Abstract
The nucleotype theory has been advanced on the basis of studies regarding genome size and composition in various plant and animal species, i.e. the influence that genome can have on the phenotype independently of the informational content of DNA. It has also been noted that during evolution various interactions between different environmental factors and genome structural and functional parameters would have occurred. In this review, changes in genome size, transposon content, and base composition occurred during the evolution of chordates were examined. Many environmental stresses, such as temperature, can act on transposons and through these on genome size. Temperature is also one of the most important elements of natural selection able to interact both with base composition and genome size. It has been evidenced that temperature exerts a direct influence on base composition and its increase would have led to an higher content of genome GC-rich components during the evolution of chordates, in particular in endotherms. Temperature would have controlled the rate of biosynthesis in G1 phase and consequently the cell cycle duration which in turn would have interacted with genome size. The combined action of temperature, base composition, and genome size would also have been very important in controlling the metabolic rate. Finally, another important aspect of the nucleotypic effect is the influence that genome size and cell cycle duration, in correlation with environmental temperature, would have exert on embryo and larval development, very important for environmental adaptation. In conclusion, studies here reviewed to confirm the existence in chordates of a mutual influence between environment and genome non-coding components that would have played an important role in the evolution of these animals especially in environmental adaptation processes.
Introduction
The idea that DNA plays a quantitative role, independently from its sequences, besides that one of protein coding, was first proposed by Commoner (Citation1964). This was explicitly formulated by Bennett (Citation1971, Citation1972) with the nucleotype theory according to which “that condition of the nucleus (most notably the DNA content) that affects the phenotype independently of the informational content of the DNA”. This theory was based on the relationships evidenced initially for many plant species and subsequently observed also in various animal species, especially chordates (Gregory Citation2005). Indeed, in these latter, associations between genome size and several structural and functional cell parameters such as size and metabolism, and also parallelisms between quantitative genome variations and environmental conditions have been evidenced. Possible interactions with environment have been hypothesized also for other genome components such as transposons (McClintock Citation1984) and base composition (Bernardi Citation2004). Since genome size, transposon percentage and base composition are known for many species of chordates, this phylum is particularly interesting to understand the meaning of interactions between genome and environment. Indeed, they are one of the most common models to study the evolutionary processes and mechanisms. In this regard, it is important to examine the variations that the aforementioned genome parameters have undergone during the evolution of chordates and potential interactions with the environment, assessing if they are random or an important constitutive phenomenon for some evolutionary processes such as, for example, speciation and adaptation. Other important aspects that have to be clarified are if variations in genome features are the consequence of environmental changes or if at the contrary, they are the drive for adaption of organisms to new habitats. Moreover, it is interesting to understand if genome–environment interactions are the same for variations in nuclear DNA content, in the transposon percentage and in base composition and if any reciprocal influences can be identified between these three parameters. In this review, we firstly examined changes that genome and environment have undergone in chordates evolution, since it is known that transposons and genome size are closely related.
Genome size and transposons
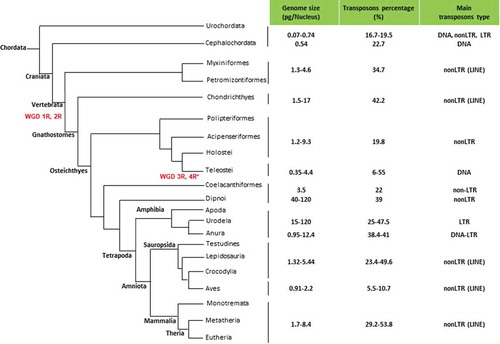
Data on genome sizes derive from Gregory (Citation2019), while those related to TE percentage are referred to Canapa et al. (Citation2015) (see ).
Figure 1. Cladogram showing evolutionary relationships between the main lineages of chordates. Whole Genome Duplication (WGD) events in vertebrate evolution are indicated in red: 1R and 2R occurred before the divergence of Vertebrata, 3R in Teleost and 4R* in salmonids
The trend of variations in genome size and in transposon percentage leads to both phylogenetic considerations and evaluations of the meaning of correlations between the aforementioned genome characters and various structural and functional parameters of cell and organism in adaptive processes.
It is assumed that the ancestral values of genome size in chordates were low and later they increased during evolution; however, the situation is more complex because during evolution of this phylum, increases and decreases in genome size occurred and a similar trend is also found within each class. Furthermore, even if it is ascertained that the main cause of genome size increase is due to TE (transposons) amplification (Canapa et al. Citation2015), other mechanisms such as whole genome duplication and polyploidy have been suggested (Gregory Citation2005).
In the primitive chordates, genome size is very low and ranges from 0.07 pg in Oikopleura to 0.74 pg in Botryllus. The transposons range from 16.7% in Ciona to 22% in the amphioxus and in the three species studied are DNA, LTR, and nonLTR.
A first substantial increase would have occurred at the origin of vertebrates and it is assumed that one of the causes was the first whole genome duplication (Meyer & Schartl Citation1999).
In cyclostomes, the genome size ranges from 1.3 pg to 2.5 pg in Petromyzontiformes and from 2.3 pg to 4.6 pg in Mixyniformes and it has been hypothesized that this difference is linked to the complexity of the development since Mixyniformes have a direct development while Petromyzontiformes present a metamorphosing stage (Hardie & Hebert Citation2003). The transposons have been studied only in Petromyzon where they represent 34.7% and are mainly nonLTR retrotransposons.
The genome size of Chondrichthyes ranges from 1.51 pg in the holocephalus Hydrolagus to over 17 pg in the elasmobranch Rhinobatus. In this subclass, the average genome size is significantly different in rays (6.7 pg) and sharks (5 pg). Furthermore, studies on the reassociation kinetics have highlighted cases of cryptic polyploidy in selachians (Olmo et al. Citation1982).
In Chondrichthyes, there is a positive correlation between genome size and nuclear and cell volume. The highest values were found in species that live in extreme conditions of temperature, light, pressure, and scarce food availability, such as deep and/or cold sea (Stingo et al. Citation1980; Hardie & Hebert Citation2003). The differences in the genome size can also be correlated with development complexity since clear differences between viviparous species and those ovoviviparous and oviparous have been evidenced (Hardie & Hebert Citation2004). Transposons have been studied only in the elepha0nt shark Callorhinchus milii where they represent 42.2% and are nonLTR retrotransposons.
The genome size of the primitive Osteichthyes ranges from 1.15 pg in Amia to 7.25 pg in Polypterus. Also in this group, there is a positive correlation between genome size and nuclear and cell volume. Transposons have been studied in Lepisosteus in which are mainly of the nonLTR type and represent 19.77% of its genome.
In vertebrates, the third whole genome duplication occurred at the origin of Teleostei, but this did not lead to genome size values higher than the average values of most living vertebrates (Meyer & Schartl Citation1999). The genome size varies from 0.35 pg in tetraodontids that have the smallest and most compact genome of all chordates to a maximum of 4.4 pg. However, if polyploidy cases are excluded, the maximum values rarely exceed 2.5 pg. The genome size is positively correlated with nuclear and cell volume (Hardie & Hebert Citation2003).
There are several parallelisms between genome size and environment: freshwater and anadromous species have larger genomes, while marine and catadromous species have smaller genomes (Hardie & Hebert Citation2004). This is in agreement with the observations made by Ebeling et al. (Citation1971) according to which the genomes of eurobiotic (eurihalyne) species are larger than those of stenobiotic (stenohalyne) species. Moreover, in the primitive Protoacanthopterygii and Paraacanthopterygii, species that live in the ocean depths and in cold environments, poorly lighted and with scarce nutrients, have larger genomes than those of the related species living in coastal waters (Ebeling et al. Citation1971). In addition to climatic characteristics, the stability of the environment, in which certain teleost species live, would be also very important. Indeed, it has been observed that polar teleosts and those of poor variable tropical environments, have small genomes and this could be related to a specific reproductive strategy (Hardie & Hebert Citation2003, Citation2004).
The percentage of transposons varies from only 6% in tetraodontids to 55% in zebrafish and, unlike most vertebrates, are mainly DNA transposons (Chalopin et al. Citation2015). On average, teleosts exhibit the widest diversity in transposons reaching 27 superfamilies in zebrafish (Sotero-Caio et al. Citation2017).
A case of correlation between transposons and environment was observed in a study on the identification and characterization of the transposable elements Rex3 in teleosts. In this paper, Carducci et al. (Citation2019a) have demonstrated for the first time, through phylogenetic analyses, the correlation between these transposable elements and environmental temperature. In particular, the results highlighted a clear sequence distinction of Rex3 elements belonging to fish living in cold waters compared to those of fish living in temperate waters, regardless of the evolutionary and taxonomic relationships of the analysed species.
The most significant increase in genome size and in the transposon percentage would have occurred during the transition from aquatic to terrestrial environment, that is, in a crucial step of evolution, and this led to the large genomes that are observed in lungfish and amphibians (Organ et al. Citation2015). It is difficult to establish whether this increase is directly related to the conquest of the land, but it is interesting that a similar situation has also been noted in the transition from aquatic to terrestrial gastropods (Vinogradov Citation2000).
Primitive sarcopterygians occupy a very important position in the evolution of vertebrates because tetrapods originated from them. The genome size is very different in crossopterygians and lungfish.
The only two living species of Latimeria have a genome of 3.5–3.6 pg and the TE percentage (mainly SINE and LINE Chalopin et al. Citation2015) is 22% (Amemiya et al. Citation2013).
In lungfish, the genome size is the highest of all vertebrates and ranges from 50 pg to over 120 pg. A polyploid karyotype has been noted in the species Protopterus dolloi. The living lungfish have several characteristics that make them paedomorphic organisms and it has been speculated that this would be related to the large genome size (Joss Citation2006; Joss & Johanson Citation2007). In primitive lungfish, genome size would have been similar to that of Latimeria (Organ et al. Citation2015) but from the Carboniferous a great expansion would have started which would have led to the current huge genomes and it would have been accompanied by an evolutionary decline with great reduction in the number of genera and species (Thomson Citation1972; Thomson & Muraszko Citation1978). This decline contrasts with the hypothesis that genome size increase may promote evolutionary diversification (Kraaijeveld Citation2010). Transposons have only been studied in Neoceratodus in which they are mainly nonLTR retrotransposons and make up 39% of its genome (Metcalfe et al. Citation2012).
In amphibians, genome size shows very different values in the three orders. In anurans it ranges from 0.95 pg to 12.4 pg; in gymnophions, in which only three species have been studied, it ranges from 3.7 pg to 13.95 pg; in urodeles it ranges from a minimum of 15 pg, higher than the maximum value observed in the other two orders, to over 120 pg in Necturus, a value similar to that recorded in lungfish. Cases of polyploidy are known in both anurans and urodeles (Schmid et al. Citation2015).
The transposon percentage in the only two anuran species analysed is around 40% and are both DNA and LTR elements; in urodeles it ranges from 25% to 47.5%, and, unlike the majority of vertebrates, they are almost exclusively LTR retroelements (Canapa et al. Citation2015; Chalopin et al. Citation2015). In both the most primitive and most recent species, the majority of transposons belongs to a few families and is mainly represented by the LTR Gypsy elements constituting more than 25% in Cryptobranchus alleganiensis (Sun & Mueller Citation2014). Several works have shown that transposons have been the main cause of genome expansion in these amphibians.
Fossil amphibians (rhipidistians, labyrinthodonts and anthracosaurs) had low values similar to the current ones of bufonids and ranids (5–10 pg) (Thomson & Muraszko Citation1978; Organ et al. Citation2011).
The expansion of the lissamphibian genome would have started in the Permian before the separation of urodeles from anurans and the expansion of the urodeles genome, due to a few LTR families, would have continued in the early and Middle Triassic (Laurin et al. Citation2015; Christoph-Liedtke Citation2016). The first phase of the expansion took place gradually and led to larger genomes in anuran; the next phase would have occurred because of the LTR retrotransposon saltatory proliferation (Sun & Mueller Citation2014) and would have led to very large genomes typical of current urodeles already in the Middle Jurassic (Laurin et al. Citation2015; Organ et al. Citation2015; Christoph-Liedtke et al. Citation2018).
An interesting aspect, common to the great expansion of genome in lungfish and amphibians, especially urodeles, is that it would have occurred in periods of drastic environmental changes such as the beginning of Carboniferous and the transition from Permian to Triassic and this is in agreement with the observation that transposon activity is often the consequence of environmental stresses (McClintock Citation1984; Capy et al. Citation2000).
A positive relationship between genome size and nuclear and cell volume has been recorded in anurans and urodeles.
Even in amphibians, there are correlations between the genome size and the environment. In general, species that live in cold environments have more DNA and a longer development; in particular, in urodeles there is a direct correlation between genome size and latitude (Litvinchuk et al. Citation2007).
In amphibians, variations in genome size show interesting relationships with reproductive strategies and embryonic development. A positive correlation is observed between genome size and cell cycle duration during the synchronous divisions of segmentation (Vinogradov Citation1999). In anurans, genome size is correlated with cell proliferation rate and with developmental rates in different embryonic stages (Chipman et al. Citation2001); in urodeles, on the other hand, there is a negative relationship between genome size and regeneration and growth rates during embryogenesis and differentiation (Sessions & Larson Citation1987). In Plethodontid urodeles genome size is positively correlated to development duration (Jockusch Citation1997).
A peculiar feature of urodeles is the relationship between genome size, paedomorphosis and neoteny: species that have a biphasic life cycle have the smallest genomes; those with large genomes are often optional neotenic and those with giant genomes are obliged neotenic (Morescalchi Citation1991; Gregory Citation2005).
Also the direct correlation between genome size and cell cycle duration (Vinogradov Citation1999) and the inverse correlation between genome size and metabolic rate observed in amphibians (Monnickendam & Balls Citation1973; Licht & Lowcock Citation1991) are interesting from an evolutionary point of view.
The genome size in reptiles ranges from 1.32 pg to 5.44 pg. The average higher values are found in cheloniids and in Tuatara. There is an inverse relationship between genome size and chromosome changing rate that is higher in Squamata and is directly related to the number of living species (Olmo Citation2005). Furthermore, genome size is also directly correlated with nucleus and cell size and is inversely correlated to the metabolic rate (Olmo Citation2003).
A difference between cheloniids/crocodiles and squamates is the presence in the latter of two types of chromosomes: microchromosomes, richer in genes, and macrochromosomes (Kasai et al. Citation2019).
The percentage of transposons ranges from 23.4% to 49.6% and are mainly LINE. Unlike what is known in other amniotes, in the Squamata, despite a little genome size variations, there is a surprisingly large variability in the amount of repetitive sequences especially transposons and in snakes there is the highest content of microsatellites originating from transposons (Pasquesi et al. Citation2018).
Birds have very low genome size ranging from 0.91 pg to 2.16 pg, with the highest value in ostrich. The percentage of transposons is the lowest among vertebrates and ranges from 5.5% to 10.7% and are mainly LINE.
Even in birds, there are microchromosomes richer in genes and macrochromosomes. Originally the genome of birds would have had a similar size to that of current mammals (3–3.7 pg) and would have undergone a reduction in Mesozoic and Caenozoic eras that would have led to current values (Organ et al. Citation2007). A similar situation is assumed to have previously characterized the pterosaurs (Organ & Shedlock Citation2009). There is an inverse relationship between genome size and metabolic rate (Vinogradov Citation1997; Gregory Citation2001) leading to hypothesize that small genomes and corresponding high metabolism would have been the premise for the development of flight (Hughes & Hughes Citation1995).
In mammals, the nuclear DNA content ranges from 2.9 pg to 3.6 pg in monotremes, from 2.99 pg to 5.02 pg in marsupials, and from 1.7 pg to 8.4 pg in placentals.
The percentage of transposons is 44.6% in platypus, from 52% to 53.8% in marsupials and from 29.2% to 46% in placental and are nonLTR retrotransposons.
An inverse relationship between genome size and metabolic rate has also been noted in mammals (Vinogradov Citation1995) and it has been hypothesized that there is also a correlation between genome size, metabolism and flight adaptation in Chiroptera, even if data are contradictory (Smith et al. Citation2013).
Base composition and methylation
The research groups of Bernardi and Vinogradov have been the main conductors of studies concerning the base composition. They have highlighted variations of this parameter during the evolution of the chordates.
Vinogradov (Citation1998a) evidenced a general positive but not linear relationship between GC content and genome size; the GC percentage shows great variability at lower values while it tends to stabilize at the level of about 46% with the increase of genome size. The regression line of reptiles and birds follows the general trend but has a steeper slope due to a greater increase in GC composition compared to the increase in nuclear DNA content, while in teleosts an inverse relationship has been noted. The higher percentage of GC is positively correlated with bendability, thermostability and the ability to change the DNA from B to Z form, property linked to chromatin opening and transcription activation, and inversely related to curvature that favours chromatin condensation. Bernardi and colleagues’ studies have shown GC% values in line with those reported by Vinogradov, although they have noted a negative correlation with genome size in most vertebrate classes (Bernardi & Bernardi Citation1990; Jabbari et al. Citation1997; Bernardi Citation2004). These researchers have shown a compartimentalization in eukaryotic genome, i.e. made up of a series of distinct fractions, each characterized by a specific and different GC average composition, the so-called isochores. Changes in base composition would have mainly depended on body temperature. In fact, during vertebrate evolution, a clear variation in GC percentage has been observed in isochore distribution, only in the transition from cold-blooded to warm-blooded. In the former, similarly to invertebrates, genome is constituted almost exclusively of AT-rich isochores, while in birds and mammals there are GC-rich isochores. An intermediate situation has been reported for reptiles, where in Squamata the isochore pattern is similar to that one observed in fish and amphibians, while in turtles and crocodiles it is more similar to that of birds and mammals (Hughes et al. Citation2002). The transition in base composition from ectothermic to endothermic vertebrates has been explained with the “thermodynamic stability hypothesis” according to which the increase of GC-rich components in endotherms would have led to a greater thermodynamic stability of DNA, RNA, and proteins. This is of great importance to cope with the impact that the increase in body temperature has on various structural and functional aspects of cell and genome (Bernardi Citation2004). The percentage in base composition has shown a positive correlation with methylation level which, however, is different between fishes and amphibians on the one hand, and birds and mammals on the other, where two-fold values have been evidenced in the former group (Bernardi Citation2004). Moreover, for reptiles, an intermediate situation has been described (Varriale & Bernardi Citation2006). In all vertebrates, methylation is inversely related to the genome size and methylation level is inversely correlated with body temperature in fish (Bernardi Citation2004; Varriale & Bernardi Citation2006). A positive correlation has been noted between the GC content and the metabolic level (Uliano et al. Citation2010; Bernà et al. Citation2012; Varriale Citation2014; Tarallo et al. Citation2016). In teleosts, it has been observed that the GC percentage increases from freshwater to marine species and within these from non-migratory to migratory species. This trend would be related to both salinity changes and higher costs in terms of energy for migratory species (Tarallo et al. Citation2016).
Relationship of genome characters with the environment
As described in the previous sections, various interactions exist between genome size, transposons and base composition, and between these and the environment. Some of these interactions concern all the aforementioned genome characters, while others are referred to some of these features. However, these relationships may not be always simple and linear, since in some cases the environmental factors would seem to be the direct cause of the changes in the size and/or genome composition, while in other cases genome characters influencing some cell structures and functions, such as cell size (Olmo Citation1983) and cell cycle (Vinogradov Citation1999) would have favored adaptation to new environmental conditions. However, not all these interactions seem to be supported by experimental data.
Recently, many researches on transposons have shown that they are among the main causes of genome expansion and that their activity is stimulated by various environmental stressors (Canapa et al. Citation2015; Carducci et al. Citation2019b), such as temperature, thermal shocks, radiations, chemicals, and viral infections (Capy et al. Citation2000; Fujino et al. Citation2011; Garcia Guerreiro Citation2012; Carducci et al. Citation2019a, b). In some cases, these stressors would act directly on transposons with a mechanism typical of defensive gene activation, since some transposons have sequences similar to the promoters of these genes (Takeda et al. Citation1999; Capy et al. Citation2000); however, in many other cases, the action would mainly depend on a temperature-dependent activation of transposase (Capy et al. Citation2000; Fujino et al. Citation2011; Carducci et al. Citation2019a).
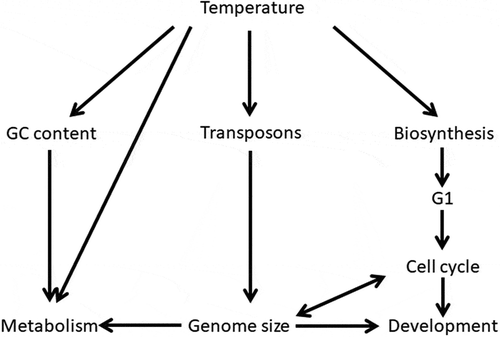
Besides to be involved in transposon activation, temperature is one of the most important elements of natural selection able to interact both with base composition and genome size (see ). Studies conducted on base composition have clearly shown that body temperature affects GC percentage. This effect is evident in structural changes of macromolecules. In accordance with the “thermodynamic stability hypothesis”, GC higher values are directly related to an increased DNA thermostability (Bernardi Citation2004) which in turn influences RNA composition and chemical and physical properties of proteins, which in species showing a higher GC percentage are richer in hydrophobic amino acids. Thus, macromolecules of endothermic organisms are thermodynamically more stable and can function optimally even in the case of an increase in body temperature (Bernardi Citation2004).
As previously described, an increase in GC content confers to DNA not only a greater thermostability, but also facilitates chromatin opening and the consequent transcriptional activation; on the contrary, a higher GC richness reflects a lower DNA curvature which favors chromatin condensation (Vinogradov Citation1998a, Citation2003, Citation2005). The GC-rich DNA, with its higher physical stability would compensate for higher gene mutation rate observed in larger genomes and would also protect against damages due to chemical mutagens, many of which have a higher affinity for GC-rich DNA and then would be more easily absorbed and neutralized by the higher GC-rich DNA regions (Vinogradov Citation1994; Bernardi Citation2004).
However, the influence of temperature would not be the direct cause of the GC content increase but it would be one of the main selective factors responsible for structural changes in macromolecules, allowing birds and mammals to cope with the increase of body temperature (Bernardi Citation2004).
Moreover, the direct relationship between GC percentage and methylation increase demonstrates that temperature, via base composition, has been able to influence an important epigenetic mechanism involved in transposon inactivation (Canapa et al. Citation2015) and in the control of different cellular functions, such as tissue-specific gene expression, cell differentiation, and development (Varriale Citation2014).
In addition, the influence of temperature on genome size could act indirectly through the control of the cell cycle duration.
This cycle mainly depends on the DNA amount that has to be duplicated. However, differences have been observed in two phases of the cell cycle: S phase is strictly related to the DNA amount, while the G1 phase duration is influenced both by DNA content and biosynthesis rate. Therefore, a higher biosynthesis rate corresponds to a shorter G1 phase and thus to a shorter cell cycle.
Since the biosynthesis rate is influenced by temperature, the G1 phase and the cell cycle can be controlled by temperature variations. Xia (Citation1995) has reported an inverse relationship between biosynthesis rate and genome size. This would explain why species, living in warm environments (especially poikilothermic species), have smaller genomes, a higher biosynthesis rate, and a shorter cell cycle compared to related species living in colder environments.
According to this author, smaller changes in G1 phase duration (and in biosynthesis rate) at specific changes in body temperature occur in species with large genomes compared to those with smaller genomes. Therefore, a large genome could be used by poikilothermic species to buffer potential damages caused by climate changes (Xia Citation1995).
The relationship between genome parameters and temperature-related metabolism (Kleiber Citation1932) is more complex.
An inverse relationship between genome size and metabolic rate has been observed in all vertebrates (Licht & Lowcock Citation1991; Gregory Citation2005; Vinogradov & Anatskaya Citation2006) in which a different trend has been evidenced in reptiles-birds compared to amphibians-mammals: indeed, an increase in genome size corresponds to a greater reduction in the metabolic rate in the former than the latter (Vinogradov & Anatskaya Citation2006); these two trends are similar to those concerning the ratio between genome size and GC content, i.e. in reptiles-birds at the same genome size the GC percentage is higher than that of amphibians-mammals (Vinogradov Citation1998a).
The GC content is proportional to chromatin condensation, thus nuclei with a greater GC quantity are larger for the same DNA amount. Therefore, it has been hypothesized that metabolic rate is influenced not by the genome size but mainly by the nuclear volume as determined by the interaction between genome size and GC content (Vinogradov & Anatskaya Citation2006).
It is assumed that metabolism is conditioned by both respiratory and nutritive exchanges which depend on surface–volume ratio of nucleus and cell (Olmo Citation1983; Vinogradov Citation1995). Consequently, genome size determining these two-dimensional cell parameters is probably one of the most important factors regulating cell metabolism (Olmo Citation1983; Hardie & Hebert Citation2003).
Relationships observed between metabolic rate and base composition are less clear. In teleosts, contradictory aspects have been evidenced in the relationships between these two parameters and the temperature (Uliano et al. Citation2010). Indeed, the GC composition has a positive correlation with metabolic rate, but both these parameters have a negative correlation with temperature. This trend cannot be generalized since a similar low metabolic rate has been found in tropical species and in organisms living in deep sea, characterized by lower temperature.
In this sub-class, genome parameters would also be influenced by salinity, especially by the ability to cope variations in saline concentration: it has been observed that GC content increases from non-migratory freshwater species to migratory marine species, similarly the genome size is higher in eurobiotic species than in stenobiotic ones (Ebeling et al. Citation1971; Hardie & Hebert Citation2003, Citation2004; Tarallo et al. Citation2016).
Tarallo et al. (Citation2016) have hypothesized that this situation is linked both to metabolic rate and to a higher necessity of oxygen for migratory species. Another possible explanation has been proposed by Vinogradov (Citation1998b, Citation1998c) which hypothesizes that non-coding DNA performs a buffering function on the intracellular concentration of solutes through non-specific bonds with proteins. Cells with a major amount of repetitive DNA, and therefore constituted by larger genomes, have a higher ability to regulate the intracellular composition of solutes balancing the variations in the composition of the extracellular solutes.
Finally, another interesting aspect of the relationship between genome size and processes involved in speciation and environmental adaptation concerns both time and mode of embryo and larval development.
It is known that the cell cycle length influences the duration of embryo and larval development (Vinogradov Citation1999; Gregory Citation2005). A positive relationship between genome size, cell cycle, and developmental duration has been detected in many classes of vertebrates, especially anamniotes, while no relationship has been reported in amniotes (Gregory Citation2001; Olmo Citation1983) concerning these aspects. These correlations have been studied particularly in amphibians. In anurans genome size is directly related to cell proliferation rate in segmentation and developmental rates in different stages (Chipman et al. Citation2001). In urodeles, besides a positive relationship between nuclear DNA content and developmental duration (Jockusch Citation1997), there is a negative relationship with the regeneration, the differentiation, and the growth rate during embryogenesis (Sessions & Larson Citation1987).
The relationship between genome size and the related developmental duration can represent a limiting factor in environment adaptation, especially concerning temperature and water availability. Indeed it has been observed that in amphibians, especially in anurans, species that live in environments characterized by scarcity of water have a small genome and a rapid development, while species that live in cold environments with water availability have larger genomes and a slow development (Goin et al. Citation1968; McMenamin & Hadly Citation2010); similarly in urodeles species living in cold environments have a larger genome and a longer duration of development compared to species living in warmer environments (Litvinchuk et al. Citation2006; Lertzman-Leofsky et al. Citation2019). Similar correlations have also been reported for some fishes (Hardie & Hebert Citation2003, Citation2004). In addition to the developmental duration, variations in the DNA content also seem to affect the developmental complexity (Gregory Citation2001, Gregory Citation2005). Genome size shows relationships with different aspects of development such as viviparity, direct or indirect development, and neoteny. In Chondrichthyes viviparous species have larger genomes (Hardie & Hebert Citation2004). A trend reported in several animals concerns the increase in genome size in relation to the transition from metamorphosis to the direct development, and facultative or obligatory neoteny. In cyclostomes, anurans, and urodeles, species having a complete metamorphosis present smaller genomes than those with direct development. Moreover, urodeles that have large genomes are facultative or obligatory neotenic species.
This trend could be common to all metazoans since also in insects it has been observed an increase in DNA content from holometabolous, to incomplete metamorphosing species (hemimetabolous) to those with direct development (ametabolous) (Gregory Citation2005). This aspect would be linked to heterochrony, a mechanism that involves changes in developmental rate and duration, and from which paedomorphosis can result, i.e. the mantainment of immature characters in adults, when development slows down or interrupts prematurely (Morescalchi Citation1991). In amphibians, one of the main causes of paedomorphosis would be an increase in genome size since this increase would reduce the growth rate, differentiation, and cell migration during embryogenesis by preventing the formation of late-developing traits (Roth et al. Citation1997; Womack et al. Citation2019).
Paedomorphosis played a very important role in the evolution of amphibians, since paedomorphic forms were already present in the labyrinthodonts and in the dyssorophoids which are the ancestors of the current lissamphibians (Morescalchi Citation1991). Paedomorphosis would have played an important role also in the Dipnoi, which have large genomes (similar to those of the facultative or obligatory neotenic urodeles) and show larval characters (Bemis Citation1984; Joss Citation2006; Joss & Johanson Citation2007).
Conclusions
This analysis shows the existence in chordates of a mutual influence between environment and the non-coding components of the genome and this supports the hypothesis that the nucleotype would have played an important role in the evolution of this phylum, mainly in environmental adaptation processes.
The parameter that seems to be most relevant in genome–environment relationships is certainly the temperature variation () that directly or indirectly, e.g. through metabolism, has an effect on transposons, genome size, and base composition. The action of genome–temperature interactions was one of the most relevant phenomena, mainly for the evolution of the poikilothermic species. However, temperature-dependent enrichment of the GC-rich isochores that has occurred in the transition from ectothermic to endothermic vertebrates was very important for the latter since it has contributed to making these vertebrates less environment-dependent.
Overall, it is the environment that exerts its effect on genome, but in certain cases the variations in the genome components, which influence the structural and functional characteristics of cell, regulate evolutionary processes, such as the environmental adaptation.
In all cases, the interactions between environmental factors and genome components are part of the typical processes of natural selection and the variations affecting genome size, the transposon percentage, and base composition represent preadaptations on which environmental variations operate.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Amemiya CT, Alfoldi J, Lee AP, Fan S, Philippe H, MacCallum I, Braasch I, Manousaki T, Schneider I, Rohner N, Organ C, Chalopin D, Smith JJ, Robinson M, Dorrington RA, Gerdol M, Aken B, Biscotti MA, Barucca M, Baurain D, Berlin AM, Blatch GL, Buonocore F, Burmester T, Campbell MS, Canapa A, Cannon JP, Christoffels, De Moro G, Edkins AL, Fan L, Fausto AM, Feiner N, Forconi M, Gamieldien J, Gnerre S, Gnirke A, Goldstone JV, Haerty W, Hahn ME, Hesse U, Hoffmann S, Johnson J, Karchner SI, Kuraku S, Lara M, Levin JZ, Litman GW, Mauceli E, Miyake T, Mueller MG, Nelson DR, Nitsche A, Olmo E, Ota T, Pallavicini A, Sumir P, Picone B, Ponting CP, Prohaska SJ, Przybylski D, Saha NR, Ravi V, Ribeiro FJ, Sauka-Spengler T, Scapigliati G, Searke SMJ, Sharpe T, Simakov O, Stadler PF, Stegeman JJ, Sumiyama K, Tabbaa D, Tafer H, Turner-Maier J, van Heusden P, White S, Williams L, Yandell M, Brinkmann H, Volff JN, Tabin CJ, Shubin N, Schartl M, Jaffe DB, Postlethwait JH, Venkatesh B, Di Palma F, Lander ES, Meyer A, Lindblad-Toh K. 2013. The african coelacanth genome provides insight into tetrapod evolution. Nature 496:311–316. DOI: 10.1038/nature12027.
- Bemis WE. 1984. Paedomorphosis and the evolution of the Dipnoi. Paleobiology 10:293–307. DOI: 10.1017/S0094837300008277.
- Bennett MD. 1971. The duration of meiosis. Proceedings of the Royal Society of London B 178:277–299.
- Bennett MD. 1972. Nuclear DNA content and minimum generation time in herbaceous plants. Proceedings of the Royal Society of London B 181:109–135.
- Bernà L, Chaurasia A, Angelini C, Federico C, Saccone S, D’Onofrio G. 2012. The footprint of metabolism in the organization of mammalian genomes. BMC Genomics 13:174. DOI: 10.1186/1471-2164-13-174.
- Bernardi G. 2004. Structural and evolutionary genomics. Lausanne, New York, Oxford, Shannon, Singapore, Tokyo: Elsevier Amsterdam. pp. 434+XV.
- Bernardi G, Bernardi G. 1990. Compositional patterns in the nuclear genome of cold-blooded verterbrates. The Journal of Molecular Evolution 31:265–281. DOI: 10.1007/BF02101122.
- Canapa A, Barucca M, Biscotti MA, Forconi M, Olmo E. 2015. Transposons, Genome size, and evolutionary insights in animals. Cytogenetic and Genome Research 147:217–239. DOI: 10.1159/000444429.
- Capy P, Gasperi G, Biémont C, Bazin C. 2000. Stress and transposable elements: Co-evolution or useful parasites? Heredity 85:101–106. DOI: 10.1046/j.1365-2540.2000.00751.x.
- Carducci F, Biscotti MA, Barucca M, Canapa A. 2019b. Transposable elements in vertebrates: Species evolution and environmental adaptation. The European Zoological Journal 86:497–503. DOI: 10.1080/24750263.2019.1695967.
- Carducci F, Biscotti MA, Forconi M, Barucca M, Forconi M. 2019a. An intriguing relationship between teleost Rex3 retroelement and environmental temperature. Biology Letters 15:20190279. DOI: 10.1098/rsbl.2019.0279.
- Chalopin D, Naville M, Plard F, Galiana D, Volff JN. 2015. Comparative analysis of transposable elements highlights mobilome diversity and evolution invertebrates. Genome Biology and Evolution 7:567–580. DOI: 10.1093/gbe/evv005.
- Chipman AD, Khaner O, Haas A, Tchernov E. 2001. The evolution of genome size: What can be learned from anuran development? Journal of Experimental Zoology Part B (Molecular and Developmental Evolution) 291:365–374. DOI: 10.1002/jez.1135.
- Commoner B. 1964. The structure and function of chromatin. Nature 202:960–968. DOI: 10.1038/202960a0.
- Ebeling AW, Atkin NB, Setzer PY. 1971. Genome sizes of teleostean fishes: Increase in some deep-sea species. The American Naturalist 105:549–562. DOI: 10.1086/282744.
- Fujino K, Hashida SN, Ogawa T, Natsume T, Uchiyama T, Mikami T, Kishima Y. 2011. Temperature controls nuclear import of Tam3 transposase in Antirrhinum. Plant Journal 65:146–155. DOI: 10.1111/j.1365-313X.2010.04405.x.
- Garcia Guerreiro MP. 2012. What makes transposable elements move in the Drosophila genome? Heredity 108:461–468. DOI: 10.1038/hdy.2011.89.
- Goin OB, Goin CJ, Bachmann K. 1968. DNA and Amphibian life history. Copeia 1968:532–540. DOI: 10.2307/1442021.
- Gregory TR. 2001. Genome size and developmental parameters in the homeothermic vertebrates. Genome 45:833–838. DOI: 10.1139/g02-050.
- Gregory TR. 2005. Genome size evolution in animals. In: Gregory TR, editor. The evolution of the genome. Boston, Heidelberg, New York, Oxford, Paris, San Diego, San Francisco, Singapore, Sydney, Tokyo: Elsevier Amsterdam. pp. 3–87.
- Gregory TR. 2019. Animal genome size database. Available: http:www.genomesize.com.
- Hardie DC, Hebert PDN. 2003. The nucleotypic effects of cellular DNA content in cartilaginous and ray-finned fishes. Genome 46:683–706. DOI: 10.1139/g03-040.
- Hardie DC, Hebert PDN. 2004. Genome size evolution in fishes. Canadian Journal of Fisheries and Aquatic Science 61:1636–1646. DOI: 10.1139/f04-106.
- Hughes AL, Hughes MK. 1995. Small genomes for better flyers. Nature 377:391. DOI: 10.1038/377391a0.
- Hughes S, Clay O, Bernardi G. 2002. Compositional patterns in reptilian genomes. Gene 295:323–329. DOI: 10.1016/S0378-1119(02)00732-1.
- Jabbari K, Cacciò S, Païs de Barros JP, Desgrès J, Bernardi G. 1997. Evolutionary changes in CpG and methylation levels in the genome of vertebrates. Gene 205:109–118. DOI: 10.1016/S0378-1119(97)00475-7.
- Jockusch EL. 1997. An evolutionary correlate of genome size change in plethodontid salamanders. Proceedings of the Royal Society of London B 264:597–604. DOI: 10.1098/rspb.1997.0085.
- Joss J, Johanson Z. 2007. Is Palaeospondylus gunni a fossil larval lungfish? Insights from Neoceratodus forsteri development. Journal of the Experimental Zoology Part B Molecular and Developmental Evolution 308:163–171. DOI: 10.1002/jez.b.21125.
- Joss JM. 2006. Lungfish evolution and development. General and Comparative Endocrinology 148:285–289. DOI: 10.1016/j.ygcen.2005.10.010.
- Kasai F, O’Brien PCM, Ferguson-Smith MA. 2019. Squamate chromosome size and GC content assessed by flow karyotyping. Cytogenetic and Genome Research 157:46–52. DOI: 10.1159/000497265.
- Kleiber M. 1932. Body size and metabolism. Hilgardia 6:315–353. DOI: 10.3733/hilg.v06n11p315.
- Kraaijeveld K. 2010. Genome size and species diversification. Evolutionary Biology 37:227–233. DOI: 10.1007/s11692-010-9093-4.
- Laurin M, Canoville A, Struble M, Organ C, de Buffrénil V. 2015. Early genome size increase in urodeles. Comptes Rendus Palevol 15:74–82. DOI: 10.1016/j.crpv.2014.12.006.
- Lertzman-Leofsky G, Mooers AO, Greenberg DA. 2019. Ecological constraints associated with genome size across salamander lineages. Proceedings of the Royal Society of London B 286(1911):20191780. DOI: 10.1098/rspb.2019.1780.
- Licht LE, Lowcock LA. 1991. Genome size and metabolic rate in salamanders. Comparative Biochemistry and Physiology Part B: Comparative Biochemistry 100:83–92. DOI: 10.1016/0305-0491(91)90089-V.
- Liedtke CH, Gower DJ, Wilkinson M, Gomez-Mestre I. 2018. Macroevolutionary shift in the size of amphibian genomes and the role of life history and climate. Nature Ecology and Evolution 2:1792–1799. DOI: 10.1038/s41559-018-0674-4.
- Liedtke HC, Müller H, Rödel MO, Menegon M, Gonwouo LN, Barej MF, Gvoždík V, Schmitz A, Channing A, Nagel P, Loader SP. 2016. No ecological opportunity signal on a continental scale? diversification and life-history evolution of african true toads (anura: bufonidae). Evolution 70:1717–1733.
- Litvinchuk SN, Rosanov JM, Borkin L. 2006. Correlations of geographic distribution and temperature of embryonic development with the nuclear DNA content in the salamandridae (Urodela, Amphibia). Genome 50:333–342.
- Litvinchuk SN, Rosanov JM, Borkin L. 2007. Correlations of geographic distribution and temperature of embryonic development with the nuclear DNA content in the Salamandridae (Urodela, Amphibia). Genome 50:333–342. DOI: 10.1139/G07-010.
- McClintock B. 1984. The significance of responses of the genome to challenge. Science 226:792–801. DOI: 10.1126/science.15739260.
- McMenamin SK, Hadly EA. 2010. Developmental dynamics of Ambystoma tigrinum in a changing landscape. BMC Ecology 10(1):10. DOI: 10.1186/1472-6785-10-10.
- Metcalfe CJ, Filée J, Germon J, Joss J, Casane D. 2012. Evolution of the Australian lungfish (Neoceratodus forsteri) genome: A major role for CR1 e L2 line elements. Molecular Biology and Evolution 29:3529–3539. DOI: 10.1093/molbev/mss159.
- Meyer A, Schartl M. 1999. Gene and genome duplications in vertebrates: The one-to-four (-to-eight in fish) rule and the evolution of novel gene functions. Current Opinion in Cell Biology 11:699–704. DOI: 10.1016/S0955-0674(99)00039-3.
- Monnickendam MA, Balls M. 1973. The relationship between cell sizes respiration rates and survival od amphibian tissues in long term organ culture. Comparative Biochemistry and Physiology Part A: Physiology 44:871–880. DOI: 10.1016/0300-9629(73)90150-3.
- Morescalchi A. 1991. Genome and the rise of terrestrial vertebrates. In: Ghiara G, editor. Symposium on the evolution of terrestrial vertebrates. Modena: Mucchi. pp. 31–50.
- Olmo E. 1983. Nucleotype and cell size in vertebrates: A review. Basic and Applied Histochemistry 27:227–256.
- Olmo E. 2003. Reptiles: A group of transition in the evolution of genome size and of the nucelotypic effect. Cytogenetic and Genome Research 101:166–171. DOI: 10.1159/000074174.
- Olmo E. 2005. Rate and chromosome changes and speciation in reptiles. Genetica 125:185–203. DOI: 10.1007/s10709-005-8008-2.
- Olmo E, Stingo V, Cobror O, Capriglione T, Odierna G. 1982. Repetitive DNA and polyploidy in selachians. Comparative Biochemistry and Physiology Part B: Comparative Biochemistry 73:739–745. DOI: 10.1016/0305-0491(82)90311-X.
- Organ CL, Canoville A, Reisz RR, Laurin M. 2011. Paleogenomic data suggest mammal-like genome size in the ancestral amniote and derived large genome size in amphibians. Journal of Evolutionary Biology 24:372–380. DOI: 10.1111/j.1420-9101.2010.02176.x.
- Organ CL, Shedlock AM. 2009. Paleogenomics of pterosaurs and the evolution of small genome size in flying vertebrates. Biology Letters 5:47–50. DOI: 10.1098/rsbl.2008.0491.
- Organ CL, Shedlock AM, Meade A, Pagel M, Edwards SV. 2007. Origin of avian genome size and structure in non-avian dinosaurs. Nature 446:180–184. DOI: 10.1038/nature05621.
- Organ CL, Struble M, Canoville A, de Buffrénil, Laurin M. 2015. Macroevolution of genome size in sarcopterygians during the water-land transition. Comptes Rendus Palevol. DOI: 10.1016/j.crpv.2015.09.003.
- Pasquesi GIM, Adams RH, Card DC, Schield DR, Corbin AB, Perry BW, Reyes-Velasco J, Ruggiero RP, Vandewege MW, Shortt JA, Castoe TA. 2018. Squamate reptiles challenge paradigms of genomic repeat element evolution set by birds and mammals. Nature Communications 9:2774. DOI: 10.1038/s41467-018-05279-1.
- Roth G, Nishikawa KC, Wake DB. 1997. Genome size secondary simplification and the evolution of the brain in salamanders. Brain, Behavior and Evolution 50:50–59. DOI: 10.1159/000113321.
- Schmid M, Evans BJ, Bogart JP. 2015. Polyploidy in Amphibia. Cytogenetic and Genome Research 145:315–330. DOI: 10.1159/000431388.
- Sessions SK, Larson A. 1987. Developmental correlates of genome size in plethodontid salamanders and their implications for genome evolution. Evolution 41:1239–1251. DOI: 10.1111/j.1558-5646.1987.tb02463.x.
- Smith JD, Bickham JW, Gregory TR. 2013. Patterns of genome size diversity in bats (order Chiroptera). Genome 56:457–472. DOI: 10.1139/gen-2013-0046.
- Sotero-Caio CG, Platt 2nd RN, Suh A, Ray DA. 2017. Evolution and diversity of transposable elements in vertebrate genomes. Genome Biology and Evolution 9:161–177. DOI: 10.1093/gbe/evw264.
- Stingo V, Du Buit MH, Odierna G. 1980. Genome size in some selachian fishes. Italian Journal of Zoology 47:129–137.
- Sun C, Mueller RL. 2014. Hellbender genome sequences shed light on genome expansion at the base of crown salamanders. Genome Biology and Evolution 6:1818–1829. DOI: 10.1093/gbe/evu143.
- Takeda S, Sugimoto K, Otsuki H, Hirochika H. 1999. A 13-pb cis-regulatory element in the LTR promoter of the tobacco retrotransposon Tto1 is involved in responsiveness to tissue culture, wounding, methyl jasmonate and fungal elicitors. Plant Journal 18:383–393. DOI: 10.1046/j.1365-313X.1999.00460.x.
- Tarallo A, Angelini C, Sanges R, Yagi M, Agnisola C, D’Onofrio G. 2016. On the genome base composition of teleosts: The effect of environment and lifestyle. BMC Genomics 17:173. DOI: 10.1186/s12864-016-2537-1.
- Thomson KS. 1972. An attempt to reconstruct evolutionary changes in the cellular DNA content of lungfish. Journal of Experimental Zoology 180(3):363–371. DOI: 10.1002/jez.1401800307.
- Thomson KS, Muraszko K. 1978. Estimation of cell size and DNA content in fossil fishes and amphibians. Journal of Experimental Zoology 205:315–320. DOI: 10.1002/jez.1402050216.
- Uliano E, Chaurasia A, Bernà L, Agnisola C, D’Onofrio G. 2010. Metabolic rate and genome GC. What we can learn from teleost fish. Marine Genomics 3:29–34. DOI: 10.1016/j.margen.2010.02.001.
- Varriale A. 2014. DNA methylation, epigenetics, and evolution in vertebrates: Facts and challenges. International Journal of Evolutionary Biology 2014:475981. DOI: 10.1155/2014/475981.
- Varriale A, Bernardi G. 2006. DNA methylation in reptiles. Gene 385:122–127. DOI: 10.1016/j.gene.2006.05.034.
- Vinogradov AE. 1994. Measurement by flow cytometry of genome AT/GC ratio and genome size. Cytometry 16:34–40. DOI: 10.1002/cyto.990160106.
- Vinogradov AE. 1995. Nucleotypic effect in homeotherms body-mass-corrected basal metabolic rate of mammals is related to genome size. Evolution 49:1249–1259. DOI: 10.1111/j.1558-5646.1995.tb04451.x.
- Vinogradov AE. 1997. Nucleotypic effect in homeotherms: Body-mass independent resting metabolic rate of passerine birds is related to genome size. Evolution 51:220–225. DOI: 10.1111/j.1558-5646.1997.tb02403.x.
- Vinogradov AE. 1998a. Genome size and GC-percent in vertebrate as determined by flow cytometry: The triangular relationship. Cytometry 31:100–109. DOI: 10.1002/(SICI)1097-0320(19980201)31:2<100::AID-CYTO5>3.0.CO;2-Q.
- Vinogradov AE. 1998b. Buffering: A possible passive-homeostasis role for redundant DNA. Journal of Theoretical Biology 193:197–198. DOI: 10.1006/jtbi.1997.0629.
- Vinogradov AE. 1998c. Variation in ligand-accessible genome size and its ecomorphological correlates in a pond snail. Hereditas 128:59–65. DOI: 10.1111/j.1601-5223.1998.00059.x.
- Vinogradov AE. 1999. Genome in toto. Genome 42:361–362. DOI: 10.1139/g98-117.
- Vinogradov AE. 2000. Larger genomes for molluskan land pioneers. Genome 43:211–212. DOI: 10.1139/g99-063.
- Vinogradov AE. 2003. DNA helix: The importance of being GC-rich. Nucleic Acid Research 31:1838–1844. DOI: 10.1093/nar/gkg296.
- Vinogradov AE. 2005. Genome size and chromatin condensation in vertebrates. Chromosoma 113:362–369. DOI: 10.1007/s00412-004-0323-3.
- Vinogradov AE, Anatskaya OV. 2006. Genome size and metabolic intensity in tetrapods: A tale of two lines. Proceedings of the Royal Society B: Biological Science 273:27–32. DOI: 10.1098/rspb.2005.3266.
- Womack MC, Metz MJ, Hoke KL. 2019. Larger genomes linked to slower development and loss of late-developing traits. The American Natural 194:654–664. DOI: 10.1086/705897.
- Xia X. 1995. Body temperature, rate of biosynthesis, and evolution of genome size. Molecular Biology and Evolution 12:834–842. DOI: 10.1093/oxfordjournals.molbev.a040260.