Abstract
Mayfly (Ephemeroptera) systematics has considerably changed over the years, but many questions have yet to be answered. The synergistic connection between traditional knowledge and new data sources, producing increasingly complex information, has become a compelling issue for modern taxonomy. Molecular tests and the use of reliable reference sequence libraries may constitute effective complements to the traditional method in guiding recognition of species and giving information about taxonomic incongruences which require further examination. In the present study, we sought to verify the current Italian mayfly nomenclatural system through DNA barcoding and relevant points to reliably manage the available amount of morpho-ecological and molecular data are discussed. We investigated COI (Cytochrome oxidase I) sequence variation in 163 individuals of Italian mayflies, 126 of which were previously assigned to 24 morphologically recognised species, and 37 could be attributed only to generic taxonomic entities (“sp.”, “cf.” or “gr.”). DNA barcoding statistical tests for species delimitation hypotheses based on genetic distances and inferred gene trees were integrated with GenBank searches and surveys of the historical literature to better understand the knowledge acquired on the status and diversity of the investigated taxa. Combined criteria to define three categories of reliability were then assessed. Concurrent data allowing unambiguous identification were attained for only eight species. High intraspecific genetic distances (> 3%) and a lack of reliable reference material or convincing taxonomic information evidenced 29 critical states, deserving further investigation. Solid species names, potential cryptic species and entities about which little is known are pointed out for a future upgrade/reorganisation of the taxonomy of Italian Ephemeroptera.
Introduction
The taxonomy of mayflies has considerably changed in the last two centuries. In Europe, different periods corresponding to improvements in knowledge can be traced through its advancement. In the 19th century, in-depth anatomical and morphological analyses were poorly considered, and a Linnean taxonomic scheme was mainly followed based on tegument colours and general aspect of the adults. The most representative scientist of that period was A. E. Eaton, with numerous studies published in the last quarter of the century (Eaton Citation1883–1888). Later on (in the first half of the 20th century), closer attention was given to the description of more detailed morphological traits (i.e. larval and genital characters: Grandi Citation1960; Landa Citation1969) but taxonomic nomenclature did not follow at the same pace, and proper species identification was often uncertain. In the second half of the 20th century, new progress was made in taxonomy from the morphological analysis based on type specimens, with closer attention to nomenclatural aspects. Müller-Liebenau introduced this process with her work on the genus Baetis (Müller-Liebenau Citation1969). Progressively, many other authors followed this approach in the last part of the century and summarised their results in comprehensive checklists (e.g. Thomas & Belfiore Citation2004; Bauernfeind & Soldán Citation2012).
In the third millennium, the massive utilisation of molecular approaches (e.g. nuclear and mitochondrial DNA sequences, microsatellite variation; see Monaghan & Sartori Citation2009 for a review) has produced a great deal of new data and further progress towards a better comprehension of mayfly systematics, phylogeography and biodiversity. However, in the long run it has also suggested that biological complexity (i.e. Rhithrogena and Baetis genera) was far greater than previously considered in the taxonomic schemes currently in use (Sroka Citation2012; Webb et al. Citation2012; Gattolliat et al. Citation2015; Vuataz et al. Citation2016). The accuracy of the present nomenclature was definitely challenged by several well-aimed studies indicating the presence of crypticism in many species (Williams et al. Citation2006; Bisconti et al. Citation2016; Leys et al. Citation2016; Rutschmann et al. Citation2017). Today, all gathered data strongly point towards the clear, large occurrence of a previously overlooked taxonomic complexity in many families and genera, and prompt new efforts to redefine part of the current systematics.
Taxonomic attribution is fundamental for appropriate divulgation of biological research, to compare different studies and improve socio-economic applications such as habitat and territory preservation (Adamowicz Citation2015). Nonetheless, a reliable and stable taxonomic framework for mayflies has yet to be defined. All faunistic data concerning the Ephemeroptera (i.e. those available today) provide only extremely old, indicative values, both from a nomenclatural point of view and in terms of the definition of specific entities. In fact, the Italian mayfly checklist, mainly based on data acquired in the last few decades of the last century (currently listing ca. 110 species: Buffagni et al. Citation2003), is outdated and approximate. As such, correct evaluation of mayfly biodiversity and future ecological applications (see for instance Gill et al. Citation2016; Morinière et al. Citation2017) is likely to be precluded.
New groundwork for an unambiguous and agreed-upon species detection method should be therefore laid down, putting in order the amount of molecular data that is progressively increasing, alongside the traditional morphological taxonomy. Instrumental and synergistic key points would be: (1) to increase the utilisation of standard molecular data; (2) to determine taxonomic entities based on clearly defined and thoroughly described morphological characters (e.g. Belfiore Citation1994; Haybach Citation1999; Sartori et al. Citation2016), while also giving proper weight to the analysis of intraspecific variation; and (3) to name taxa based on reliable reference material (e.g. type specimens).
The DNA barcoding approach, utilised for species recognition in numerous animal groups (Paz & Crawford Citation2012; Daneliya & Väinölä Citation2014; Evangelista et al. Citation2014), can effectively complement the traditional taxonomy and preliminarily point out possible nomenclatural incongruences or the existence of cryptic differentiation within a species. This method requires a correct taxonomic assignment of the specimens to generate reliable barcode (based on the Cytochorme C Oxydase mitochondrial gene sequence, COI) reference data. Ideally, the assignment of organisms to a species is correct only when the analyses are conducted with the species holotype as reference (see for instance Webb et al. Citation2012). Nevertheless, this is not always possible due to the limited availability of holotypes. Alternative solutions could provide decreasing levels of reliability depending on the specimens used as reference (i.e. paratypes, or other specimens sampled in the type locality). In the case of non-type reference material, the species name can be confidently attributed based on the specimen origin (e.g. nearby localities), the concurrence of unambiguous morphological traits, the known geographical distribution and the absence of similar co-occurring species. This approach could improve the problem of using badly assigned material in generating reference libraries.
In a preliminary work (Cardoni et al. Citation2015), we concluded that DNA barcoding is a powerful approach for taxonomic research in Mediterranean mayflies, ameliorating the available biodiversity inventories. Nevertheless, the practical and unambiguous disclosure of this approach is still hampered by the availability of current faunal lists that are not in line with the emerging molecular complexity, especially with the potential crypticism that has been reported, or is suspected to occur, based on the detected divergence of intraspecific lineages. The general scarcity of Mediterranean Mayfly (with unambiguous species name) COI sequences in the world reference databases that could drive a correct identification is an additional hindering factor.
In this work, the DNA barcoding reference of Italian Ephemeroptera was extended in terms of regions, species and intraspecific samples; whenever possible, specimens with unambiguous identification at type localities or nearby sites were included in the field collection. The accuracy of the nomenclature currently in use was tested against molecular and statistical DNA analyses, and confronted with literature surveys.
The gathered data suggest a reappraisal of the level of nominal reliability. Some useful criteria to challenge and eventually rearrange the nomenclature system currently in use have been proposed, such as combining taxonomical, faunistical and biogeographical issues with DNA barcoding data. This will be an advantage for future projects based on DNA barcoding, and will lay down the experimental groundwork for a sound and durable taxonomy of mayflies.
Material and methods
Sampling and laboratory analyses
One hundred and sixty-three specimens of mayflies at the nymph stage were collected in 18 rivers across Peninsular Italy and the main islands in November 2014, and April and July 2016 (Supplementary Figure S1). The sampling was carried out with a clear strategy, targeted to the collection of selected species in their type locality or nearby areas, and to gradually expand the available reference data set of the Italian mayflies (Cardoni et al. Citation2015; ). A priori species identification was performed based on morphology and current taxonomic literature. DNA barcoding was then used to test the potential congruence of species identification and genetic variability. DNA extractions, Cytochrome Oxidase subunit I region amplification and general sequence analyses were performed as in Cardoni et al. (Citation2015).
Table I. List of specimens investigated in this study, with sampling sites and GenBank accession numbers. HMV: the High Marecchia Valley locality (Marecchia River runs through Emilia Romagna and Tuscany). Map available as .
DNA barcoding statistical tests
The absence of stop codons in the obtained sequences was verified with MEGA 7.0.14 (Kumar et al. Citation2016) and multiple alignments were generated with Clustal X (Thompson et al. Citation1997). Efficacy of taxon discrimination in our data set was assessed by means of (1) sequence similarity, (2) genetic distance and (3) phylogenetic/clustering approaches.
Operational taxonomic units (OTUs) were generated using Mothur (Schloss et al. Citation2009) with different similarity thresholds to compare the obtained OTUs with the species identified based on morphology. The R-package SPIDER (Brown et al. Citation2012) was used to calculate Kimura two-parameter (K2P) genetic distances within and among congeneric species (dis.dna function), and to test the occurrence of the “barcoding gap” (maxInDist and nonConDist function) – that is, the assumption that the maximum intraspecific genetic distance is smaller than the minimum between species. RAxML (Stamatakis Citation2006) was used to generate maximum likelihood dendrograms under the GTRCAT model, on 252 sequences of Italian mayflies and on all COI sequences of the conspecific European mayflies available in GenBank (874 sequences); node support was evaluated with 1000 bootstrap replicates. Tree topology was inspected to check that individuals belonging to the same species were clustered together.
Finally, the Poisson tree process (PTP: Zhang et al. Citation2013) was performed as an additional test to delimitate putative species through Bayesian inference (BI) with 100 000 MCMC (Markov Chain Monte Carlo) generations on 252 COI sequences of Italian Ephemeroptera.
Taxonomic reliability and nomenclatural congruence
All publicly available European conspecific COI sequences were downloaded from GenBank and integrated in our data set (entries and relative information provided as Supplementary Table SI). As a first step, intra- and interspecific K2P genetic distances were calculated for the whole data set (626 sequences). We adopted 0–3% as the indicative range to group individuals into the same species, based on the inter-/intraspecific K2P divergence boundaries generally observed in Ephemeroptera (Ball et al. Citation2005; Gattolliat et al. Citation2012, Citation2015; Kjærstad et al. Citation2012; Webb et al. Citation2012), and considering Hebert et al. (Citation2003). Subsequently, taxonomic literature information was surveyed (e.g. type or paratype locality of the a priori identified species). Considering the origin of the investigated material (type material or type locality), the reported distribution and taxonomic history of the studied entities, we defined three categories of nomenclatural consistency for the groups defined with the K2P distance range:
A. “reliable species names”: individuals resulting from analyses conducted on (in order of decreasing reliability): (1) the type material; (2) material collected at nearby sites, with no concurring syntopic similar species; (3) members of species with a circumscribed distribution and with solid diagnostic characters. These criteria imply no explicit taxonomic problems ();
B. “difficult name attribution, potential occurrence of cryptic species”: individuals to which the criteria stated in A cannot be attributed, indicating a possible non-correspondence with the given species name, despite the presumed cospecificity of the investigated samples with a consistent GenBank reference (K2P < 0.03); intraspecific genetic distance > 0.03, indicating the possible occurrence of cryptic species. These criteria suggest a highly likely necessity for taxonomic revision;
C. “entities with unclear taxonomic status”: the identification cannot be reliably achieved due to the large occurrence of taxonomic problems, with few or no barcoding sequences available for comparison. These criteria imply further investigation is required at morphological, ecological and molecular levels before the current species name is accepted or rejected.
Table II. List of morphospecies grouped by the criteria stated for the nomenclature classes (A, B, C). The column “Genebank sequences for comparison” is related to the sequence numbers in Supplementary Table SI.
Results
Data set description
With the morphological keys currently in use, 24 morphospecies (13 genera, four families) were recognised. The data set included six endemisms [Electrogena hyblaea Belfiore, Citation1994; from Sicily, Rhithrogena sibillina Metzler, Tomka & Zurwerra, Citation1985; from central Italy, Rhithrogena nuragica Belfiore, Citation1987 from Sardinia; and Rhithrogena johannis, Belfiore, Citation1990, Choroterpes borbonica, Belfiore, Citation1988, and Habroleptoides pauliana (Grandi, 1959) from the Appennines and Sicily]. Electrogena hyblaea, R. sibillina, R. johannis and R. nuragica were sampled in their type locality. Current taxonomic knowledge did not allow the identification of 37 specimens at the species level. These specimens were attributed to generic taxonomic entities (labelled with “sp.”, “cf.” or “gr.”: see ).
COI sequences were obtained for all 163 investigated samples. The multiple alignment was unambiguous (no gaps occurred in the whole data set and no stop codons were observed in the translated protein sequences). The final matrix consisted of 639 characters (primer sequences excluded). The reference library of Italian Ephemeroptera (Cardoni et al. Citation2015) was extended with 83 new COI sequences (GenBank accession numbers reported in ).
Evaluation of the molecular data set
The 100% identity threshold generated 127 OTUs; of these, 102 corresponded to single individuals and 25 grouped individuals assigned to the same morphospecies (Supplementary Table SII). The 99–98% identity thresholds reduced the number of OTUs to 50 and 40, respectively. The 97% identity threshold generated 38 OTUs; of these, 35 grouped conspecific samples, matching ca. 95% of the morphospecies included in our data set, and one included Rhithrogena sp. cf. fiorii and Rhithrogena gr. hercynia; two species displayed multiple OTUs [Epeorus assimilis Eaton, 1885 and Baetis fuscatus (Linnaeus, 1761)]. Larger species admixtures were generated with lower thresholds.
The overall intraspecific K2P distance observed in our data set (163 COI sequences) ranged between 0.00 [e.g. Baetis alpinus (Pictet, Citation1843)] and 0.112 (B. fuscatus), whereas the congeneric interspecific values ranged between 0.077 (e.g. R. sibillina – Rhithrogena sp. from Emilia) and 0.293 (Baetis vernus Curtis, Citation1834 – Baetis. sp. cf. melanonyx). The occurrence of barcoding gaps was detected in all entities where more than two specimens were analysed, with a range between K2P = 0.047 (R. gr. hercynia) and K2P = 0.238 (B. alpinus) (Supplementary Table SIII).
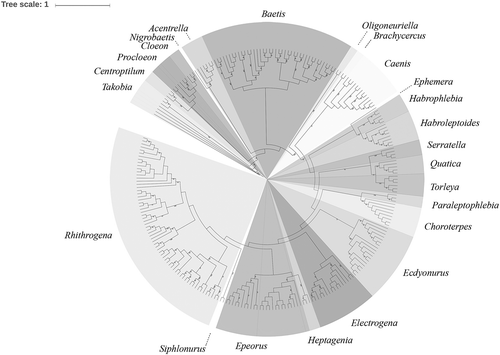
Maximum likelihood inference based on the 252 Italian mayfly COI sequences (corresponding to 64 morphospecies), generated by data from this and the previous work (Cardoni et al. Citation2015), grouped all sequences into clades corresponding to the morphologically identified species (). Moreover, the RaxML phylogram integrated with all European conspecific sequences from GenBank, relative to the Italian Ephemeroptera here and previously investigated species (874 total sequences), confirmed the previous clades and showed a clustering well in line with the acknowledged mayfly taxonomy (Supplementary Figure S2). However, in both dendrograms some “minor clades” with low bootstrap support were produced, corresponding to the morphospecies characterised by high K2P distances (). Finally, the PTP analysis conducted on the total Italian data set (252 sequences) predicted 81 putative species (Supplementary Figure S3).
Data gathering and grouping
Four hundred and sixty two GenBank sequences belonging to the same morphospecies investigated here (163 COI sequences) were used to integrate the K2P genetic distance analysis in an expanded data set (626 total sequences). We found intraspecific K2P distances < 3% in 13 species (95 individuals) and K2P values > 3% in 22 species (529 individuals) (). The remaining two species were represented only by one specimen. Graphic visualisations of the K2P distances are reported in Supplementary Figure S4.
Based on the criteria defined in the Material and methods section, the three categories of nomenclatural consistency defined eight, 19 and 10 species or undefined taxa, respectively, as reported in .
Discussion
A correct and truly effective application of DNA barcoding requires the use of a referenced, official benchmark data set to provide the correct (or the most plausible) species identification through sequence comparison. The construction of a comprehensive sequence reference library is a very long process requiring progressive and complementary approaches (field and lab work, specimen first identification, DNA analyses and data processing, historical and geographical surveys). To overcome the difficulties that often arise due to the lack of traditional taxonomic information, some authors (Gill et al. Citation2016; Morinière et al. Citation2017) introduced the use of BINs (Barcode Index Numbers; Ratnasingham & Hebert Citation2013) for temporary species assignation. However, in many cases multiple BINs were assigned to the same identified species, suggesting, as in the present work (see below), the likely occurrence of potential cryptic species. Indeed, this confirms how far we are from a real representation of mayfly diversity. It stresses also once more the necessity of a more reliable nomenclature.
In our work, we found robust congruence among estabilished taxonomy, morphology, literature information and molecular data in only eight species. Contrasting or incomplete taxonomic information, and high genetic distances suggesting the potential occurrence of cryptic species, were found in 22 cases. These, and seven further cases for which there is scarce information from both taxonomic and molecular literature, coupled with some cases of difficult K2P-level intepretation, highlight the general necessity of additional sampling efforts and investigations to achieve reliable taxonomic assessments.
DNA barcoding for a mayfly benchmark taxonomy
Sequence similarity and the genetic distance analysis were concordant in indicating 3% as a general boundary to separate different species or taxonomic units, assigning 95% of the identified morphospecies to distinct OTUs. The remaining 5% corresponds to four entities (Rhithrogena sp. cf. fiorii, R. gr. hercynia, Epeorus assimilis and Baetis fuscatus) included in the B category (difficult name attribution). The occurrence of barcoding gaps was observed across all congeneric morphospecies (Supplementary Table SIII).
The RAxML reconstruction on the overall 874 sequences was generally congruent with the acknowledged mayfly taxonomy (Supplementary Figure S2). No COI sequences of a single species clustered into a different species’ cluster, and all clades proved to be monospecific, mostly with high bootstrap support (> 70%). The high number of putative species exceeding those morphologically identified with the PTP analysis suggests the hypothesis of cryptic species largely occurring in the investigated data set. Remarkable evidence (BI > 0.987) was provided by E. assimilis, B. fuscatus, B. alpinus, Takobia mutica [Takobia Novikova and Kluge 1987 = Alainites Waltz and McCafferty 1994 (in Waltz et al. Citation1994): Kluge & Novikova Citation2014] and Habrophlebia eldae Jacob & Sartori, Citation1984. Conversely, the separation of the two Electrogena grandiae (Belfiore, Citation1981) samples (BI = 0.390) was probably due to uneven sampling (Zhang et al. Citation2013).
Taxonomic issues
The currently available estimate of the diversity of this order in Italy is not sufficiently accurate.
The obtained molecular results combined with the literature survey showed serious deficiencies in the current taxonomy of mayflies (i.e. underestimated diversity and species names not correctly assigned), for which we believe a critical revision is required. Therefore, we propose some criteria to evaluate the reliability of the current species names that can be used for reconsidering the taxonomic lists. This could be useful also for future practical applications in the light of the newly emerging bio-taxonomic tools (Deiner et al. Citation2016).
Based on the criteria stated for the A category (), the names of eight species of the investigated data set could be reliably ascertained. Of these, five were sampled in their type locality (Rhithrogena adrianae Belfiore, Citation1983, R. nuragica, R. johannis, R. sibillina, E. hyblaea) and one was investigated in our previous study (E. grandiae, Cardoni et al. Citation2015). In addition, for all these species there are no relevant taxonomic problems. Based on the low K2P distance (< 0.028) calculated with the type material (Latium, Belfiore Citation1981; Cardoni et al. Citation2015), the three specimens we sampled in the Marche region and preliminarily attributed to E. grandiae could be unambiguously assigned to this species. Moreover, the low divergence between material from the type locality (Sicily, Belfiore Citation1990) and peninsular sequences (High Marecchia Valley) of R. johannis indicates that all of the samples belong to the same species. The samples of R. sibillina were collected very close to the species type locality (Metzler et al. Citation1985). The high genetic (> 98%) and morphological similarity to Rhithrogena reatina (type locality Latium; Sowa & Belfiore Citation1984) support the synonymy R. reatina Sowa & Belfiore Citation1984 = R. sibillina Metzler et al. Citation1985 syn. nov.
Choroterpes borbonica (type locality R. Mingardo, southern Campania: Belfiore Citation1988) has a relatively restricted distribution (southern Apennines and Sicily), clear diagnostic characters and low intraspecific genetic variability (). Finally, we assigned Epeorus yougoslavicus (Šámal, Citation1935) (type locality: Macedonia, Šámal Citation1935) to category A, due to the low intraspecific genetic distance displayed by the several (14) sequences available for comparison, and considering the marked distinctiveness of the species.
Fifty-nine total COI sequences referred to these eight species are now made available in GenBank to serve as benchmarks for future species attributions. Of these, five species (41 sequences) were previously not represented in GenBank (E. hyblaea, C. borbonica, E. yougoslavicus, R. adrianae and R. johannis).
The largest part of the investigated data set (82 individuals, a priori attributed to 19 morphospecies) was assigned to category B (; Supplementary Figure S4). All these entities displayed taxonomic problems based on the standards proposed for the A category, challenging the correctness of their current names. In accordance, they evidenced marked intraspecific genetic distances (K2P range: 0.050–0.253) with high numbers of conspecific sequences available in GenBank. Strong lineage divergence might suggest the existence of potential cryptic species (Ball et al. Citation2005; Jackson et al. Citation2014), as also supported by the PTP results (Supplementary Figure S3). Exhaustive molecular studies and detailed comparison with reliable reference material are therefore necessary to definitely assess the new nomenclatural status of these entities, described as follows.
The Sicilian specimens identified as Takobia sp. were characterised by six tracheal gills as Takobia albinatii Sartori & Thomas, Citation1989 (type locality from Corsica: Sartori & Thomas Citation1989). Nevertheless, they were clearly separated from the Corsican T. albinatii specimens and Sardinian samples of T. sp. cf. albinatii (K2P ≥ 0.196 and ≥ 0.210, respectively). High divergence was also observed between the Corsican and Sardinian samples (K2P > 0.188), supporting the hypothesis of a species complex, possibly with an evolutionary history similar to that of Baetis gr. rhodani (Bisconti et al. Citation2016).
The high COI divergence (> 0.090) within B. fuscatus could also suggest cryptic species. The B. fuscatus neotype is known in Sweden (Brinck & Müller-Liebenau Citation1965), but no COI sequences from that area are available in GenBank. Therefore, the nomenclatural status of these entities remains unresolved.
The Italian specimens of Baetis sp. cf. lutheri are morphologically different from the types of B. lutheri Müller-Liebenau, Citation1967 (loc. typ. Germany) (C. Belfiore unpublished data), and their COI sequences are highly divergent from those available in Genbank (K2P = 0.173–0.253); therefore, they may represent another new species possibly deserving the species rank. Furthermore, divergence values of 0.074–0.094 separated the specimens assigned to B. sp. cf. melanonyx (High Marecchia Valley) from B. melanonyx (Pictet, Citation1843) collected in Switzerland, at about 50 km from the type locality of this species (Savoy near Samoens, France: Müller-Liebenau Citation1969) (Leys et al. Citation2016).
In B. alpinus we observed two lineages (K2P > 0.052). Based on the sequence comparisons, one lineage has a wide distribution (Sicily, Latium and other European regions: K2P < 0.023) and the other was only found in Central Italy (sequence nos. 74–75, Supplementary Table SI: K2P = 0.015). It is reasonable to hypothesise that the two lineages might represent different species. Further molecular studies on expanded data sets will certainly contribute to understanding the well-known taxonomic complexity of this group, as already pointed out in previous works in which different lineages were found in sympatric coexistence (Leys et al. Citation2016, Citation2017). This is a typical example in which names should be assigned by analysis of types or by conventional, arbitrary choices.
Three samples labelled Electrogena sp. and one Rhithrogena sp. (from Emilia Romagna and HMV: ), very similar to E. grandiae and R. adrianae, respectively, showed high genetic divergences from the type locality material of these two species. Furthermore, Rhithrogena sp. was also highly divergent from another species of the diaphana group (Rhithrogena sp. cf. adrianae from the HMV area: K2P > 0.120), and, in contrast, it showed high affinity with Rhithrogena sp. 31 from the Alps (99% similarity; Vuataz et al. Citation2011). Rhithrogena sp. cf. adrianae from HMV is also highly divergent from R. adrianae collected in the type locality (K2P = 0.145–0.151).
In Sicily and Corsica, the COI divergence recorded in H. eldae might indicate the possible occurrence of two different entities, both differing also from the continental H. eldae (holotype described from Bulgaria: Jacob & Sartori Citation1984) here investigated (see Supplementary Figure S4).
The Italian samples of Baetis buceratus Eaton, 1870, B. vernus and Rhithrogena semicolorata (Curtis, Citation1834) showed low intraspecific divergence but were highly divergent from their European conspecifics. Specimens of B. vernus with high COI divergence were also observed in Germany and assigned to different BINs (13.94% intraspecific divergence by Morinière et al. Citation2017). Since the type localities of these three species are located in the United Kingdom (Curtis Citation1834; Kimmins Citation1960), it would be reasonable to assume that the Italian samples belong to different species. However, no sequences from the United Kingdom are available to clear up affinity/divergence patterns with the continental species, except for R. semicolorata whose only sequence available from United Kingdom (see Vuataz et al. Citation2016) confirms the genetic divergence from Italian specimens. The same conclusion is conceivable for both the single Italian investigated sample assigned to Nigrobaetis digitatus (Bengtsson, 1912) and Torleya major (Klapálek, Citation1905) specimens whose type localities are in Sweden (Müller-Liebenau Citation1969) and Bosnia Herzegovina (Klapálek Citation1905), respectively, highly divergent from all other European members of these species. Furthermore, the highly similar (K2P < 0.005) Sicilian samples of Electrogena lateralis (Curtis, Citation1834), very divergent from French, British and Swiss sequences, could belong to a different biological entity. In Ecdyonurus venosus (Fabricius, 1775), the current unavailability of the type locality hinders the nominal recognition of the three lineages identified (roughly corresponding to Italy, Germany and France; Supplementary Figure S4), separated by medium-low K2P distances (0.030–0.050). Finally, the possible occurrence of crypticism in E. assimilis (intraspecific K2P distance > 0.089), already discussed in Cardoni et al. (Citation2015), and in Baetis pavidus Grandi, 1951 (K2P > 0.109), is supported by the high genetic divergence observed from the European conspecific samples. For the 60 specimens (10 morphospecies) included in category C (; Supplementary Figure S4), the lack of data from the type localities, the taxonomic uncertainties and the very limited sequence availability in GenBank prevent any inferences being made regarding their status. For instance, with regard to Acentrella sinaica (Bogoescu, Citation1931) there are too few reference sequences available (see ; Supplementary Table SI) and all collections are so far from the type localities that no considerations about its status can be made. In addition, naming the Italian entities with seven gills identified within Takobia, divergent from the European ones (K2P > 0.162) but with high affinity with one COI sequence from France (seq. no. 11, Supplementary Table SI), is currently difficult due to the K2P distance bieng hardly interpretable (close to the 3% conventional limit), and due to the type unavailability (Müller-Liebenau Citation1969).
Somewhat unexpected is the presence of Rhithrogena gr. hercynia in Sardinia. It was collected in the R. nuragica type locality (Gennargentu: Belfiore Citation1987). The two taxa are clearly distinguished by morphological characteristics (i.e. Supplementary Figure S5), corroborated by a very high interspecific genetic diversity at the COI locus (K2P > 0.100). Interestingly, the R. gr. hercynia samples showed 99% sequence similarity with R. gratianopolitana Sowa, Degrange & Sartori, 1986 from Germany (Morinière et al. Citation2017). In any case, it is a species new to Sardinia. We also compared another species of R. hercynia group, R. sp. cf. fiorii from Latium, with R. gr. hercynia samples from Sardinia and R. gratianopolitana. The high similarity detected (K2P = 0.027–0.032 and K2P = 0.023–0.030, respectively) requires further data (i.e. nuclear markers and thorough morphological analyses) to clarify the taxonomical status of this second Rhithrogena in Sardinia. The same investigation is necessary for other cases such as Rhithrogena sp. from Emilia Romagna, Habroleptoides sp. and Caenis gr. macrura. Specifically, in the latter group, which also includes Caenis sp. cf. luctuosa (from Sicily), three different Italian entities were found to be highly different from each other (K2P > 0.139), in turn highly separated from European sequences belonging to the same group.
Nineteen COI sequences were deposited in GenBank to increase the current availablity of the entities included in category C. Of these, nine sequences represent three taxa for which no data were previously available on GenBank: Quatica ikonomovi (Puthz, 1971), Habroleptoides pauliana and R. sp. cf. fiorii. In future works, comprehensive surveys are required, especially focusing on the type localities; detailed morpho-ecological descriptions will obviously be essential complements to clearly understand their species status.
In conclusion, the taxonomy of Italian mayflies appears more complicated than what is currently recognised. This can affect the actual evaluation of territorial mayfly biodiversity with implications for the compilation of Red Lists and running water quality assessments. We concur with the opinion that the synergy of disciplines which were once studied separately, as taxonomy and molecular biology, will contribute to elucidate some of the outstanding issues. In this context, type material represents a fundamental reference to drive species recognition, and as such it should be adequately considered in subsequent DNA barcoding campaigns. This integrated approach (cf. Schlick-Steiner et al. Citation2010) may take a long time, but it can provide a stronger and more reliable nomenclature as compared to the traditional methods used so far. This is expected to bring clarity and precision to the classification of nominal species. Thus, by using correct species names, official data reference becomes more stable and univocal even for further studies and future applications based on molecular approaches with innovative techniques (i.e. next-generation sequencing platforms with environmental DNA for biomonitoring assessment). This study has shown that there are taxonomic uncertainties in numerous species, evidenced by the applied molecular and statistical approaches. Given a sampling strategy that was mainly concentrated on the selected species, it will be necessary in further studies to increase the amount of samples and species. It will also be necessary to take advantage of a synergy of multiple data for each taxon, including ecological, morphological, historical and additional genetic descriptors.
Supplemental Material
Download MS Word (14 KB)Supplemental Material
Download MS Word (36.7 KB)Supplemental Material
Download MS Excel (38 KB)Supplemental Material
Download PDF (1.5 MB)Supplemental Material
Download PDF (1.3 MB)Supplemental Material
Download PDF (1.5 MB)Supplemental Material
Download PDF (7.6 MB)Supplemental Material
Download TIFF Image (8.3 MB)Disclosure statement
No potential conflict of interest was reported by the authors.
Supplemental data
Supplemental data for this article can be accessed here.
References
- Adamowicz SJ 2015. International Barcode of Life: Evolution of a global research community. Genome 58:151–162. DOI:10.1139/gen-2015-0094.
- Ball S, Hebert PDN, Burian S, Webb J 2005. Biological identifications of mayflies (Ephemeroptera) using DNA barcodes. Journal of the North American Benthological Society 24:508–524. DOI:10.1899/04-142.1.
- Bauernfeind E, Soldán T 2012. The mayflies of Europe. Ollerup: Apollo Books. p. 781.
- Belfiore C 1981. On the Italian species of the Ecdyonurus lateralis group (Ephemeroptera, Heptageniidae). Aquatic Insects 3:171–178. DOI:10.1080/01650428109361059.
- Belfiore C 1983. Notes on Italian Heptageniidae (Ephemeroptera). Rhithrogena fiorii Grandi, 1953 and R. adrianae sp. n. Aquatic Insects 5:69–76. DOI:10.1080/01650428309361129.
- Belfiore C 1987. Heptageniidae from Corsica and Sardinia. Rhithrogena nuragica n. sp., R. eatoni Esben Petersen 1912, and R. insularis Esben Petersen 1913 (Ephemeroptera). Annales de Limnologie- International Journal of Limnology 23:87–94. DOI:10.1051/limn/1987013.
- Belfiore C 1988. A new species of Leptophlebiidae from Southern Italy: Choroterpes borbonica n. sp. (Ephemeroptera). Fragmenta Entomologica 21:61–65.
- Belfiore C 1990. A new species of Heptageniidae from Sicily: Rhithrogena johannis sp. n. (Ephemeroptera). Fragmenta Entomologica 22:11–17.
- Belfiore C 1994. Taxonomic characters for species identification in the genus Electrogena Zurwerra and Tomka, with a description of Electrogena hyblaea sp.n. from Sicily (Ephemeroptera, Heptageniidae). Aquatic Insects 16:193–199. DOI:10.1080/01650429409361555.
- Bisconti R, Canestrelli D, Tenchini R, Belfiore C, Buffagni A, Nascetti G 2016. Cryptic diversity and multiple origins of the widespread mayfly species group Baetis rhodani (Ephemeroptera: Baetidae) on northwestern Mediterranean islands. Ecology and Evolution 6:7901–7910. DOI:10.1002/ece3.2465.
- Bogoescu C 1931. Une nouvelle espece d’Acentrella Bgtss. (Ephemeroptere) Acentrella sinaica n.sp. Bulletin de la Section Scientifique de l’Academie Roumanie 14:197–202.
- Brinck P, Müller-Liebenau I 1965. On the status of Ephemera fuscata Linnaeus, 1761 (Insecta, Ephemeroptera). Proposed designation of a neotype. Z.N. (S.) 1620. Bulletin of Zoological Nomenclature 22:119–122.
- Brown SDJ, Collins RA, Boyer S, Lefort MC, Malumbres-Olarte J, Vink CJ, Cruickshank RH 2012. Spider: An R package for the analysis of species identity and evolution, with particular reference to DNA barcoding. Molecular Ecology Resources 12:562–565. DOI:10.1111/j.1755-0998.2011.03108.x.
- Buffagni A, Belfiore C, Erba S, Kemp JL, Cazzola M 2003. A review of Ephemeroptera species distribution in Italy: Gains from recent studies and areas for future focus. In: Gaino E, editor. Research update on Ephemeroptera & Plecoptera. Perugia, Italy: Università di Perugia. pp. 279–280.
- Cardoni S, Tenchini R, Ficulle I, Piredda R, Simeone MC, Belfiore C 2015. DNA barcode assessment of Mediterranean mayflies (Ephemeroptera), benchmark data for a regional reference library for rapid biomonitoring of freshwaters. Biochemical Systematics and Ecology 62:36–50. DOI:10.1016/j.bse.2015.07.035.
- Corse E, Meglecz E, Archambaud G, Martin JF 2017. A from-benchtop-to-desktop workflow for validating HTS data and for taxo-nomic identification in diet metabarcoding studies. Molecular EcologyResources 17:146–159.
- Curtis J 1834. Descriptions of some nondescript British species of Mayflies of anglers. London and Edinburgh Philosophical Magazine and Journal of Science 4:120–125.
- Daneliya ME, Väinölä R 2014. Five subspecies of the Dorogostaiskia parasitica complex (Dybowsky) (Crustacea: Amphipoda: Acanthogammaridae), epibionts of sponges in Lake Baikal. Hydrobiologia 739:95–117. DOI:10.1007/s10750-013-1671-x.
- Deiner K, Fronhofer EA, Mächler E, Walser JC, Altermatt F 2016. Environmental DNA reveals that rivers are conveyer belts of biodiversity information. Nature Communications 7:12544. DOI:10.1038/ncomms12544.
- Drotz MK, Savolainen E, Saura A, Ståhls G 2012. The genetic population structure of lotic and lentic mayflies of the Baetis vernus group (Ephemeroptera: Baetidae). The Canadian Entomologist 144:679–690. DOI:10.4039/tce.2012.69.
- Eaton AE 1883–1888. A revisional monograph of recent Ephemeridae or Mayflies. 2nd ser. Transactions of the Linnean Society London (Zoology). pp. 1–352.
- Evangelista DA, Bourne G, Ware JL 2014. Species richness estimates of Blattodea s.s. (Insecta: Dictyoptera) from northern Guyana vary depending upon methods of species delimitation. Systematic Entomology 39:150–158. DOI:10.1111/syen.12043.
- Gattolliat JL, Cavallo E, Vuataz L, Sartori M 2015. DNA barcoding of Corsican mayflies (Ephemeroptera) with implications on biogeography, systematics and biodiversity. Arthropod Systematics Phylogeny 73:3–18.
- Gattolliat JL, Vuataz L, Sartori M 2012. First contribution to the mayflies of Jordan. Zoology in the Middle East 56:91–110. DOI:10.1080/09397140.2012.10648945.
- Gill BA, Kondratieff BC, Casner KL, Encalada AC, Flecker AS, Gannon DG, Ghalambor CK, Guayasamin JM, Poff NL, Simmons MP, Thomas SA, Zamudio KR, Funk WC 2016. Cryptic species diversity reveals biogeographic support for the ‘mountain passes are higher in the tropics’ hypothesis. Proceedings. Biological Sciences Royal Society 283:20160553. DOI:10.1098/rspb.2016.0553.
- Grandi M 1949. Contributi allo studio degli “Efemeroidei” italiani. XIII. Baetis parva sp.n. Baetis principii sp.n. Bollettino dell’Istituto di Entomologia dell’Università di Bologna 17:287–300.
- Grandi M 1960. Ephemeroidea. Fauna d’Italia 3. Bologna: Calderini. p. 328.
- Haybach A 1999. Beitrag zur Larvaltaxonomie der Ecdyonurus-venosus-Gruppe. Deutschland. Lauterbornia 37:113–150.
- Hebert PDN, Cywinska A, Ball SL, DeWaard JR 2003. Biological identifications through DNA barcodes. Proceedings of the Royal Society B: Biological Sciences 270:313–321. DOI:10.1098/rspb.2002.2218.
- Ikonomov P 1961. Eintagsfliegen (Ephemeroptera) Mazedoniens. Fam. Ephemerellidae. Acta Musei Macedonici Scientiarum Naturalium 8:53–74.
- Jackson JK, Battle JM, White BP, Pilgrim EM, Stein ED, Miller PE, Sweeney BW 2014. Cryptic biodiversity in streams: A comparison of macroinvertebrate communities based on morphological and DNA barcode identifications. Freshwater Science 33:312–324. DOI:10.1086/675225.
- Jacob U 1974. Zur Kenntnis zweier Oxycypha-Arten Hermann Burmeisters (Ephemeroptera, Caenidae). Reichenbachia 15:93–97.
- Jacob U, Sartori M 1984. Die europäischen Arten der Gattung Habrophlebia Eaton (Ephemeroptera, Leptophlebiidae). Entomologische Abhandlungen Staatliches Museum für Tierkunde Dresden 48:45–52.
- Kimmins DE 1960. The Ephemeroptera types of species described by A.E. Eaton, R. McLachlan and F. Walker, with particular reference to those in the British Museum (Natural History). Bulletin of the British Museum (Natural History). Entomology 9:269–318.
- Kjærstad G, Webb JM, Ekrem T 2012. A review of the Ephemeroptera of Finnmark – DNA barcodes identify Holarctic relations. Norwegian Journal of Entomology 59:182–195.
- Klapálek F 1905. Ephemeridarum species quatuor novae. Casopis Ceske Spolecnosti Entomologicke 2:75–79.
- Kluge NJ, Novikova EA 2014. Systematics of Indobaetis Müller-Liebenau & Morihara 1982, and related implications for some other Baetidae genera (Ephemeroptera). Zootaxa 3835:209–236. DOI:10.11646/zootaxa.3835.2.3.
- Kumar S, Stecher G, Tamura K 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33:1870–1874. DOI:10.1093/molbev/msw054.
- Landa V 1969. Jepice-Ephemeroptera. Fauna CSSR, Ceskoslovevska. Akademie 18:1–352.
- Leys M, Keller I, Räsänen K, Gattolliat JL, Robinson CT 2016. Distribution and population genetic variation of cryptic species of the Alpine mayfly Baetis alpinus (Ephemeroptera: Baetidae) in the Central Alps. BMC Evolutionary Biology 16:77. DOI:10.1186/s12862-016-0643-y.
- Leys M, Keller I, Robinson CT, Räsänen K 2017. Cryptic lineages of a common alpine mayfly show strong life-history divergence. Molecular Ecology 26:1670–1686. DOI:10.1111/mec.2017.26.issue-6.
- Lucentini L, Rebora M, Puletti ME, Gigliarelli L, Fontaneto D, Gaino E, Panara F 2011. Geographical and seasonal evidence of cryptic diversity in the Baetis rhodani complex (Ephemeroptera, Baetidae) revealed by means of DNA taxonomy. Hydrobiologia 673:215–228. DOI:10.1007/s10750-011-0778-1.
- Metzler M, Tomka I, Zurwerra A 1985. Beitrag zur Kenntnis der europäischen Rhithrogena-Arten: Nivata, hybrida und hercynia, sowie Beschreibung von zwei neue Arten. Folia Entomologica Hungarica 46:117–135.
- Monaghan MT, Sartori M 2009. Genetic contributions to the study of taxonomy, ecology, and evolution of mayflies (Ephemeroptera): Review and future perspectives. Aquatic Insects 31:19–39. DOI:10.1080/01650420902734145.
- Morinière J, Hendrich L, Balke M, Beermann AJ, König T, Hess M, Koch S, Müller R, Leese F, Hebert PDN, Hausmann A, Schubart CD, Haszprunar G 2017. A DNA barcode library for Germany′ s mayflies, stoneflies and caddisflies (Ephemeroptera, Plecoptera & Trichoptera). Molecular Ecology Resources 00:1–15.
- Müller-Liebenau I 1967. Zur Frage der Baetis ” venustulus”, zugleich Beschreibung der neuen Art Baetis lutheri n.sp. (Insecta, Ephemeroptera). Gewässer und Abwässer 44/45:48–64.
- Müller-Liebenau I 1969. Revision der europäischen Arten Gattung Baetis Leach, 1815 (Insecta, Ephemeroptera). Gewässer und Abwässer 48/49:1–214.
- Múrria C, Bonada N, Vellend M, Zamora-Muñoz C, Alba-Tercedor J, Sainz-Cantero CE, Garrido J, Acosta R, El Alami M, Barquín J, Derka T, Álvarez-Cabria M, Sáinz-Bariain M, Filipe AF, Vogler AP 2017. Local environment rather than past climate determines community composition of mountain stream macroinvertebrates across Europe. Molecular Ecology 26:6085–6099. DOI:10.1111/mec.2017.26.issue-21.
- Paz A, Crawford AJ 2012. Molecular-based rapid inventories of sympatric diversity: A comparison of DNA barcode clustering methods applied to geography-based vs clade-based sampling of amphibians. Journal of Biosciences 37:887–896. DOI:10.1007/s12038-012-9255-x.
- Pictet FJ 1843. Histoire naturelle des Insectes Névroptères. Famille des Ephémérines. Genève: Chez J. Kessmann et Ab. Cherbuliez. p. 300.
- Ratnasingham S, Hebert PDN 2013. A DNA-based registry for all animal species: The Barcode Index Number (BIN) system. PLoS ONE 8:e66213. DOI:10.1371/journal.pone.0066213.
- Rutschmann S, Detering H, Simond S, Funkf DH, Gattolliat JL, Hughesi SJ, Raposeiro PM, DeSalle R, Sartori M, Monaghan MT 2017. Colonization and diversification of aquatic insects on three Macaronesian archipelagos using 59 nuclear loci derived from a draft genome. Molecular Phylogenetics and Evolution 107:27–38. DOI:10.1016/j.ympev.2016.10.007.
- Sàmal J 1935. Nova evropska jepice Iron. Vestnik Ceskoslovenské Spolecnosti Zoologické 2:87–91.
- Sartori M, Kubiak M, Michalik P 2016. Deciphering genital anatomy of rare, delicate and precious specimens: First study of two type specimens of mayflies using micro-computed X-ray tomography (Ephemeroptera; Heptageniidae). Zoosymposia 11:28–32. DOI:10.11646/zoosymposia.11.1.
- Sartori M, Thomas AGB 1989. Contribution à la connaissance du genre Baetis Leach, 1815 en Corse (Ephemeroptera; Baetidae). B. albinatii nov.sp. du groupe muticus (L.). Annales de Limnologie- International Journal of Limnology 25:131–137. DOI:10.1051/limn/1989013.
- Schlick-Steiner BC, Steiner FM, Seifert B, Stauffer C, Christian E, Crozier RH 2010. Integrative taxonomy: A mul- tisource approach to exploring biodiversity. Annual Review of Entomology 55:421–438. DOI:10.1146/annurev-ento-112408-085432.
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Wan Horn DJ, Weber CF 2009. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Applied Environmental Microbiology 75:7537–7541. DOI:10.1128/AEM.01541-09.
- Sowa R, Belfiore C 1984. Rhithrogena reatina sp. n., a new species of the hybrida group from Central Italy (Ephemeroptera, Heptageniidae). Bollettino dell’Associazione Romana di Entomologia 38:5–8.
- Sroka P 2012. Systematics and phylogeny of the West Palaearctic representatives of subfamily Baetinae (Insecta: Ephemeroptera): Combined analysis of mitochondrial DNA sequences and morphology. Aquatic Insects 34:23–53. DOI:10.1080/01650424.2012.718081.
- Ståhls G, Savolainen E 2008. MtDNA COI barcodes reveal cryptic diversity in the Baetis vernus group (Ephemeroptera Baetidae). Molecular Phylogenetics and Evolution 46:82–87. DOI:10.1016/j.ympev.2007.09.009.
- Stamatakis A 2006. Raxml-vi-hpc: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. DOI:10.1093/bioinformatics/btl446.
- Thomas A, Belfiore C 2004. Fauna Europaea: Ephemeroptera. Fauna Europaea version 1.1. Available: http://www.fauna-eu.org/. Accessed Apr 2017 6.
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG 1997. The ClustalX windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 25:4876–4882. DOI:10.1093/nar/25.24.4876.
- Vuataz L, Rutschmann S, Monaghan MT, Sartori M 2016. Molecular phylogeny and timing of diversification in Alpine Rhithrogena (Ephemeroptera: Heptageniidae). BMC Evolutionary Biology 16:194. DOI:10.1186/s12862-016-0758-1.
- Vuataz L, Sartori M, Wagner A, Monaghan MT 2011. Toward a DNA taxonomy of Alpine Rhithrogena (Ephemeroptera: Heptageniidae) using a mixed Yule-coalescent analysis of mitochondrial and nuclear DNA. PloS ONE 6:e19728. DOI:10.1371/journal.pone.0019728.
- Wagner A, Vuataz L, Sartori M 2017. Electrogena brulini sp. nov. and E. vipavensis Zurwerra & Tomka, 1986 syn. nov. revealed by integrative taxonomy of E. gridellii (Grandi, 1953) (Ephemeroptera: Heptageniidae). Zootaxa 4362:359–384. DOI:10.11646/zootaxa.4362.3.3.
- Waltz RD, McCafferty WP, Thomas A 1994. Systematics of Alainites n.gen., Diphetor, Indobaetis, Nigrobaetis n.stat., and Takobia n.stat. (Ephemeroptera, Baetidae). Bulletin de la Société d’Histoire Naturelle de Toulouse 130:33–36.
- Webb JM, Jacobus LM, Funk DH, Zhou X, Kondratieff B, Geraci CJ, DeWalt RE, Baird DJ, Richard B, Phillips I, Hebert PDN 2012. A DNA barcode library for North American Ephemeroptera: Progress and prospects. PLoS ONE 7:e38063. DOI:10.1371/journal.pone.0038063.
- Williams HC, Ormerod SJ, Bruford MW 2006. Molecular systematics and phylogeography of the cryptic species complex Baetis rhodani (Ephemeroptera, Baetidae). Molecular Phylogenetics and Evolution 40:370–382. DOI:10.1016/j.ympev.2006.03.004.
- Yanai Z, Sartori M, Dor R, Dorchin N 2017. Molecular phylogeny and morphological analysis resolve a long‐standing controversy over generic concepts in Ecdyonurinae Mayflies (Ephemeroptera: Heptageniidae). Systematic Entomology 42:182–193. DOI:10.1111/syen.2017.42.issue-1.
- Zhang J, Kapli P, Pavlidis P, Stamatakis A 2013. A general species delimitation method with applications to phylogenetic placements. Bioinformatics 29:2869–2876. DOI:10.1093/bioinformatics/btt499.