Abstract
The Mesopotamian spiny eel, Mastacembelus mastacembelus, an endemic freshwater fish restricted to the Euphrates and Tigris river drainages of Mesopotamian, and its related taxa in Afrotropica and South Asia present suitable elements for studies of the evolutionary history of mastacembelids across these heterogeneous areas. To understand and clarify the geographic variation of the Mesopotamian spiny eel from the Persian Gulf basin, we investigated the molecular and morphological characteristics of individuals collected from its whole distribution range in the Tigris (Karkheh, Karoon and Zab), Zohreh and Persis (Mond and Helleh) main river drainages in Iran. The morphological analysis revealed a separation of the studied populations; however, the genetic distances were relatively small and the molecular tree did not present a distinct branching pattern for the main river drainages based on the COI mitochondrial marker. Six haplotypes were identified: Helleh, Mond, Karkheh, Karoon, Zab and Zohreh tributaries each showed a separate haplotype. The COI marker indicates that the Mesopotamian eel has greater affinity with the African species than with the Southeast Asian species. Estimation of divergence times indicates the Mesopotamian spiny eel apparently diverged about 1 million years ago (Mya) (95% highest posterior density [HPD]: from 390,000 years ago to 2.1 Mya) in different river drainages of the Persian Gulf basin.
Introduction
Freshwater spiny eels/mastacembelids (Teleostei: Synbranchiformes: Mastacembelidae) are a predominantly riverine family of freshwater fish distributed in the Old World from the tropical parts of Africa to Southeast Asia, containing the three genera Macrognathus, Sinobdella and Mastacembelus and 93 species (Coad Citation2015; Hastings et al. Citation2015; Fricke et al. Citation2021).
To explore the diversity, monophyly and colonization history of Mastacembelus eels, the first molecular phylogenetic analysis of this group was performed by Brown et al. (Citation2010), using a multigene dataset of mitochondrial and nuclear genes applying several methods. The study highlighted the Africa–Asia split of mastacembelid eels at ~19 Mya, which is considerably younger than the split between the two continents, suggesting a dispersal scenario for their current distribution. Also based on the ancestral range estimation, it has been demonstrated that mastacembelids and their constituent genera originated and diverged preliminary in Asia, with Mastacembelus suggested to have reached Africa (the Afrotropical Mastacembelus lineage) shortly before its divergence from the Mesopotamian/Middle East spiny eel, Mastacembelus mastacembelus, between 27.5 and 12 Mya (Lavoué Citation2019). However, Day et al. (Citation2017) concluded the monophyly of an African Mastacembelus radiation plus the Mesopotamian spiny eel, M. mastacembelus (with uncertain position), and highlighted the origination of the spiny eel in Asia and its colonization of Africa c. 15.4 Mya (95% highest posterior density [HPD]: 23.9–8.8 Mya), presumably through the Middle East.
The Mesopotamian spiny eel is interesting in term of phylogeography, and it has not been well studied in the Middle East. The morphological status of the Mesopotamian spiny eel populations from Karakay Reservoir, Tohma Stream and Tigris River were investigated by Çakmak and Alp (Citation2010) using morphometric and meristic traits. Tutar (Citation2015) compared three populations of the species from the Turkish part of the Tigris River drainage based on the combined mitochondrial DNA (12S rRNA, 16S rRNA, tRNAPhe and tRNAVal genes). However, morpho-molecular variations of the species have not been investigated among the Iranian populations so far, although Iran includes a large part of its distribution. Iranian populations of this species inhabit different river tributaries and habitats with different ecological conditions that have prompted us to initiate a more detailed investigation, both morphologically and molecularly. Hence, this study aims to understand and clarify the geographic variations of the Mesopotamian spiny eel from the Persian Gulf basin, using integrated molecular and morphological characteristics of the individuals collected from its whole distribution range in the Iranian part of the Persian Gulf basin.
Materials and methods
Sampling
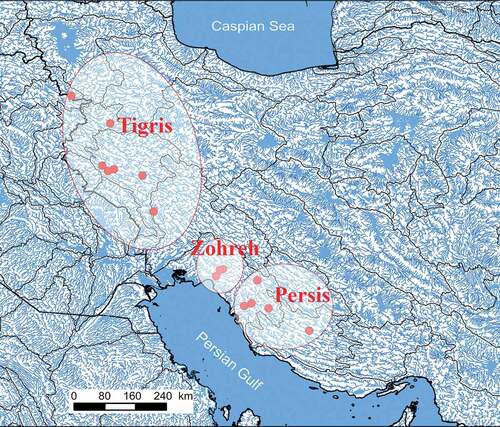
A total of 151 specimens of Mesopotamian spiny eel were collected from its whole distribution range in the Tigris, Zohreh and Persis (Mond and Helleh) river drainages (Persian Gulf basin) in Iran using an electrofishing device (ELEMAX, SHX 2000) during fieldwork in 2016 and 2017 (; ). An attempt to obtain samples in the Kor River basin was not successful (see Discussion). After anesthesia with 1% clove oil solution, the pectoral fin (or, from some specimens, tissue from below the dorsal fin) on the right side of each specimen was removed and preserved in 96% ethanol for the molecular analyses. A total of 126 adult individuals were fixed in 10% formaldehyde for morphological studies. Voucher specimens were deposited in the Zoological Museum, Collection of Biology Department, Shiraz University (ZM-CBSU).
Table I. Sampling sites of Mesopotamian spiny eel and details of specimens
Molecular analyses
DNA extraction, polymerase chain reaction and sequencing
A total of seven specimens of the Mesopotamian spiny eel from the Persis (Mond and Helleh) river drainages, four specimens from the Zohreh River drainage and 14 specimens from the Tigris River drainage were selected for molecular analyses (). Total genomic DNA was extracted from the preserved tissues using the standard salt extraction method (Bruford et al. Citation1992). We amplified the DNA barcode region of the mitochondrial COI marker using a polymerase chain reaction (PCR) procedure with the universal primers FishF1 and FishR1 (Sharina & Kartavtsev Citation2010) and the following amplification protocol: initial denaturation at 94°C for 5 min; 35 amplification reaction cycles consisting of denaturation at 94°C for 60s, annealing at 55°C for 60s, and extension at 72°C for 60s per cycle; and a final extension cycle at 72°C for 5 min. PCR products were evaluated with a 100 bp ladder on 1% agarose gel electrophoresis. Sequencing was carried out using a forward primer by Macrogen Service Centre (Seoul, South Korea). The obtained sequences were deposited in the national center for biotechnology information (NCBI) GenBank under specific accession numbers (see ).
Phylogenetic analyses
In addition to the samples sequenced here, sequences of one Mastacembelus species from southeastern Asia (LT577001.1 M. armatus) and another from Africa (LT576985.1 M. taiaensis) were retrieved from NCBI with M. armatus as the outgroup. We also included the only COI sequence of Mesopotamian spiny eel from the Middle East in the NCBI (with the accession number LT577004, from Turkey). The sequences were edited and aligned using BioEdit v. 7.0.0 (Hall Citation1999) and checked for the presence of unexpected stop codons using Mega 7 software (Kumar et al. Citation2016). The model suggested for the COI was determined using jModelTest 2.1.10 and GTR+G + I selected according to the Akaike information criterion. Bayesian inference (BI) was done using MrBayes 3.1.2 (Ronquist & Huelsenbeck Citation2003) with 10 million generations with four Markov chain Monte Carlo sampling and with a sampling frequency of 100; 25% of the trees were discarded as burn-in. A maximum likelihood (ML) (Felsenstein Citation1985) gene tree was inferred using RAxML 7.2.5 with 10,000 bootstrap replicates (Stamatakis Citation2006) and the GTR+G + I model to examine the robustness of the Bayesian results. Mean genetic distances among populations were calculated using the Kimura two-parameter model (K2P) implemented in Mega v. 7.0, and this software was also used to extract variable nucleotide sites from the sequence alignment.
Estimation of divergence times
Divergence times of Iranian populations were estimated using the time tree provided by Day et al. (Citation2017). The sequences examined here (belonging to other species) were retrieved from the NCBI, mostly from Day et al.’s (Citation2017) study. We included African and Asian species as monophyletic, and then three nodes from the previously published time tree were considered to calibrate the tree: mean age of the family at 29 Mya, mean age for divergence of Mastacembelus at 21 Mya and its divergence in Africa at 15.5 Mya. Rates among branches were assumed to follow a lognormal distribution. We specified a normal prior distribution for the mean age of the nodes with a standard deviation (SD) of 3 for the family, 2 for the genus and 1.5 for the African clade using BEAST v. 1.7.2 software. The analysis was run for 100 million generations under the GTR+G + I model, with empirical base frequencies, Yule process and a random starting tree with parameters sampled every 10,000 steps, and Tracer v. 1.4 was used to assess convergence and effective sample size (ESS) following a preburn-in of 10%. Finally, we generated maximum clade credibility trees with mean node heights using TreeAnnotator v. 1.7.4, and the time tree was visualized in FigTree v. 1.4.4. We considered Macrognathus zebrinus the outgroup and included one sequence per haplotype from our 25 new sequences.
Morphological analyses
The morphological analyses were carried out using 126 specimens covering the whole distribution range of the Mesopotamian spiny eel in the Tigris River drainage (n = 71), the Mond and Helleh river drainages (n = 25) and the Zohreh River drainage (n = 30), which all drain into the Persian Gulf. Morphometric characters were measured with digital calipers and recorded to 0.01 mm. Methods for counts and measurements follow Plamoottil and Abraham (Citation2013) and include the following 30 characters: total length (TL), standard length (SL), head length (HL), head depth (HD), head width (HW), mouth width (MW), eye diameter (ED), interorbital distance (InOD), preorbital distance (Prod), postorbital distance (POD), predorsal distance (Prdd), preanal distance (Prad), length of dorsal fin (Ldf), length of anal fin (Laf), length of pectoral fin (Lpcf), length of caudal fin (LCF), distance between pectoral and anal fin (Dpcaf), maximum body depth (MaxBD), minimum body depth (MinBD), length of base of pectoral fin (BLPF), length of spinous anal fin ray (LSAF), length of base of soft anal fin ray (LBSAF), length of base of soft dorsal fin ray (LBSODF), length of soft dorsal fin ray (LSDF), height of soft dorsal fin (Hsdf), height of soft anal fin (HSAF), height of anal fin spine (HAS), depth of dorsal fin (Ddf), depth of anal fin (DAF) and length of base of spinous dorsal fin (LBSpF). In addition, five meristic characters were counted under a stereomicroscope as follows: the numbers of dorsal fin spines (DFS), anal fin spines (AFS), pectoral fin rays (PFR), anal fin rays (AFR) and dorsal fin rays (DFR).
To eliminate any size effects, the morphometric data were standardized according to the equation (Mmp/SL)*100, where Mmp: measured morphometric parameter, and SL: standard length, except for head characters (HD, HW, ED, Prod, POD, InOD) that divide into HL (head length). Meristic characters are independent of fish size and were used as raw data. Canonical discriminant analysis (CDA) was carried out to determine the success of classification based on the morphometric measurements and meristic counts separately, using IBM SPSS v. 26. Principal component analysis (PCA) was also performed, to assess individual relationships within and between populations, based on the morphometric and meristic characters separately. To examine the suitability of the data for PCA, the Kaiser–Meyer–Olkin (KMO) measure of sampling adequacy and Bartlett’s test of sphericity were performed (KMO: 0.72; P < 0.001) using IBM SPSS v. 26. Then, PCA analysis was performed to visualize individual relationships using PAST 3.
Results
Phylogenetic analyses
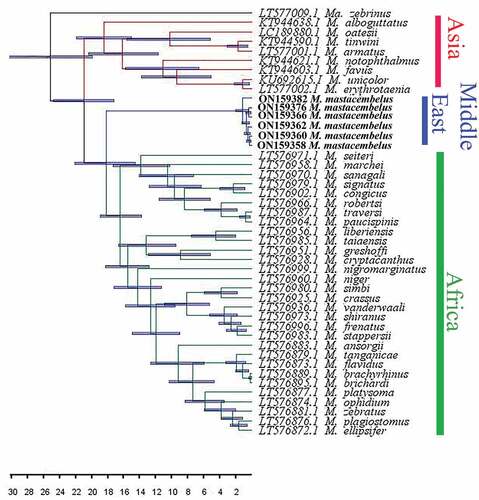
The COI sequences including 536 nucleotides were obtained from 25 specimens of the Mesopotamian spiny eel, of which 526 nucleotides were constant and 10 were parsimony informative. Two different phylogenetic analyses (BI, ML) for Mastacembelus spp. produced nearly identical topologies; thus, only the topology from the ML analysis is presented, along with the statistical values from the ML and BI algorithms ().
Figure 2. Phylogenetic estimates for Mastacembelus mastacembelus based on the COI mitochondrial marker. The topology is taken from the ML, and numbers above branches are bootstrap values/posterior probabilities. ML: maximum likelihood.
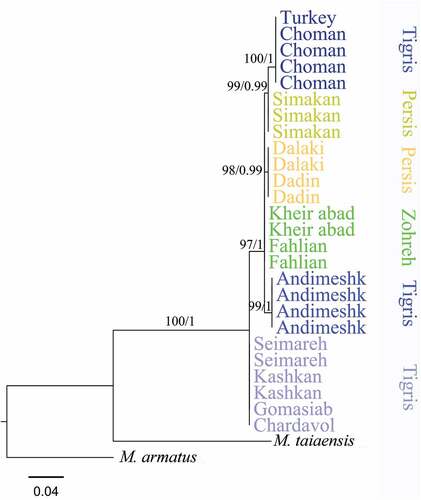
Some molecular variations were recovered in the Iranian Mesopotamian spiny eel. lists the 10 variable nucleotide sites found in the COI barcode region. Specimens of Zohreh and Persis are more similar to each other than to the Tigris, and Tigris specimens including Karkheh, Karoon and Zab tributaries showed more molecular variations (). Specimens of Karkheh tributary (Gomasiab, Seimareh, Kashkan, Chardavol) showed a separation from the rest of the samples (Zohreh, Mond, Zab, Helleh, Karoon tributaries) (). Specimens of the three main river systems (Tigris, Zohreh, Persis) were not nested in three separate groups; however, six haplotypes were identified in total and Helleh, Mond, Karkheh, Karoon, Zab and Zohreh tributaries each showed a separate haplotype.
Table II. Variable sites of Mesopotamian spiny eel relative to KY609156.1 Mastacembelus armatus complete mitochondrial genome
We found genetic distances (K2P) of between 0.2% and 1.3% for the COI among the tributaries (). The maximum genetic distance was observed between specimens of Kharkeh tributary and Choman. The genetic distances among Tigris, Zohreh and Persis populations varied from 0.2% to 0.7%.
Table III. Average genetic distance between pairs of tributaries based on Kimura two-parameter (K2P). Bold values report maximum and minimum distance values between tributaries
Divergence time
Inclusion of Macrognathus as outgroup and Asian and African species as monophyletic clades suggested M. mastacembelus is the sister of the African mastacembelids (). The diversification of Mastacembelus in the Middle East is relatively recent compared with diversification in Africa, and the Mesopotamian spiny eel diverged apparently about 1 Mya (95% HPD: from 390,000 years ago to 2.1 Mya) in different river drainages of the Persian Gulf basin.
Morphological analyses
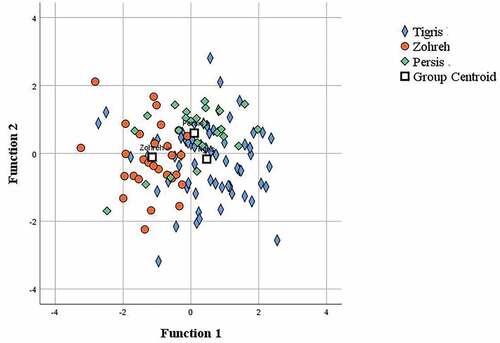
All traits were subjected to the CDA, which showed the random assignment of individuals to their basin was 85.7% based on the morphometric characters () and 57.9% for meristic characters (). The classifications were 85.9% for Tigris, 92% for Persis and 80% for Zohreh populations based on the morphometric characters and 54.9% for Tigris, 70% for Zohreh and 68% for Persis based on the meristic characters. Discriminant functions (DF) 1 and 2 together explained 100% of the variance, with DF1 accounting for 68.7% and DF2 31.3% of all variance for the morphometric characters; and with DF1 explaining 82.2% and DF2 17.8% of all variance for the meristic characters. The test of functions for DF 1 and 2 was significant for both morphometric and meristic data (Wilks’s lambda, P < 0.001).
Figure 4. Scatter plot of the canonical discriminant analysis based on morphometric variables of the Mesopotamian spiny eel from three river drainages of Iran.
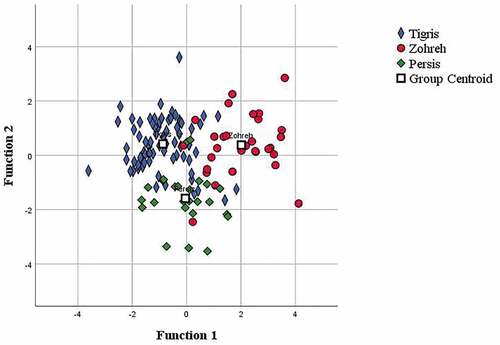
Figure 5. Scatter plot of the canonical discriminant analysis based on meristic variables of the Mesopotamian spiny eel from three river drainages of Iran.
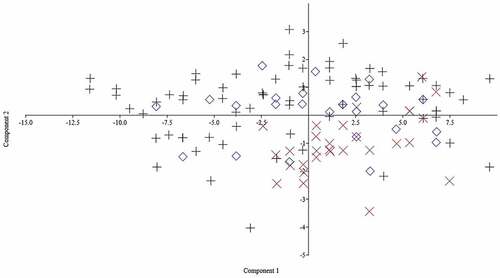
The PCA carried out on the morphometric measurements extracted 13 factors with eigenvalues > 1, explaining 94.39% of the variance. The first principal component (PC) described 37.47% of the variation and the second 14.96%. Therefore, the first PC was the most important component contributing to separation among the basins. The most significant loadings on PC 1 were HW and MW and on PC 2 they were HD and HW, all from head characters. With regard to factor loading, values above 0.30 are considered significant (Lombarte et al. Citation2012); in the present study significant factors on PC1 were HW and MW and significant factors on PC2 were HD, HW and LBSpF. A visual inspection of the plotted PC1 and PC2 scores showed a high degree of overlap among the three basins ().
Figure 6. Principal components analysis (PCA) of morphometric characters of Mesopotamian spiny eel from three river drainages of Iran. Plus symbol: Tigris; multiplication symbol: Zohreh; diamond: Persis.
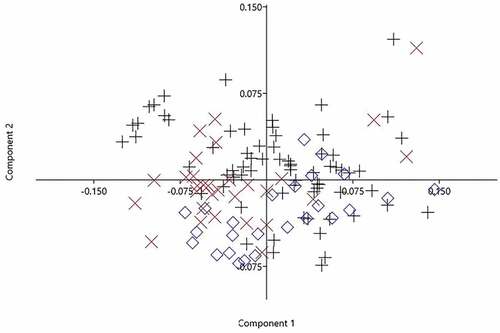
PCA on the meristic measurements extracted three factors with eigenvalues > 1, explaining 97.85% of the variance. The first PC described 88.06% of the variation and the second 5.97%. The most significant loadings on PC 1 were AFR and DFR, and the most significant loading on PC 2 was DFS ().
Discussion
The Middle Eastern species M. mastacembelus is restricted to the inland waters of the Middle East, occurring in a wide range of isolated water bodies in the Euphrates and Tigris river drainages/Mesopotamia, in Iran, Turkey, Syria and Iraq (Çakmak & Alp Citation2010; Coad Citation2015). The Mesopotamian spiny eel and its relatives in Afrotropica and south Asia present suitable elements for studies of the evolutionary history of mastacembelids across these heterogeneous areas. Day et al. (Citation2017) included the monophyly of an African Mastacembelus radiation plus the Middle Eastern species, M. mastacembelus, based on five molecular markers and found that the position of this taxon is uncertain (as sister to the African radiation or nested within it, although support for the former scenario was stronger).
Mastacembelus is suggested to have diverged sometime during the Early Miocene, coinciding with the closure of the Tethys Sea c. 18–20 Mya (Day et al. Citation2017). During the Miocene and Pliocene, the major Levantine river systems and Euphrates were connected to each other (Alwan et al. Citation2016). Probably via headwater capture, the genus entered from the Tigris–Euphrates river system to rivers of southern Turkey and the Levant and subsequently disappeared from the Levant and North Africa. Mastacembelus is not found in eastern Iran. According to the above scenario and distribution pattern of Mastacembelus, this genus may have reached the Tigris–Euphrate basin across the Iranian land mass followed by range loss in eastern Iran, as many rivers in eastern Iran are saline and regularly dry up. However, past and present species distribution modeling is suggested.
Presently available data on the distribution of Mesopotamian spiny eel reveals that in Iran the species is restricted to Tigris, Zohreh and Persis drainages. We did not record M. armatus from the Kor River drainage in our extensive survey, in contrast to findings by Mokhayer (Citation1981). Coad (Citation2015) noted that the record of M. armatus by Mokhayer (Citation1981) from the Kor River basin is probably a misidentified M. mastacembelus. There were also no observations of the Mesopotamian spiny eel from the Kor River in our fieldwork or in the literature in recent decades.
Although significant differences in morphometric traits were detected among some populations of the Mesopotamian spiny eel collected from Karakaya Reservoir, Tohma Stream in Euphrates and Tigris river in Diyarbakır by Çakmak and Alp (Citation2010), molecular differences among populations of the species have not previously been investigated, except for a comparison by Tutar (Citation2015) of the three populations from the Turkish part of the Tigris basin based on the combined mitochondrial DNA (16S rRNA, 12S rRNA and tRNA genes), in which no differences were found. According to our CDA results, there was a significant difference in morphometric characters of eels from different basins which could be used with great accuracy to distinguish the Tigris, Zohreh and Persis populations. According to the PCA results, although visual inspection of plotted PC1 and PC2 scores showed a high degree of overlap among the three basins, some characters of the head (HW, HM) showed significant differences among basins (sampled eels from the Zohreh basin have smaller measurements for these characters than those from the two other basins) that may be interpretable from an adaptive perspective as being caused by different ecological conditions.
The genetic distance analysis showed relatively low genetic distances among Iranian tributaries. In concordance with the genetic distance analysis, the phylogenetic trees did not show any congruent pattern including separate clusters of the Tigris, Zohreh and Persis (although all tributaries formed separate groups), revealing that the morphometric differences may be independent of COI genetic distances, highlighting a need for further molecular marker studies in the future. In any case, age estimation of the splitting of Mesopotamian spiny eel from African species, observation of different haplotypes and molecular variations of this species in the inland waters of Iran, along with the morphological differences among major river systems, may indicate a long presence of this genus in Iranian waters and may confirm the possible scenario of dispersal from Southeast Asia to Africa through Iran. More accurate conclusions will be reached when more studies are conducted on the populations of this species in the Middle East. Another reason for the morphological and molecular variations of this species in Iran may be related to the high climatic and habitat diversity from southern to western Iran. To reconstruct the distribution route of the genus Mastecembelus, population genetic studies with a sufficient number of samples and past climatic modeling are suggested.
In conclusion, from the results of the molecular divergence time of masacemblids based on COI, it can be concluded that the Mesopotamian spiny eel diverged apparently about 1 million years ago (Mya) in different river drainages of the Persian Gulf basin.
Separation among the populations of the Mesopotamian spiny eel from the different basins of Iran was shown based on morphometry; however, the genetic distances were relatively low based on COI for the main river drainages. Access to more data on this species in Mesopotamia and more comprehensive analyses would be helpful to indicate the route of Mestacembelus dispersal from Southeast Asia to Africa.
Acknowledgements
The project was financially supported by Shiraz University and was approved by the Ethics Committee of the Biology Department of the university (SU 9331009). We thank R. Sadeghi. H. Mehraban, M. Masoudi, H. Darvishnia, G. Sayyadzadeh and H. Zareian for their help in sampling. We also thank F. Zarei for help in preparing .
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Alwan N, Esmaeili HR, Krupp F. 2016. Molecular Phylogeny and Zoogeography of the Capoeta damascina species complex (Pisces: Teleostei: Cyprinidae). PLoS ONE 11:e0156434. DOI: 10.1371/journal.pone.0156434.
- Brown KJ, Ruber L, Bills R, Day JJ. 2010. Mastacembelid eels support Lake Tanganyika as an evolutionary hotspot of diversification. BMC Evolutionary Biology 10:188. DOI: 10.1186/1471-2148-10-188.
- Bruford MW, Hanotte O, Brokfield JFY, Burke T. 1992. Single-locus and multilocus DNA fingerprinting, and Hoelzel AR, editor. Molecular genetic analysis of populations: A practical approach. New York: Oxford University Press. pp. 225–269.
- Çakmak E, Alp A. 2010. Morphological differences among the mesopotamian spiny eel, Mastacembelus mastacembelus (Banks and Solander 1794), populations. Turkish Journal of Fisheries and Aquatic Sciences 10:87–92. DOI: 10.4194/trjfas.2010.0113h.
- Coad WB. 2015. Review of the Spiny Eels of Iran (Family Mastacembelidae). Iranian Journal of Ichthyology 2:1–12. DOI: 10.22034/iji.v2i1.7.
- Day JJ, Fages A, Brown KJ, Vreven EJ, Stiassny MLJ, Bills R, Friel JP, Rüber L. 2017. Multiple independent colonizations into the Congo Basin during the continental radiation of African Mastacembelus spiny eels. Journal of Biogeography 44:2308–2318. DOI: 10.1111/jbi.13037.
- Felsenstein J. 1985. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39:783–791. DOI: 10.2307/2408678.
- Fricke R, Eschmeyer WN, van der Laan R, editors. 2021. Eschmeyer’s catalog of fishes: Genera, species, references. Online version, updated June 2 2020. San Francisco, CA: California Academy of Sciences. http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp.
- Hall TA. 1999. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium 41:95–98. DOI: 10.14601/Phytopathol_Mediterr-14998u1.29.
- Hastings PA, Walker HJ, Galland GR. 2015. Fishes: A guide to their diversity. USA: University of California Press, United States.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33(7):1870–1874. DOI: 10.1093/molbev/msw054.
- Lavoué S. 2019. Origins of Afrotropical freshwater fishes. Zoological Journal of the Linnean Society 188:345–411. DOI: 10.1093/zoolinnean/zlz039.
- Lombarte A, Gordoa A, Whitfield AK, James NC, Tuset VM. 2012. Eco morphological analysis as a complementary tool to detect changes in fish communities following major perturbations in two South African estuarine systems. Environmental Biology of Fishes 94(4):601–614. DOI: 10.1007/s10641-011-9966-0.
- Mokhayer B. 1981. Habitat of spiny eels in Iranian water with a note on their helminthic infection. Journal of the Veterinary Faculty of the University of Tehran 36:35–47. In Farsi.
- Plamoottil M, Abraham NP. 2013. Rediscovery of Mastacembelus malabaricus after one and half century. Research Journal of Animal, Veterinary and Fishery Sciences 1:6–11.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574. DOI: 10.1093/bioinformatics/btg180.
- Sharina S, Kartavtsev Y. 2010. Phylogenetic and taxonomic analysis of flatfish species (Teleostei, Pleuronectiformes) inferred from the primary nucleotide sequence of cytochrome oxidase 1 gene (Co-1). Russian Journal of Genetics 46:356–361. DOI: 10.1134/S1022795410030130.
- Stamatakis A. 2006. RAxML-VI-HPC: Maximum likelihood based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. DOI: 10.1093/bioinformatics/btl446.
- Tutar E. 2015. Combined mitochondrial DNA analysis of the Mesopotamian spiny eel, Mastacembelus mastacembelus (Banks & Solander 1794), and its phylogenetic position. International Journal of Aquatic Biology 3:314–322. DOI: 10.22034/ijab.v3i5.110.