (97%, 99%) respectively. Xpert could distinguish between M. tuberculosis and non-tuberculosis myobacteria (NTM) in clinical samples with high accuracy. Of 139 specimens with NTM, cross-reactivity was observed in only one specimen. A more recent Cochrane report examined the newer Xpert MTB/RIF Ultra© test (Cepheid Inc, Sunnyvale, CA) and compared it to the Xpert MTB/RIF© test. The operating parameters for the classic test were effectively unchanged in this updated meta-analysis: sensitivity and specificity of 84.7% (78.6 to 89.9) and 98.4% (97.0 to 99.3). In comparison, for the newer test (Ultra), the pooled sensitivity and specificity from seven studies (2,834 participants) against culture were 90.9% (86.2 to 94.7) and 95.6% (93.0 to 97.4). Overall, the available evidence shows high accuracy for TB detection for both tests, with higher sensitivity and lower specificity for the Xpert Ultra.Citation52 The laboratory should therefore be alert with the transition to the Ultra assay for the possibility of observing NAAT-positive, culture-negative samples. Of note, this evidence is mostly from high-burden countries and involves the use of spontaneous sputum samples. Similar data from low-incidence settings and with the use of induced sputum samples are lacking. However retrospective studies demonstrated that in low TB-burden settings where there is very limited TB transmission, the specificity of Ultra is very high (99.3%, 95% CI 96-99).Citation53 Operational data, although limited, also suggest that Xpert MTB/RIF is able to significantly reduce the time to diagnosis and treatment.Citation74
NAATs are currently recommended for use only on respiratory specimens, although upon special request they can be used on other specimens (eg, cerebrospinal fluid) from laboratories that have validated their assays for those sample types. As specified in Chapter 3: Diagnosis of Tuberculosis Disease and Drug-resistant Tuberculosis, NAATs should not be used for monitoring TB treatment response or for infection-control purposes after the start of treatment.
In some cases, results may be "indeterminate" because of inhibitors in the specimen or a very low bacterial load. Appropriate controls should be included when applicable to rule out inhibition by the specimen. Special care should be taken to avoid cross-contamination of NAAT samples. Laboratories should ensure that there is a clean environment and should follow proper molecular-testing practices in the preparation of solutions used in NAAT tests to effectively prevent contamination. There should be a physical separation of the laboratory areas used to prepare molecular reagents, handle the DNA template and conduct post-amplification detection. It is advisable not to conduct molecular assays in the containment level 3 laboratory, where mycobacterial cultures grow, as this increases the opportunities for contamination.
Laboratory-developed PCR methods can be less costly than commercially available methods but require advanced technical skills. Such methods can be used for detection of MTBCCitation75 in specimens not recommended for testing with a commercial kit, such as formalin-fixed tissue blocks or extra-pulmonary samples. The analytical sensitivity of such tests should be reported with the results. Before implementing an MTBC molecular assay (laboratory-developed or commercial), laboratories should consult the CLSI guideline, Molecular Diagnostic Methods for Infectious Diseases, for guidance on the validation and implementation of a new molecular diagnostic test.Citation76 Validation of any new or adapted test methods should be completed to evaluate the performance characteristics and technical competence of the test. All test methods should be verified as being appropriate and adequate before being undertaken. Quality-control and -assurance programs should be established to monitor the performance of NAAT assays and avoid false-positive and false-negative results.
A.3.4. Mycobacterial culture
Culture remains the gold standard for a positive laboratory diagnosis of TB. MTBC typically has a faster growth rate in liquid media than on solid agar. Also, liquid cultures are 15-30% more sensitive than solid cultures.Citation77 Three automated liquid culture systems are approved by Health Canada: Becton-Dickinson (Bactec MGIT [mycobacterial growth indicator tube]); bioMérieux (BacT/ALERT); and Trek Diagnostic Systems Inc. (Myco-ESP culture System II). These are fully automated systems that use either fluorometric or colorimetric detection of mycobacterial growth and permit a higher throughput of specimens for testing. For pulmonary TB, the sensitivity of three sputum cultures exceeds 90%, although six specimens are required to achieve 100% sensitivity. Three sputum cultures are recommended for the diagnosis of a new case, as this represents the best balance between high sensitivity and efficiency.Citation78
At least one liquid medium should be inoculated from each clinical specimen for culturing of AFB and, depending on the sample, labs may also do a solid medium culture. Cultures should be kept for a minimum of six and up to eight weeks for observation of growth. Positive cultures of MTBC should be retained for at least one year, in case additional testing is required.Citation28,Citation79
It is important to remember that occasionally, cultures can be falsely positive for MTBC, primarily because of cross-contamination within the laboratory, although “mix-up” by the submitter has also been documented.Citation80,Citation81 When clinical suspicion is low, a report of a single positive culture, especially with a negative smear and a long detection time, should raise the possibility of a false-positive result. The laboratory reporting this culture should investigate and review all positive cultures initially processed on the same day or within proximity to the culture. If there are other positive cultures that could be the source of cross-contamination, genetic analyses on the isolate alongside other isolates from the same lab processed around the same date should be done.Citation82
A.3.5. Identification of mycobacterial species from culture
Mycobacterial identification based on biochemical and/or physical characteristics is labor-intensive and slow, and may not adequately identify the organism. Rapid identification of a growing culture as MTBC (vs NTM) and further subspeciation of the MTBC members is necessary for clinical and public health purposes.
Rapid culture identification can take different approaches, largely based on either genotypic characteristics or antigen detection/protein spectrometry. Immunochromatographic tests targeting DNA or protein of M. tuberculosis have been shown to be a sensitive and specific way to identify MTBC members. Similarly, MALDI-ToF analysis can be used to conclusively place an organism within either the MTBC or the NTM group. Neither of these approaches will allow sub-speciation of complex members.
Genotypic-based techniques, such as NAAT targeting specific differentiating gene targets (eg, the regions-of-difference), targeted sequencing (eg, of gyrB gene) or whole genome sequencing (WGS) can allow sub-speciation between complex members. Confirmation of a growing organism as M. bovis (or M. bovis Bacille Calmette-Guérin [BCG]) is particularly important, given the intrinsic resistance of these organisms to pyrazinamide and the need to look for an underlying cause of BCG infection (bladder cancer therapy or young infant post-vaccination).
A.3.6. Susceptibility testing for antituberculous drugs
Agar proportion is still considered the gold standard for MTBC drug susceptibility testing (DST).Citation83 However, because of the labor-intensive nature and lengthy incubation time for the assay, the more rapid liquid media detection methods using continuous monitoring systems are now recommended and DST results are often available within 10-14 days from the time of receipt of the culture. The most current CLSI guidelines should be consulted for the following testing parameters.Citation84
Laboratories that perform DST should generally be accredited reference laboratories, to ensure adequate volume of activity to maintain expertise.
For all new M. tuberculosis isolates, susceptibility to first-line antibiotics should be tested. First-line antibiotics are isoniazid (INH), rifampin (RMP), ethambutol (EMB) and pyrazinamide (PZA).
DST of second-line antibiotics should be set up when resistance to first-line anti-tuberculous drugs is detected, regardless of whether the DST on those first-line drugs is repeated.
Second-line drugs for which testing is currently available (at the time of writing) in reference labs in Canada include amikacin, fluoroquinolones (levofloxacin and/or moxifloxacin), rifabutin, ethionamide, p-aminosalicylic acid and linezolid.
Laboratories should test at least one drug from each class; in particular, at least one fluoroquinolone should be tested; the selection of which fluoroquinolone to test should be based on consultation with physicians who manage patients with drug-resistant TB.
Other drugs that are used for the treatment of MDR TB include bedaquiline, clofazimine, cycloserine, delamanid, and imipenem/meropenem. For bedaquiline, delamanid and clofazimine, testing is not available in Canada but the authors urge Canadian reference labs to meet this need, given that the drugs are being used and standards are available. Although cycloserine and imipenem/meropenem are viable treatment options, the CLSI does not recommend testing of cycloserine or imipenem/meropenem.
A.3.7. Molecular prediction of anti-tuberculous drug resistance
The molecular prediction of anti-tuberculous drug resistance in M. tuberculosis has become an important tool in the rapid identification of MDR TB. These molecular methods can decrease the time it takes to detect resistance using phenotypic methods and accelerate the time to adjustment of therapy. Molecular testing for determinants of drug resistance provides presumptive results and the use of these tests does not eliminate the need for conventional DST.
These methods should be validated just as any other method would be, and used only in conjunction with phenotypic susceptibility testing. However, until now, there are no defined positive controls to represent the presence of MDR-strains of M. tuberculosis. One recently developed option is safe BCG (a tuberculosis vaccine) strains marked with known mutations that confer resistance to either INH, RMP, moxifloxacin or bedaquiline.Citation85 BCG is already resistant to PZA.
The methods to predict resistance include laboratory-developed PCR, approved commercial line-probe assays, real-time PCR-based assays, targeted sequencing and WGS. With the exception of WGS, all are limited to specific, predetermined targets in the genome; as a result, resistance-associated mutations and/or insertions/deletions outside these targets can be missed. Regarding targeted methods, two genotypic methods are endorsed by the WHO: 1) line-probe assays (LPAs) and 2) the GeneXpert MTB/RIF test.
LPAs have been developed and evaluated to perform DST from smear-positive sputum samples directly or to perform rapid DST on culture isolates. Two LPA tests are commercially available: the Inno-LiPARif.TB line probe assay (Innogenetics, Belgium) and the GenoType MTBDRplus assay (Hain Lifescience, Germany). The GenoTypeMTBDRplus assay is approved by Health Canada.Citation86 It can be used for testing sputum or for testing an M. tuberculosis culture. A meta-analysis showed that the GenoType MTBDRplus assay has a pooled sensitivity of 98.1% and specificity of 98.7%.Citation87 The accuracy for INH was variable, with lower and inconsistent sensitivity (84.3%) and high specificity (99.5%). LPAs are endorsed by the WHO for rapid diagnosis of INH and RMP resistance from sputum smear-positive samples. However, the use of LPAs does not eliminate the need for phenotypic DST.
As previously described, the Xpert MTB/RIF assay can provide a rapid diagnosis of TB and can also detect rpoB mutations, providing a sensitivity of about 94% and a specificity of 98% for RMP resistance.Citation52 However, these estimates are from high-burden settings. The predictive value for RMP resistance will depend on the prevalence of drug-resistant TB in a given setting. In the aforementioned meta-analysis of the test for diagnosis of TB, an analysis was also performed for RMP resistance detection (11 studies, 2,340 participants). The pooled median sensitivity and specificity were 94% (87%, 97%) and 98% (97%, 99%) respectively for RMP resistance. Although the specificity is high, the prevalence of RMP resistance is low in Canada. The management of patients with an isolated RMP resistance report in an individual with low risk of MDR-TB is discussed in Chapter 8: Drug-Resistant Tuberculosis. This issue likely also applies with the latest generation of the test, the Xpert Ultra, where the pooled sensitivity and specificity for RMP resistance were 94.9% (88.9 to 97.9) and 99.1% (97.7 to 99.8).Citation52
In contrast to targeted approaches, WGS provides information on the entire genome, which can then be used to not only predict resistance, but also predict susceptibility to different antibiotics, based on known genotype-phenotype associations.Citation88
WGS has been demonstrated to have high sensitivity and specificity overall for prediction of phenotypic drug resistance for first-line drugs in particular.Citation89 An analysis of more than 10,000 genomes with associated phenotypic data from 16 countries conducted by the CRyPTIC consortium found sensitivities of WGS-based predictions for INH, RMP, EMB and PZA were 97.5, 97.1, 94.6, and 91.3%, with specificities of 98.8, 99.0, 93.6, and 96.8%, respectively.Citation89 Importantly for a low-resistance setting such as Canada, the modeled negative predictive value of this approach remained above 95% even with resistance prevalence levels reaching 34% (PZA) to 57% (RMP). A sub-analysis was also done, restricted to datasets not enriched for resistance; when capitalizing on the association between INH resistance and resistance to other drugs, pan-susceptibility to first-line drugs was 99.7% concordant with phenotypic DST. Similar findings have subsequently been reported in studies from the NetherlandsCitation90 and New York State.Citation91 Discordant results in these studies were largely attributed to clerical errors/mislabeling, mutations where the minimum inhibitory concentration was close to threshold (eg, often seen with EMB), or inability to reproduce results on Mycobacteria Growth Indicator Tube (MGIT).Citation91
In the Netherlands, a change to WGS for sensitivity prediction was estimated to reduce phenotypic testing for TB by 90%,Citation90 as isolates predicted to be pan-sensitive no longer undergo phenotypic testing.Citation89 To catch the aforementioned discrepancies, as well as novel mutations and/or mutations that have not yet been catalogued; however, it would be good practice for Canadian laboratories considering the transition to WGS-based predictions to continue to test a minimum percent of isolates (eg, 5%) predicted to be pan-susceptible on WGS, using phenotypic DST.
In addition to reducing phenotypic testing demands, another advantage of using WGS for resistance prediction is a potentially reduced turnaround time. This is seen in 2 scenarios. If the WGS result predicts resistance, the laboratory can proceed immediately to both first- and second-line phenotypic DST, without needing to do these tests sequentially. If the WGS results predicts pan-susceptibility to front-line drugs, the result can be emitted immediately, with no need to do phenotypic DST.
Ideally, to reduce turnaround time, WGS would be conducted directly on sputum; however, obtaining sufficient M. tuberculosis genomic DNACitation92,Citation93 and sequencing depth for resistance analysis remains challenging. To enable identification of most low-frequency variants, a target of ∼100x depth of coverage is recommended for positive cultures. In New York State, sequencing is done on early-positive-MGIT culture, where a minimum concentration of 0.2 ng/microL was required to achieve <7% failure on initial WGS; under these conditions, mean depth was 133x with results reported on average 9 days earlier for first-line drugs and 32 days earlier for second-line drugs compared to DST, with a turnaround time of 15 days from first positive and including weekly batching of specimens.
A.4. Interferon-gamma release assays
Interferon-gamma release assays (IGRAs) are tests that have been developed for identifying TB infection (see Chapter Citation4: Diagnosis of Tuberculosis Infection). They detect cell-mediated immune responses to specific antigens found in MTBC organisms (including virulent M. bovis) but absent from M. bovis BCG, and most non-tuberculosis myobacteria (the antigens are exceptionally present in M. kansasii, M. szulgai and M. marinum). Detection of a response to these antigens indicates present or previous infection with M. tuberculosis. There are two assays currently approved for use in Canada, the QuantiFERON-TB Gold In-Tube Plus assay (QFT-Plus) (Cellestis/Qiagen, Carnegie, Australia) and the T-SPOT.TB (T-SPOT) (Oxford Immunotec, Abingdon, UK).
IGRAs use whole blood samples and may be performed by any licensed laboratory in Canada. They do not require specialized TB and mycobacteriology laboratory expertise or a CL3 laboratory facility. The assays do, however, require standard clinical laboratory expertise in specimen collection, transportation, and performing the assay.
Laboratories should ensure that specimen collection and transportation are standardized. This is because pre-analytical steps, such as tube shaking, delay between blood draw and incubation, and the exact duration of incubation, are known to affect the results.Citation94,Citation95 If portable incubators are used, it is important to make sure that such incubators can accurately stabilize the temperature at 37 °C. Strict quality assurance is necessary to detect unusual patterns in results (such as a spike in the number of indeterminate results due to low mitogen response or high negative control responses), and it is important to run both positive and negative controls with each assay. Specific details of each test can be obtained from the manufacturers’ instructions.
A.4.1. Assay performance, quality assurance and results interpretation: Key technical information
A.4.1.1. QFT-Plus
QuantiFERON Gold Plus is the new version of the QuantiFERON Gold TB test. This test has four tubes in total: negative control, TB antigen tube 1 (TB1), TB antigen tube 2 (TB2) and mitogen control. TB1 contains peptides from ESAT-6 and CFP-10, which are designed to elicit an immune response from CD4+ T-helper lymphocytes. TB2 contains an additional set of proprietary peptides to ESAT-6, CFP-10, which together elicit interferon-gamma (IFN-γ) secretion from both CD4+ and CD8+ T lymphocytes.
Blood may be collected directly into the 4 QFT-Plus test tubes or into a single lithium heparin tube, and then transferred to QFT-Plus tubes. Blood specimens collected in lithium-heparin tubes may be stored at 2-8 °C for up to 48 hours prior to transfer to QFT-Plus tubes.
Result reporting and interpretation for QFT-GIT: Refer to manufacturer’s package insert (reproduced as ). The results of the QFT-Plus assay are defined as positive if either or both of the TB antigen tubes (TB1 and/or TB2) are positive.
While the QFT assay positive cutoff is IFN-γ 0.35 IU/mL for either TB1-Nil or TB2-Nil, it is important to provide to clinicians who have requested this test the actual numerical value of the result (quantitative value), as well as the interpretation (positive, negative, indeterminate). It is recommended that IFN-γ values of 0.20-1.00 IU/mL for QFT be interpreted cautiously. High rates of IGRA conversions, reversions and imperfect reproducibility have been reported in the literature with results in this range. Yet results in this range have also been found to be clinically significant in specific higher risk populations. Hence the positive predictive value of results in this range will vary according to clinical risk factors (see Chapter Citation4: Diagnosis of Tuberculosis Infection).
Guidance should be provided for an indeterminate result as per the manufacturer’s instructions:
high Nil (high background interferon production) — does not allow an interpretation to be made
low Mitogen (lack of response to antigen stimulation) — does not allow an interpretation to be made
Unreliable or indeterminate results may be due to:
technical failure, including improper protocol
excessive levels of circulating IFN-γ or the presence of heterophile antibodies
greater than 16 hours between time of blood draw and incubation at 37 °C
storage of blood outside ambient temperature range (17 -25 °C)
insufficient mixing of blood collection tubes
incomplete washing of the ELISA plate.
If an indeterminate result is suspected as a result of technical protocol issues (eg, plate washing), repeat testing.
A.4.1.2. T-Spot
The T-SPOTCitation96–98 assay uses the ELISPOT technique, which involves incubating a defined number of peripheral blood mononuclear cells (PBMCs) with CFP-10 and ESAT-6 antigens. The T cells that have previously been sensitized to M. tuberculosis antigens as a result of infection secrete IFN-γ in response to in vitro stimulation with CFP-10 and ESAT-6.
T-SPOT requires standard blood specimen collection into lithium heparin tubes. After collection, PBMC are separated and enumerated.
T-SPOT requires an inoculum of 1 × 106 viable PBMCs per patient (2.5 × 105 per well, for a total of four wells (nil, ESAT-6 antigen, CFP-10 antigen) and positive control (phytohaemagglutinin, or PHA). PBMCs added to the wells are affixed to a well membrane. Cells responding to the antigens release IFN-γ in the vicinity of their location on the membrane, which is subsequently visualized using anti-IFN-γ antibodies. Results are quantified by counting spots produced where IFN-γ is released. For T-Spot result interpretation, see .
Figure 1. Algorithm for interpretation of T-SPOT®.TB assays.
T-SPOT product insert can be consulted for additional information and details: https://www.tspot.com/wp-content/uploads/2021/04/TB-PI-US-0001-V9.pdf.
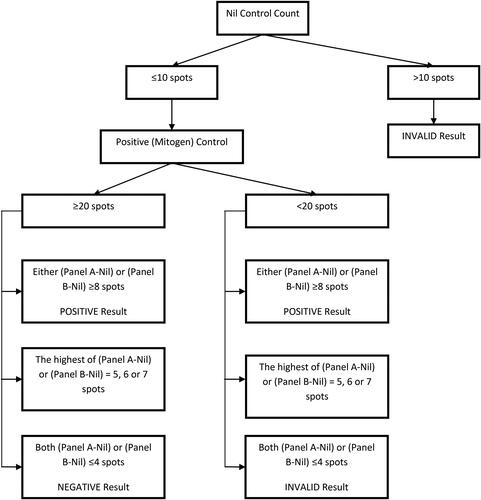
Typical results have few or no spots in the nil control.
A nil control spot count in excess of 10 spots should be considered as “indeterminate.”
If a high numbers of spots or a dark background is observed in the nil control wells, the assay reagents and culture media should be checked for contamination.
Greater than 20 spots should be counted in the positive control.
When the positive control is less than 20 spots, it is considered “indeterminate” (unless panel A or B are “reactive” as per the result reporting, see ); check to ensure that recommended incubation conditions were used. Weak PHA responsiveness may reflect anergy in the patient.
A.5. Reporting criteria and turnaround times
The following are suggested for each laboratory reporting system:
Established turnaround times and reporting parameters for each testing methodology () should be readily available in the laboratory standard operating procedures.
Reports should be dated and indicate the laboratory microbiologist overseeing the analyses.
Reported information should be disseminated by a digital laboratory information system that is accessible to the submitting clinician, via secure telephone, facsimile or e-mail, within 24 hours of test completion.
Preliminary results (e.g., new AFB positive) and critical changes (eg, NTM reclassified as M. tuberculosis) should be communicated immediately to the submitting physician, ideally by phone.
The submitting physician and public health should be notified as soon as suspected or confirmed resistant TB are detected.
Reports on non-standardized testing (such as antimicrobials not recommended by the CLSI for susceptibility testing) should indicate these limitations.
Turnaround times for microscopy and NAATs should be monitored periodically (monthly) to check compliance and evaluated annually.
Table 1. Number of bacteria seen on microscopy and laboratory interpretation.a
Table 2. Summary of suggested turnaround times (refer to individual section for more information).Citation 28
Table 3. QuantiFERON®-TB Gold Plus interpretive criteria (manufacturer’s recommendations).
