Abstract
The Mediterranean Sea is a hot spot for marine biodiversity, and this is particularly evident taking into consideration the diversity observed in many animal groups, among them the Molluscs. In the last decade, several works have revealed a high rate of cryptic diversity characterizing the Molluscan fauna of the Mediterranean Sea and an increasing number of endemic and/or new species inhabiting this semi-enclosed basin have been recorded or described. The DNA-barcoding method is considered an essential step in the integrative taxonomy applications, to unravel cryptic diversity and for species identification. Here we report the case of DNA-barcoding technique applied to identify a nudibranch (Heterobranchia) collected from the Adriatic Sea, in the Bay of Kotor (Montenegro), for which a standard morphological identification was not possible. Mediterranean specimen belonging to Pruvotfolia pselliotes (Labbé, 1923) is for the first time molecularly identified and its COI DNA sequence compared with the one of an individual collected from the type locality. In addition, this is the first verified report of this species from the Adriatic Sea. Finally, the potential of using DNA-barcoding is here discussed, together with the habitat and the geographical distribution of this uncommon species.
Introduction
The Mediterranean Sea is a semi-enclosed marine basin considered a hot spot for marine biodiversity (Coll et al. Citation2010; Danovaro et al. Citation2010; Lejeusne et al. Citation2010). In this context, Mollusca is one of the more represented group with about 2,200 accepted species (Coll et al. Citation2010), approximately 550 of which belonging to the Heterobranchia subclass (Trainito & Doneddu Citation2014). The study of Mediterranean Heterobranchia diversity is particularly interesting due to the continuous discovery of new and/or cryptic species, often endemic of this semi-closed marine basin (Furfaro & Trainito Citation2017; Martín-Hervás et al. Citation2019; Furfaro & Mariottini Citation2020). Within this group, the order Nudibranchia includes organisms with soft bodies and lacking shells, whose identification can be often difficult or not possible based on images or external anatomy. Moreover, samples immediately preserved in ethanol for molecular studies can unavoidably undergo the loss of their colours, which in some species is of diagnostic importance. DNA barcoding can support the specific identification of specimens collected with destructive sampling methods, or in case where taxonomically important characters are not properly visible. This molecular approach is routinely used to study biodiversity, and nowadays is considered an essential step in the integrative taxonomy applications to many animal taxa (e.g. Mastrototaro et al. Citation2019, Citation2020; Furfaro & Mariottini Citation2020). This paper reports the DNA-barcoding applied to the identification of the nudibranch Pruvotfolia pselliotes (Labbé, 1923) in the Mediterranean Sea, with the first documented record of the species in the Adriatic Sea. This Facelinidae nudibranch has been previously observed in the Atlantic Ocean, from south England to Ghana, including Cape Verde, the Canary Islands, and the Mediterranean Sea (Ortea & Urgorri Citation1981; Moro et al. Citation1995; Rolán Citation2005; Edmunds Citation2015). In this latter basin, P. pselliotes has been reported from Maltese waters (Mifsud & Cachia Citation2011), the Levantine Sea (Karhan et al. Citation2010) and the Marmara Sea (Albayrak & Çağlar Citation2016). The species has been also observed along the coasts of Spain and off Split (Croatia), according to the OPK platform (https://opistobranquis.info/ca), although without scientific evidences. In fact, the few Mediterranean records reported thus far refer mostly to photos and in vivo observations, while a proper morphological or molecular analysis has not been carried out thus far. Considering the high number of Mediterranean cryptic species and the lack of genetic data from populations of P. pselliotes inhabiting this basin, here we report for the first time results from molecular comparison between Mediterranean specimen and the one from the Atlantic type locality. Finally, since a clear-cut morphological identification is often not possible, the potential of using DNA-barcoding is here discussed, together with the habitat and the geographical distribution of the species.
Materials and methods
Sampling was carried out by scuba diving during January 2018 in the Bay of Kotor, Montenegro (42°28.791ʹN, 18°42.708ʹE) (), at 40 m depth. The Bay is composed by three major basins, connected by two narrow straits with a maximum depth of 67 m and a penetration of the sea to the inland for over 20 km (Bortoluzzi et al. Citation2017). The sampling site is located in the central sector of Morinj-Risan-Kotor Bay, characterized by several depressions of circular to elliptical shape that display a well-developed, funnel-shape vertical section, representing groundwater discharge such as spring outlets (Bortoluzzi et al. Citation2017). The nudibranch collected was immediately preserved in 95% ethanol (EtOH), and deposited with voucher MO18-46 in the Zoology Laboratory of the University of Bari.
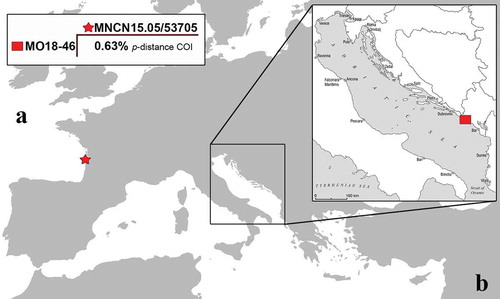
Figure 1. Map showing the sampling localities. The red star highlights the type locality of Pruvotfolia pselliotes from where the GenBank specimen was collected, while the red square indicates the sampling locality of the Mediterranean specimen here studied. In the upper left box is reported the COI divergence (p-distance) between the two compared specimens.
DNA was extracted from a small piece of tissue by using the “salting out” procedure (Aljanabi & Martinez Citation1997). Amplifications were performed by PCR using universal primers: LCO1490 and HCO2198 (Folmer et al. Citation1994) for the barcode fragment of the Cytochrome Oxidase subunit I (COI). PCR conditions were: 5 min of initial DNA denaturation step at 94°C; 35 cycles of 94°C/30 s (DNA denaturation step), 48°C/60 s (annealing step), 72°C/60 s (elongation step); and 7 min of final extension at 72°C (Furfaro et al. Citation2016). All amplicons were sequenced at the European Division of Macrogen Inc. (Amsterdam, The Netherlands). Sequences from each DNA strain were assembled and edited with Staden Package 2.0.0b9 (Staden et al. Citation2000). BLASTN (Altschul et al. Citation1990) search was conducted in the GenBank database to confirm the identity of the sequenced fragment and to exclude contamination. Consensus sequence was aligned together with GenBank (https://www.ncbi.nlm.nih.gov/nucleotide/) sequence using the Muscle algorithm implemented in MEGA 6.0 (Tamura et al. Citation2013). Since the COI mitochondrial marker is the most used for species barcoding in Heterobranchia, a comparison between the new sequence here reported and the one already deposited in GenBank was carried out. The number of COI base differences per site from averaging over the sequence pair was calculated, and the mean uncorrected p-distances were obtained using MEGA 6.0 software.
Results
Collected specimen showed an elongated body, a series of single cerata, lamellate rhinophores shorter than the elongated oral tentacles and a transparent oral veil, features commonly shared by representatives of the Facelinidae family. Furthermore, it presented a translucent white to yellow body colour pattern with the apical portion of the oral tentacle and the rhinophores being from yellowish to light brown. The brown ramifications of the digestive gland are visible through the transparent coating of cerata. Unfortunately, the bad weather conditions of the sea during the sampling did not allow to take a picture of the specimen collected and did not permit the more detailed in vivo observation of the diagnostic characters before it was stored in alcohol solution. The DNA barcoding was then considered the more suitable method to obtain a clear-cut identification of the specimen collected. The COI sequence obtained from the voucher MO18-46 were 650 bp (base pairs of nucleotides) has been deposited in GenBank with the accession number LR813620. The only one specimen belonging to P. pselliotes present in GenBank (voucher: MNCN15.05/53705) was deposited by Carmona et al. (Citation2011) and was collected from Cap Ferret in France, Atlantic Ocean (COI accession number: HQ616762). The resulting alignment was 634 bp with four variable sites. The comparison between the new COI sequence here reported and the one present in the GenBank showed 0.63% of COI p-distance value ().
Discussion
Taxonomic identification of rare or uncommon species is often based on one single specimen, due to the substantial difficulty of collecting biological material. Immediate storage in EtOH for future molecular studies allows DNA to be correctly preserved but inevitably causes the loss of some morphological traits, as well as the strong contraction of the specimens, which can be hard to identify basing only on morphology. Albeit it has been found from 6 to 120 m depth in the Mediterranean Sea (Karhan et al. Citation2010; Mifsud & Cachia Citation2011), its records are very few and often uncertain. Zenetos et al. (Citation2016) have reviewed the Adriatic opisthobranch fauna, providing an updated checklist of the taxa occurring in this distinguished basin of the Mediterranean Sea, in particular adding 15 species to the previous Montenegrin Heterobranchia inventory counting only 26 taxa. These authors also mentioned that the coastline of Montenegro was indeed poorly studied in the past and the few references focused on the molluscan biodiversity inhabiting this country provided limited information (Stjepčević Citation1967; Stjepčević & Parenzan Citation1980, Citation1982). The last new records by Petović and Lipej (Citation2017) and Petović (Citation2018) added 4 nudibranch species to the Montenegrin heterobranchs checklist. Although the Heterobranchia fauna of Montenegro has been recently revised and updated (Jovanović et al. Citation2019), P. pselliotes was not found thus far. This study reports the first documented record of P. pselliotes in the Adriatic Sea, adding an additional species to the Montenegrin list, and the first application of DNA barcoding to Mediterranean specimen. The genetic comparison between Mediterranean specimen and the one from Cape Ferret, ca. 300 km far from the type locality (Le Croisic, France, Atlantic Ocean), in fact, resulted in 0.63% of COI p-distance that is within the range of intraspecific divergence commonly used for species identification in Heterobranchia (<3%, see Furfaro et al. Citation2018). Considering the bathymetric range of distribution of P. pselliotes, from shallow to mesophotic seabed, as well as its Atlantic-Mediterranean areal, it is expected that further records of the species will occur all over the Mediterranean Sea. The ongoing exploration of mesophotic environments and the concomitant increasing use of the DNA-barcoding approach could support new occurrences of this species, as well as of other rare or unknown marine taxa.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Albayrak S, Çağlar S. 2016. Mollusca fauna of the Sea of Marmara. In: Özsoy E, Çağatay MN, Balkıs N, Balkıs N, Öztürk B, editors. The Sea of Marmara; Marine biodiversity, fisheries, conservation and governance. Vol. 42. Istanbul (Turkey): Turkish Marine Research Foundation (TUDAV). pp. 503–516.
- Aljanabi SM, Martinez I. 1997. Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Research 25(22):4692–4693. DOI: 10.1093/nar/25.22.4692.
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. Journal of Molecular Biology 215:403–410. DOI:10.1016/S0022-2836(05)80360-2.
- Bortoluzzi G, Giglio F, Ligi M, Del Bianc F, Ferrante V, Gasperini L, Ravaioli M. 2017. The seafloor geomorphology of Boka Kotorska Bay. IMEKO International Conference on Metrology for The Sea, Naples (Italy), pp 246–251.
- Carmona L, Gosliner TM, Pola M, Cervera JL. 2011. A molecular approach to the phylogenetic status of the aeolid genus Babakina Roller, 1973 (Nudibranchia). Journal of Molluscan Studies 77(4):417–422. DOI: 10.1093/mollus/eyr029.
- Coll M, Piroddi C, Steenbeek J, Kaschner K, Ben Rais Lasram F, Aguzzi J, Ballesteros E, Bianchi CN, Corbera J, Dailianis T, Danovaro R, Estrada M, Froglia C, Galil BS, Gasol JM, Gertwagen R, Gil J, Guilhaumon F, Kesner-Reyes K, Kitsos MS, Koukouras A, Lampadariou N, Laxamana E, López-fé de la Cuadra CM, Lotze HK, Martin D, Mouillot D, Oro D, Raicevich S, Rius-Barile J, Saiz-Salinas JI, San Vicente C, Somot S, Templado J, Turon X, Vafidis D, Villanueva R, Voultsiadou E. 2010. The biodiversity of the Mediterranean Sea: Status, patterns and threats. PLoS One 5(8):e11842.
- Danovaro R, Corinaldesi C, D’Onghia G, Galil B, Gambi C, Gooday AJ, Lampadariou N, Luna GM, Morigi C, Olu K, Polymenakou P. 2010. Deep-sea biodiversity in the Mediterranean Sea: The known, the unknown, and the unknowable. PLoS One 5(8):e11832. DOI: 10.1371/journal.pone.0011832.
- Edmunds M. 2015. Opisthobranchiate Mollusca from Ghana: Facelinidae. Journal of Conchology 42(2):125–161.
- Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology 3:294–299.
- Furfaro G, Mariottini P. 2020. A new Dondice Marcus Er. 1958 (Gastropoda: Nudibranchia) from the Mediterranean Sea reveals interesting insights into the phylogenetic history of a group of Facelinidae taxa. Zootaxa 4731(1):1–22. DOI: 10.11646/zootaxa.4731.1.1.
- Furfaro G, Picton B, Martynov A, Mariottini P. 2016. Diaphorodoris alba Portmann & Sandmeier, 1960 is a valid species: Molecular and morphological comparison with D. luteocincta (M. Sars, 1870) (Gastropoda: Nudibranchia). Zootaxa 4193(2):304–316. DOI: 10.11646/zootaxa.4193.2.6.
- Furfaro G, Salvi D, Mancini E, Mariottini P. 2018. A multilocus view on Mediterranean aeolid nudibranchs (Mollusca): Systematics and cryptic diversity of Flabellinidae and Piseinotecidae. Molecular Phylogenetic and Evolution 118:13–22. DOI:10.1016/J.YMPEV.2017.09.001.
- Furfaro G, Trainito E. 2017. A new species from the Mediterranean Sea and North-Eastern Atlantic Ocean: Knoutsodonta pictoni n. sp. (Gastropoda Heterobranchia Nudibranchia). Biodiversity Journal 8(2):725–738.
- Jovanović M, Mačić V, Trkov D, Orlando-Bonaca M, Lipej L. 2019. Review of heterobranch molluscs fauna in the Boka Kotorska Bay, Montenegro. Acta Adriatica 60:115–126. DOI:10.32582/AA.60.2.1.
- Karhan SÜ, Kalkan E, Yokes MB, Balkis H, Dalyan C. 2010. On a collection of opisthobranchs (Mollusca, Gastropoda) from the Levantine coast of Turkey. Rapports et procés-verbaux des réunions Commission internationale pour l’exploration scientifique de la Mer Méditerranée 39:555.
- Lejeusne C, Chevaldonné P, Pergent-Martini C, Boudouresque CF, Pérez T. 2010. Climate change effects on a miniature ocean: The highly diverse, highly impacted Mediterranean Sea. Trends in Ecology & Evolution 25(4):250–260. DOI: 10.1016/j.tree.2009.10.009.
- Martín-Hervás MR, Carmona L, Jensen KR, Licchelli C, Vitale F, Cervera JL. 2019. Description of a new pseudocryptic species of Elysia Risso, 1818 (Heterobranchia, Sacoglossa) in the Mediterranean Sea. Bulletin of Marine Science 96(1):127–143.
- Mastrototaro F, Montesanto F, Salonna M, Grieco F, Trainito E, Chimienti G, Gissi C. 2019. Hitch-hikers of the sea: Concurrent morphological and molecular identification of Symplegma brakenhielmi (Tunicata: Ascidiacea) in the western Mediterranean Sea. Mediterranean Marine Science 20:197–207.
- Mastrototaro F, Montesanto F, Salonna M, Viard F, Chimienti G, Trainito E, Gissi C. 2020. An integrative taxonomic framework for the study of the genus Ciona (Ascidiacea) and description of a new species, Ciona intermedia. Zoological Journal of the Linnean Society, in press. DOI: 10.1093/zoolinnean/zlaa042.
- Mifsud C, Cachia C. 2011. New additions and corrections, with annotations, to the check-list of the marine Mollusca of the Maltese Islands. Triton 23:10–18.
- Moro L, Ortea J, Bacallado JJ, Valdes A, Pérez Sanchez JM. 1995. Nuevos aeolidaceos (Gastropoda, Nudibranchia) para la fauna de Canarias. Revista de la Academia Canaria de Ciencias 7:63–75.
- Ortea JA, Urgorri V. 1981. Runcina ferruginea Kress 1977, et Pruvotfolia pselliotes (Labbe, 1923) dans les eaux ibériques. Vie et Milieu 31(2):149–151.
- Petović S. 2018. Additions to the checklist of the malacofauna of the Boka Kotorska Bay (south-east Adriatic Sea). Studia Marina 31(1):23–36.
- Petović S, Lipej L. 2017. New Mediterranean record of the rare sea slug Thecacera pennigera (Montagu, 1815), (Nudibranchia,Polyceridae). Mediterranean Marine Science. 15:198–212.
- Rolán E. 2005. Malacological Fauna From The Cape Verde Archipelago. Part 1. Polyplacophora and Gastropoda. Conchbooks, Germany. p. 455.
- Staden R, Beal KF, Bonfield JK. 2000. The Staden package, 1998. Methods in Molecular Biology 132:115–130.
- Stjepčević J. 1967. Macro-mollusca of boka kotorska bay. Studia Marina 2:1–64.
- Stjepčević J, Parenzan P. 1980. Il Golfo delle Bocche di Cattaro. Condizioni generali e biocenosi bentoniche con carta ecologica delle sue due baie interne di Kotor (Cattaro) e Risan (Risano). Studia Marina 9−10:3–148.
- Stjepčević J, Parenzan P. 1982. Survey on benthic Mollusca population of the inner part of the Boka Kotorska Bay. Studia Marina 11−12:3–28.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular Biology and Evolution 30:2725–2729. DOI:10.1093/MOLBEV/MST197.
- Trainito E, Doneddu M. 2014. Nudibranchi del Mediterraneo; Il Castello. Milan (Italy). p. 192.
- Zenetos A, Vesna Mačić V, Andrej Jaklin A, Lipej L, Poursanidis D, Cattaneo-Vietti R, Beqiraj S, Betti F, Poloniato D, Kashta L, Katsanevakis S, Crocetta F. 2016. Adriatic ‘opisthobranchs’ (Gastropoda, Heterobranchia): Shedding light on biodiversity issues. Marine Ecology 37(6):1239–1255. DOI: 10.1111/maec.12306.
