Abstract
The fin whale Balaenoptera physalus is a large rorqual species occurring worldwide, mainly in temperate and subpolar zones. In contrast to many baleen whales, not all the fin whale populations show the same model of migration. In fact, migratory behaviours of this latter species range from long seasonal migration between high and low latitudes to a complete non-migratory behaviour. A resident fin whale population was described in the Mediterranean Sea, which is also frequented by North Atlantic individuals entering through the Strait of Gibraltar in winter to feed. Between 2020 and 2021 three individuals initially identified as fin whales died along the Tyrrhenian coasts (Mediterranean Sea, Italy). Their mitochondrial DNA control region (mtDNA CR) was analysed and compared to fin whale haplotypes previously described in North Atlantic Ocean and Mediterranean Sea to identify their geographical origin. Our results show that two individuals most likely belong to the Mediterranean fin whale population, while an individual was recognised as a putative fin-blue whale hybrid (Balaenoptera physalus x Balaenoptera musculus) with a North Atlantic origin. The discovery of the first fin-blue whale hybrid in the Mediterranean Sea was confirmed by the analysis of a biparentally inherited marker, the α-lactalbumin (α-lac) nuclear gene, demonstrating that the morphological analysis alone does not allow to correctly identify hybrids, especially if intermediate characters of both parental species are not clearly distinguishable.
Introduction
The fin whale Balaenoptera physalus (Linnaeus 1758) is the second-largest whale in the world, reaching lengths of up to 27 m in the Southern Hemisphere (Mackintosh Citation1942) and 23 m in the Northern Hemisphere (Nichol & Heise Citation1992; Loy et al. Citation2019). The difference in body size between fin whales inhabiting the Southern and Northern Hemispheres validated the initial separation in two subspecies (Tomilin Citation1946): the Northern fin whale B. p. physalus (Linnaeus 1758) and the southern fin whale B. p. quoyi (Fischer 1929). Two additional subspecies were later proposed, the North Pacific fin whale B. p. velifera Cope 1869, recently separated from the North Atlantic fin whale (B. p. physalus) on the basis of genetic evidence (Archer et al. Citation2019) and the pygmy fin whale B. p. patachonica Burmeister 1865, inhabiting the Southern Hemisphere and characterised by a smaller size, dark body and black baleen (Clarke Citation2004). For this latter, however, the taxonomic status is still debated due to the lack of a genetic validation (Pérez-Alvarez et al. Citation2021).
In the open sea, B. physalus is often mistaken with other species of rorquals of similar size, such as blue whale B. musculus, sei whale B. borealis, Bryde’s whale B. edeni and Omura’s whale B. omurai (Jefferson et al. Citation2015; Committee on Taxonomy Citation2021). The characters that most help fin whale identification are the V-shaped head, the size and position of the dorsal fin and, above all, the body colouration. The fin whale is in fact characterised by a dark body with a light belly, several lighter V-shaped chevrons behind the head and an asymmetrical head colour, the left lower jaw is mainly dark while the right one is white (Jefferson et al. Citation2015). In the late nineteenth century, the capture during commercial whaling operations of individuals with unusual size and morphological characters suggested the occurrence of hybridization between fin and blue whales (Cocks Citation1887), later confirmed by genetic analysis (Árnason et al. Citation1991; Spilliaert et al. Citation1991; Bérubé & Aguilar Citation1998; Pampoulie et al. Citation2021).
Fin whales are common in the waters of the whole world, including the polar ones of both hemispheres, specifically at latitudes higher than 20°N in the North Pacific Ocean, 30°N in the North Atlantic Ocean and 20°S in the Southern Hemisphere. It is rarer in the tropics, except in areas with cold waters, like Peru, and it is almost absent in the equatorial region (Hamilton et al. Citation2009; Edwards et al. Citation2015). The equatorial hiatus observed in the global distribution suggests that fin whales do not perform the long seasonal migration typical of baleen whales (Edwards et al. Citation2015). The traditional Mysticete migration model predicts a movement between summer feeding grounds, located at high latitudes, and winter breeding grounds at lower latitudes (Kellogg Citation1929). However, several exceptions have been noticed over time to this pattern, from complete migration to non-migratory behaviour (Geijer et al. Citation2016). Resident populations of B. physalus, composed of individuals staying in the same area all year round, were described in the East China Sea, Norther Sea of Japan (Mizroch et al. Citation2009), Sea of Cortez (Tershy et al. Citation1993; Bérubé et al. Citation2002) and Mediterranean Sea (Bérubé et al. Citation1998; Geijer et al. Citation2016; Notarbartolo di Sciara et al. Citation2016).
In the Mediterranean Sea, the existence of an isolated and resident fin whale population was verified by the analysis of six microsatellite loci and a sequence of the mitochondrial DNA control region (mtDNA CR) (Bérubé et al. Citation1998). Results obtained from the mtDNA CR showed the presence of three private haplotypes in the Mediterranean Sea, and a significant degree of differentiation between North Atlantic and Mediterranean Sea samples, supporting the presence of a recently diverged Mediterranean population (Bérubé et al. Citation1998). However, a low male-mediated gene flow was also hypothesised because of the lack of divergence, highlighted by nuclear loci, between some North Atlantic areas and the Mediterranean Sea (Bérubé et al. Citation1998; Palsbøll et al. Citation2004). The movement of fin whales through the Strait of Gibraltar was well documented and seems to occur seasonally (Gauffier et al. Citation2018). In addition, the isotopic analysis performed on baleen plates (Bentaleb et al. Citation2011) and the analysis of recorded fin whale songs (Castellote et al. Citation2012a) allowed the recognition of two distinct populations in the Mediterranean Sea. The first (MED whales) consists of resident individuals inhabiting the basin all year round, while the second (NENA whales) is composed of North Atlantic individuals that move into the westernmost region of the Mediterranean Sea (Notarbartolo di Sciara et al. Citation2016).
Fin whales are present throughout the Mediterranean Sea but they are very abundant in the western region, especially in the Ligurian-Corsican-Provençal Basin and Gulf of Lion, an area characterised by a high biological productivity (Notarbartolo di Sciara et al. Citation2003). The intense maritime traffic of the Mediterranean Sea is affecting the survival of fin whales. Collisions with vessels are a significative cause of death (Panigada et al. Citation2006) and underwater noise negatively affects the species (Castellote et al. Citation2012b). In addition, Mediterranean fin whales could also be threatened by the ingestion of plastic debris (Fossi et al. Citation2012), exposure to chemical contaminants (Fossi et al. Citation2003), dolphin morbillivirus (DMV) infections (Mazzariol et al. Citation2016) and negative effects of ongoing climate changes (Gambaiani et al. Citation2009). The Mediterranean population of B. physalus is currently listed as “vulnerable” under criteria C2a(ii) by the IUCN Red List of Threatened Species (ver. 2021-3) (Panigada & Notarbartolo di Sciara Citation2012). The high degree of anthropization of the Mediterranean Sea and the lack of information about the ecology of this population, require an increase in research efforts in order to implement conservation and management actions.
The present study investigates the geographical origin of three fin whales, recently stranded along the coasts of the Tyrrhenian Sea (Mediterranean Sea, Italy), comparing a sequence of the mtDNA CR with those previously obtained for North Atlantic and Mediterranean samples (Bérubé et al. Citation1998; Caputo & Giovannotti Citation2009; Cabrera et al. Citation2019). The analysis of the mtDNA CR, a maternally inherited marker, revealed the misidentification of one of the fin whale individuals. In fact, the analysis of a biparentally inherited marker, the α-lactalbumin (α-lac) nuclear gene, together with some morphometric and morphological characters, allowed the identification of a fin-blue whale hybrid among the sampled individuals.
Materials and methods
All tissue samples analysed in this study were provided by the Mediterranean Marine Mammals Tissue Bank (MMMTB) of the University of Padova (Italy) and were collected from three individuals, identified as B. physalus, who died in the Tyrrhenian Sea between 2020 and 2021. The individual ID531 was found in a hidden inlet, Cala del Rio (Capri, Naples, Italy), on November 7, 2020; the individual ID536 was found in the Port of Sorrento (Sorrento, Naples, Italy) on January 15, 2021; the individual ID553 stranded on a beach near Albinia (Grosseto, Italy) on September 2, 2021. The tissue samples were shipped in 70% ethanol and stored at −20°C until genetic analysis. Genomic DNA was isolated from samples using a digestion protocol with Proteinase K followed by a standard phenol-chloroform extraction (Sambrook Citation1989). The quantity and quality of DNA extracted were assessed using a Nanodrop-ND1000 spectrophotometer (NanoDrop Technologies) and a 1% agarose gel electrophoresis.
A fragment of the mtDNA CR was amplified by Polymerase Chain Reaction (PCR) using the Forward primer MT4 (5’-CCT CCC TAA GAC TCA AGG AAG-3’) (Palsbøll et al. Citation1995) and the Reverse primer Bp16071 (5’-CCT CAG TTA TGT TAT GAT CAT GGG C-3’) (Drouot et al. Citation2004), as proposed by Cabrera et al. (Citation2019), to ascertain the geographical origin of the stranded fin whales. The amplification was performed in a 25 µl solution containing 5 µl of 5X MyTaq™ Reaction Buffer (BioLine), 2.5 μl of F + R primer solution [5 μM], 0.3 μl of MyTaq™ DNA Polymerase (BioLine), 3 μl of DNA template [20–40 ng/μl] and 14.2 μl of ddH2O. The PCR cycling profile consisted of an initial denaturation at 95°C for 5 min, followed by 30 cycles of 95°C for 45 s, 54°C for 45 s, and 72°C for 90 s, with a final extension step at 72°C for 7 min. PCR products were visualized by 2% agarose gel stained with GelRed™ (Biotium) and then sent to BMR Genomics (Padova, Italy) for sequencing. At this facility, PCR products were purified by exoSAP-IT™ (USB Corp.) and Sanger sequenced in both directions on an ABIPRISM 3730XL automated sequencer (Applied Biosystems). All mtDNA CR sequences obtained were aligned on BLAST (Altschul et al. Citation1990) to check for their accuracy. After the discovery of an inconsistent result for the samples ID531, its sequence was aligned using CLUSTALW (Larkin et al. Citation2007) with those identified for B. musculus and fin-blue whale hybrids caught in Icelandic waters (Pampoulie et al. Citation2021; downloaded from https://osf.io/hfjgx/?view_only=deab3655a50243e0bcc5ce138bd05872). Instead, sequences of individuals ID536 and ID553 were aligned with mtDNA CR haplotypes described for B. physalus in the North Atlantic Ocean and Mediterranean Sea (Bérubé et al. Citation1998; Cabrera et al. Citation2019). North Atlantic and Mediterranean haplotypes from Bérubé et al. (Citation1998) are available in Genbank (Accession numbers AF119956-AF120003) while, mtDNA CR sequences and information about sampling locations from Cabrera et al. (Citation2019) are downloadable from Datadryad.org under accession: https://doi.org/10.5061/dryad.qt528n0.
Morphometric measurements, including body length, were downloaded from the Italian Stranding Database (http://mammiferimarini.unipv.it/) to validate the result obtained for the individual ID531 using the mtDNA CR. The length from the tip of the snout to the anterior insertion of the dorsal fin, the height of the dorsal fin and the length from anterior insertion of the flipper to tip of the flipper (see Tomilin Citation1957; Yablokov et al. Citation1972; Robineau Citation2005; Cagnolaro et al. Citation2015). Several photographs were also obtained and used to detect distinctive morphological characters between fin and blue whales as the body colouration and the colour of baleen. In addition, a nuclear DNA (nuDNA) marker, the α-lac gene, was also analysed using a PCR-RFLP (Restriction Fragment Length Polymorphism) procedure previously described by Bérubé and Aguilar (Citation1998). Primers LacII.F (5’- CCA AAA TGA TGT CCT TTG TC −3’) and Lac1.R (5’-CTC ACT GTC ACA GGA GAT GT −3’) were used to amplify a fragment of the α-lac gene (Bérubé & Aguilar Citation1998). The PCR was carried out in a 25 µl solution containing 5 µl of 5X MyTaq™ Reaction Buffer (BioLine), 2.5 μl of F + R primer solution [5 μM], 0.4 μl of MyTaq™ DNA Polymerase (BioLine), 5 μl of DNA template [40 ng/μl] and 12.1 μl of ddH2O. Amplification conditions were as follows: an initial denaturation at 95°C for 5 min, followed by 35 cycles of denaturation at 95°C for 45 s, annealing at 54°C for 30 s, and extension at 72°C for 90s, with a final extension at 72°C for 7 min. After checking the PCR product on a 2% agarose gel, an aliquot of them was Sanger sequenced (see above for details) and another was digested at 37°C for 30 min using the FokI restriction enzyme (Bérubé & Aguilar Citation1998). The digestion was performed in a 25 μl volume containing 2.5 μl of 10X rCutSmart™ Buffer (New England Biolabs), 0.25 μl of FokI (New England Biolabs), 5 μl of PCR product and 17.25 μl of ddH2O. Restriction fragments were separated by electrophoresis on 2% agarose gel and the restriction pattern observed was compared with those obtained by Bérubé and Aguilar (Citation1998) for a fin whale, a blue whale and a fin-blue whale hybrid. In addition, chromatograms obtained by Sanger sequencing were checked for the presence of double peaks and used to infer two sequences of the α-lac gene for individual ID531, one for each presumed allele. The two α-lac gene sequences obtained were aligned using CLUSTALW (Larkin et al. Citation2007) with those available in GenBank (Accession numbers AY398656, AY398655) for B. physalus and B. musculus (Rychel et al. Citation2004).
Results
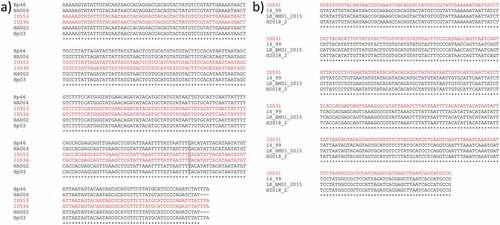
A sequence of ~300 bp of the mtDNA CR was obtained from all samples analysed but, after the alignment on BLAST (Altschul et al. Citation1990), only two (ID536 and ID553) of them were attributed to B. physalus while the third (ID531) belongs to B. musculus. This result suggests that the individual ID531 classified as a fin whale is actually a fin-blue whale hybrid. The alignment of our sequences against those recently published for B. physalus (Bérubé et al. Citation1998; Cabrera et al. Citation2019), B. musculus and fin-blue whale hybrids (Pampoulie et al. Citation2021) highlights that studied individuals carry mtDNA CR haplotypes already observed (). Specifically, the sequence of the individual ID536 resulted identical to the haplotypes Bp03 (Bérubé et al. Citation1998) and NATL002 (Cabrera et al. Citation2019) while the sequence of the individual ID553 matched the haplotypes Bp46 (Bérubé et al. Citation1998) and NATL004 (Cabrera et al. Citation2019) ()). The putative fin-blue whale hybrid ID531 showed a mtDNA CR sequence identical to that of two blue whales (14–99, LK-BM01-2015) and a fin-blue whale hybrid (H2018-2) caught in Icelandic waters (Pampoulie et al. Citation2021) ()). The latter was a hybrid adult male captured in 2018 and genetically identified as a backcross between a first-generation hybrid mother and a fin whale father (Pampoulie et al. Citation2021). The mtDNA CR sequences obtained in the present study were uploaded in GenBank under the accession numbers OM859042, OM859043 and OM859044.
Figure 1. ClustalW multiple sequence alignment of ID531, ID536, ID553 mtDNA CR sequences (in red) with (a) identical haplotypes described for B. physalus by Bérubé et al. (Citation1998) and Cabrera et al. (Citation2019), and (b) identical sequences of B. musculus and a fin-blue whale hybrid from Pampoulie et al. (Citation2021). Grey colour indicates a mutation site, asterisk (*) indicates positions which have a single, fully conserved residue and dash (-) indicates a gap due to the presence of shorter sequences in the alignment.
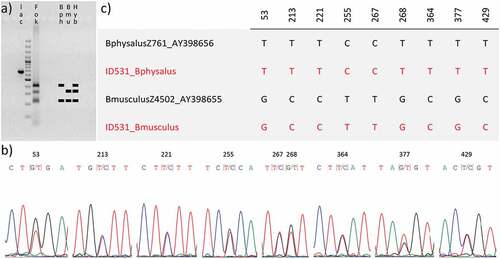
A fragment of the α-lac gene was amplified to confirm the hybrid status of individual ID531, thus generating a PCR product of ~600 bp in length ()). The digestion with the FokI endonuclease produced three restriction fragments ()). The comparison of this digestion pattern with those obtained by Bérubé and Aguilar (Citation1998) for a fin-blue whale hybrid, a fin whale and a blue whale ()), confirms the hybrid origin of the individual ID531. The digestion pattern of the hybrid shows a ~200 bp fragment, present in both fin and blue whales, and two restriction fragments of ~300 bp and ~400 bp in length characteristic of blue and fin whales, respectively ()). Accordingly, Sanger sequencing chromatograms revealed the presence of nine double peaks ()) indicating a heterozygous genotype in which the two alleles are inherited from parents of different species. The alignment of the two sequences inferred from chromatograms showed that they match those described for the two species, B. physalus and B. musculus ()), thus again confirming the hybrid origin of the analysed individual.
Figure 2. Analyses of the ~600 bp fragment of the α-lactalbumin nuclear gene. (a) Results of the PCR (lac) and RFLP analysis (Fok) compared with a representation of digestion patterns observed by Bérubé and Aguilar (Citation1998) for the fin whale (Bph), the blue whale (Bmu) and the fin-blue whale hybrid (Hyb). (b) Double peaks observed in sequencing chromatograms. (c) Diagnostic sites obtained after the alignment of our inferred sequences (in red) against sequences from GenBank.
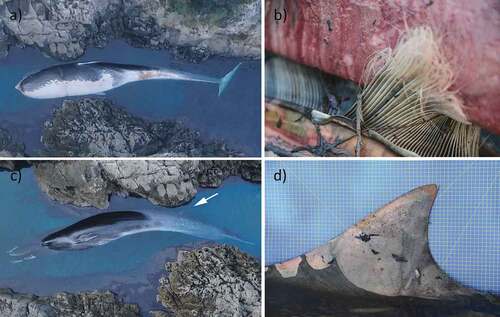
Morphometric measurements and morphological characters of the individual ID531 showed that colouration and body proportions of this whale were closer to those of a fin whale than a blue whale (Tomilin Citation1957; Yablokov et al. Citation1972; Robineau Citation2005; Cagnolaro et al. Citation2015). The individual ID531 was a young female with a body length of 14.20 m. The body colouration was asymmetrical as in fin whales, being the right side lighter than the left one however, on the left side of the animal, the grey colour of the back extends more towards the belly, so when observed laterally the individual appeared uniformly dark as a blue whale ()). The colour of baleen plates was yellowish anteriorly in the frayed portion and not uniformly dark as in blue whales ()). The length from anterior insertion of the flipper to tip of the flipper was 1.30 m, corresponding to 9.15% of the body length, and therefore shorter than that of a blue whale (about 15% of the body length; see Tomilin Citation1957; Robineau Citation2005; Sears & Perrin Citation2009; Cagnolaro et al. Citation2015). The dorsal fin was moderately concave and pointed ()) and is 0.29 m high, corresponding to 2.04% of the body length. The size of the dorsal fin was probably a character intermediate between that of a fin whale (about 2.5% of the body length; Tomilin Citation1957; Jefferson et al. Citation1993) and a blue whale (about 1% of the body length; Jefferson et al. Citation1993; Robineau Citation2005). The length from the tip of the snout to the anterior insertion of the dorsal fin was 10.43 m and corresponded to 73.5% of the body length ()). The position of the dorsal fin was more similar to that of the blue whale (dorsal fin at 3/4 back on body; Geraci & Lounsbury Citation2005) rather than that of the fin whale (dorsal fin at 2/3 back on body; Geraci & Lounsbury Citation2005).
Discussion
The three rorquals analysed in this study were, at first, identified as fin whales based on the sole analysis of morphological characters. The genetic analysis performed highlights, on the contrary, the presence of a fin-blue whale hybrid (B. physalus x B. musculus) among them. The mtDNA CR sequences of the two individuals genetically identified as B. physalus (ID536 and ID553) were compared with the largest dataset of mtDNA CR haplotypes collected so far for the North Atlantic area (Cabrera et al. Citation2019). Cabrera et al. (Citation2019) analysed the first 285 bp of the mtDNA CR in 828 North Atlantic samples (115 from the Mediterranean Sea) recording the presence of 80 haplotypes. The sequence of the individual ID536 was attributed to the haplotype NATL002 while the sequence of the individual ID553 was attributed to the haplotype NATL004. Observing the sampling locations of all the individuals analysed by Cabrera et al. (Citation2019) is evident that the haplotype NATL002 is widespread both in North Atlantic Ocean and in the Mediterranean Sea, while the haplotype NATL004 is abundant in the Mediterranean basin and extremely rare in the North Atlantic Ocean being observed here in only three individuals (Cabrera et al. Citation2019). The comparison between sequences of the haplotypes NATL002 and NATL004 (Cabrera et al. Citation2019) and those described previously by Bérubé et al. (Citation1998) shows that they are identical to haplotypes Bp03 and Bp46 respectively. Bérubé et al. (Citation1998) sequenced the first 288 bp of the mtDNA CR in 69 individuals sampled in the Ligurian Sea (Mediterranean Sea) finding the haplotype Bp03 in both North Atlantic Ocean and Mediterranean Sea, while the haplotype Bp46 was classified as private of the latter basin (Bérubé et al. Citation1998). The mismatch in the frequency of haplotypes observed in North Atlantic and Mediterranean areas is certainly linked to a difference in the number of individuals analysed in the above-mentioned works (Bérubé et al. Citation1998; Cabrera et al. Citation2019). However, the haplotypes assigned to individuals ID536 and ID553 are among the most common in the Mediterranean Sea (Bérubé et al. Citation1998; Cabrera et al. Citation2019), leading us to hypothesize that these two individuals most probably belongs to the resident Mediterranean fin whales population. In addition, a preliminary analysis of the dorsal fin, using a photo-identification method, highlighted that the individual ID536 had already been observed in June 1994 during a survey in the Pelagos Sanctuary (Tethys Research Institute Citation2021). The Pelagos Sanctuary is a Marine Protected Area (MPA) established for the conservation of Mediterranean Marine mammals which includes the Ligurian Sea and parts of the Corsican and Tyrrhenian Sea (Notarbartolo di Sciara et al. Citation2008). Both genetic (Bérubé et al. Citation1998) and acoustic analyses (Castellote et al. Citation2012a) confirmed that this area was frequented by MED whales, instead NENA individuals enter through the Strait of Gibraltar but remain confined in the westernmost part of the Mediterranean Sea. The discovery of a fin whale already sighted 26 years ago in the Pelagos sanctuary seems to confirm that it most likely belong to the MED whales population even if the individual shows the haplotype NA002, the most abundant in both North Atlantic Ocean and Mediterranean Sea. The individual ID553 was found on a beach near Albinia (Grosseto, Italy), a locality included in the Pelagos sanctuary and carries the haplotype NA004, very rare in the North Atlantic Ocean, so also in this case the belonging of the individual to the MED whales population seems more probable. The mtDNA CR sequence of individual ID553 also resulted identical to that described for a fin whale stranded on a beach near Ancona (Adriatic Sea, Italy) in the 2007 and already assigned to the haplotype Bp46 (Caputo & Giovannotti Citation2009).
The discovery of fin whales carrying haplotypes common in the Mediterranean resident population along the Tyrrhenian coasts (ID536 and ID553), and even in the Adriatic Sea (Caputo & Giovannotti Citation2009), is in accordance with the ability of MED whales to widely move in the Mediterranean Sea. MED whales are considered as “nomadic opportunists” because their seasonal feeding and breeding grounds are related with the presence of favourable environmental conditions in different Mediterranean areas rather than to a fixed migratory pattern (Geijer et al. Citation2016; Notarbartolo di Sciara et al. Citation2016). Fin whales are usually aggregated in the Ligurian-Corsican-Provençal Basin and Gulf of Lion during the summer because the high levels of productivity in this area and disperse towards the Southern Mediterranean basin in winter when trophic conditions become less favourable (Notarbartolo di Sciara et al. Citation2016). The summer trophic conditions of the Central and Southern Tyrrhenian Sea make it attractive to an ever increasing number of fin whales (Mussi et al. Citation1999; Arcangeli et al. Citation2014) showing that this basin is not only a transit area for individuals moving from the Ligurian Sea, but also an important opportunistic feeding ground. The use of the Tyrrhenian Sea by an increasing number of fin whales would also explain the recent stranding of the individuals analysed in this work. This feeding behaviour is also common to other species considered “vagrant” in the Mediterranean Sea (Maio et al. Citation2016).
The taxonomic identification of the individual ID531 was found to be more problematic. The initial misidentification is certainly due to its morphology which makes it very similar to a B. physalus. The morphological character that mostly distinguishes B. physalus from B. musculus is the colouration (Jefferson et al. Citation2015): the fin whale has a dark dorsal region with a light belly and an asymmetrical head colour, while the blue whale has a mottled dark-grey body. In addition, the fin whale has a V-shaped head, an asymmetrical colour of the baleen which are light in the anterior right side, a taller (about 2.5% of the body length; Tomilin Citation1957; Jefferson et al. Citation1993) and more falcate dorsal fin located at 2/3 back on body (Geraci & Lounsbury Citation2005), while the blue whale as a U-shaped head, dark baleen and a small dorsal fin (about 1% of the body length; Jefferson et al. Citation1993; Robineau Citation2005) located at 3/4 back on body (Geraci & Lounsbury Citation2005). The flippers are long in both species but in B. musculus they reach up to 15% of the body length (Sears & Perrin Citation2009). The majority of morphological and morphometric characters observed in the individual ID531, e.g. the body colouration, the baleen colour and the size of flippers, are distinctive characters of the fin whale, while the position of the dorsal fin along the back is a distinctive character of the blue whale. The individual ID531 therefore has the general appearance of a fin whale and for this reason it was not immediately recognised as a hybrid, but our analysis of mitochondrial and nuclear markers allowed to clarify its peculiar origin.
The first reports of putative hybrids between fin and blue whales date back to the end of the nineteenth century when individuals sharing morphological characters of both parental species were caught during commercial whaling operations (Cocks Citation1887). However, genetic analysis of some of them confirmed hybridization between these species of rorquals only many years later (Árnason et al. Citation1991; Spilliaert et al. Citation1991; Bérubé & Aguilar Citation1998; Pampoulie et al. Citation2021). Our findings show that the individual ID531 was a hybrid between a female blue whale and a male fin whale. In fact, the sequence of mtDNA CR, a maternally inherited marker, suggests that the mother of the hybrid is a blue whale (B. musculus) while the analysis of the α-lac nuclear gene, which follows a mendelian inheritance pattern, confirms that the father is a fin whale (B. physalus). The mtDNA CR sequence of the individual ID531 is identical to those observed in two blue whales and in one fin-blue whale hybrid sampled in Icelandic waters (Pampoulie et al. Citation2021). Interestingly, the presence of blue whales or fin-blue whale hybrids represents the first record for the Mediterranean Sea, indicating that a North Atlantic origin is most likely for the individual ID531. North Atlantic fin whales would in fact pass through the Strait of Gibraltar to feed in the Mediterranean Sea during the winter months (Gauffier et al. Citation2018, Citation2020).
In the recent study of Pampoulie et al. (Citation2021), the genetic analysis of eight hybrids showed that seven of them had a blue whale mother. In our study, the discovery of a hybrid resulting from the successful mating between a female blue whale and a male fin whale supports the occurrence of unidirectional hybridization between these species of rorquals (Árnason et al. Citation1991; Spilliaert et al. Citation1991; Bérubé & Aguilar Citation1998; Pampoulie et al. Citation2021). Probably, the unidirectional hybridization occurs when females of a rare species mate with males of a common species, a phenomenon described by the “sexual selection hypothesis” (Wirtz Citation1999). Initially, the females are more discriminating and reject males of the different species, but if the search of a conspecific male is not successful, females become less discriminating and mate with males of the common species (Wirtz Citation1999). As a result of intense whaling in the past, the abundance of North Atlantic blue whales is lower than that of fin whales (North Atlantic Marine Mammal Commision Citation2020), bringing blue whale females to exceptionally mate with fin whale males and give birth to hybrids offspring (Pampoulie et al. Citation2021). However, the capture in 1983 of a male, later identified as a cross between a fin whale mother and a blue whale father, does not completely support this hypothesis (Árnason et al. Citation1991) suggesting that different factors could promote interspecific mating in the wild.
Hybridization in cetaceans is promoted when the species involved have morphological, behavioural, and ecological characteristics in common. In particular, sexual dimorphism, body length, geographic range size and vocalization frequency are the main traits positively related with the propensity of cetaceans to hybridize (Crossman et al. Citation2016). Blue and fin whales are the largest animals in the world and the size and morphology of the adults of both species are quite similar. Moreover, they have overlapping distribution ranges, being present in temperate and polar waters of both hemispheres (Jefferson et al. Citation2015). Sightings and tagging studies highlighted that fin and blue whales sometimes share the same feeding grounds. In spring, both species were recorded in the Azorean waters (Silva et al. Citation2013) and aggregations of fin and blue whales was also observed in autumn in the Porcupine Seabight, southwest of Ireland, probably to feed on abundant northern krill (Meganyctiphanes norvegica) (Baines et al. Citation2017). In the Porcupine Seabight, Baines et al. (Citation2017) observed in addition to a feeding behaviour also a chasing behaviour in fin whales that was interpreted as a social interaction preparatory to the mating season. When cetaceans aggregate in the same area they can also engage in mating-like behaviours probably as a form of social play (Brown & Norris Citation1956; Herzing & Johnson Citation1997) or to practice in order to have greater reproductive success during the mating season (Mann Citation2006), thus increasing the probability of interspecific hybridization (Crossman et al. Citation2016).
Even if the reasons that promote interspecific mating between fin and blue whales are not well known, hybridization is certainly favoured by the karyotype (2n = 44) common to most cetaceans (Árnason Citation1974, Citation1982) which reduces the risk of incorrect pairing of homologous chromosomes during meiosis, promoting the fertility of hybrid offspring. The evidence that fin-blue whale hybrids can successfully mate was given by the capture in 1986 of a pregnant hybrid female (Árnason et al. Citation1991; Spilliaert et al. Citation1991) and by the genetic identification of a backcross adult male (Pampoulie et al. Citation2021). However, the fertility seems confirmed only for female hybrids while males are probably sterile (Árnason et al. Citation1991). The hybridization between fin and blue whales is well documented in the wild but its impact on populations of either species is unclear. It was estimated that in Mammalian species hybridization leads mainly to negative consequences as the partial or total replacement of parental species genotypes by hybrid genotypes, a phenomenon known as “genetic swamping” that could promote the extinction of small and isolated populations or entire species (Adavoudi & Pilot Citation2022). The negative consequences of hybridization could be more pronounced in blue whales because their North Atlantic population account for about 3,000 individuals, a very low number compared to fin whales which amount to over 50,000 individuals (North Atlantic Marine Mammal Commission Citation2020). In rare cases, hybridization can also have positive consequences such as the spread of novel adaptive variation and even the creation of new species increasing the biodiversity (Adavoudi & Pilot Citation2022). The hybrid speciation in cetaceans has been reported in wild by Amaral et al. (Citation2014) which, using morphological and genetic analysis, described the Clymene dolphin Stenella clymene as the result of natural hybridization between the striped dolphin S. coeruleoalba and the spinner dolphin S. longirostris.
In conclusion, the mtDNA CR sequencing performed in this study on individuals morphologically described as B. physalus stranded along the Tyrrhenian coast, allowed us to identify their most probable geographical origin. Individuals ID536 and ID553 belonged to the resident Mediterranean fin whale population, while individual ID531 most probably came from the North Atlantic Ocean. These results indicate that Tyrrhenian waters could be an important feeding ground not only for Mediterranean fin whales but also for Atlantic visitors (Maio et al. Citation2016). Genetic analyses have also proved essential to reveal that individual ID531 was a fin-blue whale hybrid, highlighting that morphological analysis alone does not allow to recognize hybrids in the wild especially when an intermediate phenotype is not so evident. The use of morphological characters alone to identify hybrids could therefore lead to an underestimation of the extent of hybridization, preventing us from detecting potential negative effects on small populations such as that of North Atlantic blue whales. The results obtained highlight the importance to increase sampling and analysis efforts in order to fill gaps in the knowledge of fin and blue whales. In addition, for the correct identification of the fin-blue whale hybrids the morphological analysis must always be supported by the genetic analysis of biparentally inherited markers. Only in this way it will be possible to assess the real extent of the hybridization and clarify its effects on both species involved.
Acknowledgements
We thank Ivan Rubino (Capri, Naples, Italy) and Cristina Otero (Stabio CERT, Italy) for photos and videos of the fin whale discovered in Capri; Francesco Pollaro (Centro Studi Ecosistemi Marini, Pollica, Salerno, Italy) for technical support; Alessandro Minelli (Padova, Italy) for taxonomic suggestions.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Adavoudi R, Pilot M. 2022. Consequences of hybridization in mammals: A systematic review. Genes 13(1):50. DOI: 10.3390/genes13010050.
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. Journal of Molecular Biology 215(3):403–410. DOI: 10.1016/S0022-2836(05)80360-2.
- Amaral AR, Lovewell G, Coelho MM, Amato G, Rosenbaum HC. 2014. Hybrid speciation in a marine mammal: The Clymene dolphin (Stenella clymene). PLoS ONE 9(1):e83645. DOI: 10.1371/journal.pone.0083645.
- Arcangeli A, Orasi A, Carcassi SP, Crosti R. 2014. Exploring thermal and trophic preference of Balaenoptera physalus in the central Tyrrhenian Sea: A new summer feeding ground? Marine Biology 161(2):427–436. DOI: 10.1007/s00227-013-2348-8.
- Archer FI, Brownell RL, Hancock-Hanser BL, Morin PA, Robertson KM, Sherman KK, Calambokidis J, Urbán RJ, Rosel PE, Mizroch SA, Panigada S, Taylor BL. 2019. Revision of fin whale Balaenoptera physalus (Linnaeus, 1758) subspecies using genetics. Journal of Mammalogy 100(5):1653–1670. DOI: 10.1093/jmammal/gyz121.
- Árnason Ú. 1974. Comparative chromosome studies in Cetacea. Hereditas 77(1):1–36. DOI: 10.1111/j.1601-5223.1974.tb01351.x.
- Árnason Ú. 1982. Karyotype stability in marine mammals. Cytogenetic and Genome Research 33(3):274–276. DOI: 10.1159/000131771.
- Árnason Ú, Spilliaert R, Pálsdóttir Á, Árnason A. 1991. Molecular identification of hybrids between the two largest whale species, the blue whale (Balaenoptera musculus) and the fin whale (B. physalus). Hereditas 115(2):183–189. DOI: 10.1111/j.1601-5223.1991.tb03554.x.
- Baines M, Reichelt M, Griffin D. 2017. An autumn aggregation of fin (Balaenoptera physalus) and blue whales (B. musculus) in the Porcupine Seabight, southwest of Ireland. Deep sea research part II. Topical Studies in Oceanography 141:168–177. DOI: 10.1016/j.dsr2.2017.03.007.
- Bentaleb I, Martin C, Vrac M, Mate B, Mayzaud P, Siret D, de Stephanis R, Guinet C. 2011. Foraging ecology of Mediterranean fin whales in a changing environment elucidated by satellite-tracking and baleen plate stable isotopes. Marine Ecology Progress Series 438:285–302. DOI: 10.3354/meps09269.
- Bérubé M, Aguilar A. 1998. A new hybrid between a blue whale, Balaenoptera musculus, and a fin whale, B. physalus: Frequency and implications of hybridization. Marine Mammal Science 14(1):82–98. DOI: 10.1111/j.1748-7692.1998.tb00692.x.
- Bérubé M, Aguilar A, Dendanto D, Larsen F, Notarbartolo di Sciara G, Sears R, Sigurjónsson J, Urban-R J, Palsbøll PJ. 1998. Population genetic structure of North Atlantic, Mediterranean Sea and Sea of Cortez fin whales, Balaenoptera physalus (Linnaeus 1758): Analysis of mitochondrial and nuclear loci. Molecular Ecology 7(5):585–599. DOI: 10.1046/j.1365-294x.1998.00359.x.
- Bérubé M, Urbán J, Dizon AE, Brownell RL, Palsbøll PJ. 2002. Genetic identification of a small and highly isolated population of fin whales (Balaenoptera physalus) in the Sea of Cortez, Mexico. Conservation Genetics 3(2):183–190. DOI: 10.1023/A:1015224730394.
- Brown DH, Norris KS. 1956. Observations of captive and wild cetaceans. Journal of Mammalogy 37(3):311–326. DOI: 10.2307/1376730.
- Cabrera AA, Hoekendijk JPA, Aguilar A, Barco SG, Berrow S, Bloch D, Borrell A, Cunha HA, Dalla Rosa L, Dias CP, Gauffier P, Hao W, Landry S, Larsen F, Martín V, Mizroch S, Oosting T, Øien N, Pampoulie C, Panigada S, Prieto R, Ramp C, Rivera-Léon V, Robbins J, Ryan C, Schall E, Sears R, Silva MA, Urbán J, Wenzel FW, Palsbøll PJ, Bérubé M. 2019. Fin whale (Balaenoptera physalus) mitogenomics: A cautionary tale of defining sub-species from mitochondrial sequence monophyly. Molecular Phylogenetics and Evolution 135:86–97. DOI: 10.1016/j.ympev.2019.02.003.
- Cagnolaro L, Cozzi B, Notarbartolo di Sciara G, Podestà M. 2015. Fauna d’Italia. Vol. XLIX. Mammalia IV. Cetacea. Bologna: Calderini.
- Caputo V, Giovannotti M. 2009. Haplotype characterization of a stranded Balenoptera physalus (Linnaeus, 1758) from Ancona (Adriatic sea, central Italy). Hystrix, the Italian Journal of Mammalogy 20(1):83–85. DOI: 10.4404/hystrix-20.1-4438.
- Castellote M, Clark CW, Lammers MO. 2012a. Fin whale (Balaenoptera physalus) population identity in the western Mediterranean Sea. Marine Mammal Science 28(2):325–344. DOI: 10.1111/j.1748-7692.2011.00491.x.
- Castellote M, Clark CW, Lammers MO. 2012b. Acoustic and behavioural changes by fin whales (Balaenoptera physalus) in response to shipping and airgun noise. Biological Conservation 147(1):115–122. DOI: 10.1016/j.biocon.2011.12.021.
- Clarke R. 2004. Pygmy fin whales. Marine Mammal Science 20(2):329–334. DOI: 10.1111/j.1748-7692.2004.tb01161.x.
- Cocks AH. 1887. The fin whale fishery of 1886 on the Lapland coast. Zoologist 11:207–222.
- Committee on Taxonomy. 2021. List of marine mammal species and subspecies. Society for Marine Mammalogy. Available: www.marinemammalscience.org. Accessed Feb 2022 11.
- Crossman CA, Taylor EB, Barrett‐Lennard LG. 2016. Hybridization in the Cetacea: Widespread occurrence and associated morphological, behavioral, and ecological factors. Ecology and Evolution 6(5):1293–1303. DOI: 10.1002/ece3.1913.
- Drouot V, Bérubé M, Gannier A, Goold JC, Reid RJ, Palsbøll PJ. 2004. A note on genetic isolation of Mediterranean sperm whales (Physeter macrocephalus) suggested by mitochondrial DNA. Journal of Cetacean Research and Management 6(1):29–32.
- Edwards EF, Hall C, Moore TJ, Sheredy C, Redfern JV. 2015. Global distribution of fin whales Balaenoptera physalus in the post‐whaling era (1980–2012). Mammal Review 45(4):197–214. DOI: 10.1111/mam.12048.
- Fossi MC, Marsili L, Neri G, Natoli A, Politi E, Panigada S. 2003. The use of a non-lethal tool for evaluating toxicological hazard of organochlorine contaminants in Mediterranean cetaceans: New data 10 years after the first paper published in MPB. Marine Pollution Bulletin 46(8):972–982. DOI: 10.1016/S0025-326X(03)00113-9.
- Fossi MC, Panti C, Guerranti C, Coppola D, Giannetti M, Marsili L, Minutoli R. 2012. Are baleen whales exposed to the threat of microplastics? A case study of the Mediterranean fin whale (Balaenoptera physalus). Marine Pollution Bulletin 64(11):2374–2379. DOI: 10.1016/j.marpolbul.2012.08.013.
- Gambaiani D, Mayol P, Isaac S, Simmonds M. 2009. Potential impacts of climate change and greenhouse gas emissions on Mediterranean marine ecosystems and cetaceans. Journal of the Marine Biological Association of the United Kingdom 89(1):179–201. DOI: 10.1017/S0025315408002476.
- Gauffier P, Borrell A, Silva MA, Víkingsson GA, López A, Giménez J, Colaço A, Halldórsson SD, Vighi M, Prieto R, de Stephanis R, Aguilar A. 2020. Wait your turn, North Atlantic fin whales share a common feeding ground sequentially. Marine Environmental Research 155:104884. DOI: 10.1016/j.marenvres.2020.104884.
- Gauffier P, Verborgh P, Giménez J, Esteban R, Salazar Sierra JM, de Stephanis R. 2018. Contemporary migration of fin whales through the Strait of Gibraltar. Marine Ecology Progress Series 588:215–228. DOI: 10.3354/meps12449.
- Geijer CKA, Notarbartolo di Sciara G, Panigada S. 2016. Mysticete migration revisited: Are Mediterranean fin whales an anomaly? Mammal Review 46(4):284–296. DOI: 10.1111/mam.12069.
- Geraci JR, Lounsbury VJ. 2005. Marine mammals ashore: A field guide for strandings. 2nd ed. Baltimore, MD: Baltimore Aquarium.
- Hamilton TA, Redfern JV, Barlow J, Balance LT, Gerrodette T, Holt RS, Forney KA, Taylor BL. 2009. Atlas of cetacean sightings for Southwest Fisheries Science Center Cetacean and ecosystem surveys: 1986 – 2005. NOAA Technical Memorandum, NOAA-TM-NMFSSWFSC-440. U.S. Dep. of Commerce.
- Herzing DL, Johnson CM. 1997. Interspecific interactions between Atlantic spotted dolphins (Stenella frontalis) and bottlenose dolphins (Tursiops truncatus) in the Bahamas, 1985-1995. Aquatic Mammals 23(2):85–99.
- Jefferson TA, Leatherwood S, Webber MA. 1993. FAO species identification guide. Marine mammals of the world. Rome: FAO. pp. 587.
- Jefferson TA, Webber MA, Pitman RL. 2015. Marine mammals of the world: A comprehensive guide to their identification. 2nd ed. Elsevier Science Publishing. pp. 616. DOI: 10.1016/C2012-0-06919-0.
- Kellogg R. 1929. What is known of the migration of some of the whalebone whales. Smithsonian Institution Annual Report of the Board of Regents 1928:467–494.
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23(21):2947–2948. DOI: 10.1093/bioinformatics/btm404.
- Loy A, Aloise G, Ancillotto L, Angelici FM, Bertolino S, Capizzi D, Castiglia R, Colangelo P, Contoli L, Cozzi B, Fontaneto D, Lapini L, Maio N, Monaco A, Mori E, Nappi A, Podestà M, Russo D, Sarà M, Scandura M, Amori G. 2019. Mammals of Italy: An annotated checklist. Hystrix, the Italian Journal of Mammalogy 30(2):87–106. DOI: 10.4404/hystrix-00196-2019.
- Mackintosh NA. 1942. The southern stocks of whalebone whales. Discovery Reports 22:197–300.
- Maio N, Giovannotti M, Caputo Barucchi V, Petraccioli A, Pollaro F, Guarino FM, Splendiani A, De Stasio R, Odierna G. 2016. Haplotype characterization of a young stranded common Minke Whale (Balaenoptera acutorostrata Lacépède, 1804): Is the Mediterranean Sea a potential calving or nursery ground for the species? Hystrix, The Italian Journal of Mammalogy 27(2):205–208. DOI: 10.4404/hystrix-27.2-11661.
- Mann J. 2006. Establishing trust: Socio-sexual behaviour and the development of male-male bonds among Indian Ocean bottlenose dolphins. In: Sommer V, Vassey PL, editors. Homosexual behaviour in animals. New York: Cambridge University Press. pp. 107–130.
- Mazzariol S, Centelleghe C, Beffagna G, Povinelli M, Terracciano G, Cocumelli C, Pintore A, Denurra D, Casalone C, Pautasso A, Di Francesco CE, Di Guardo G. 2016. Mediterranean fin whales (Balaenoptera physalus) threatened by dolphin morbillivirus. Emerging Infectious Diseases 22(2):302–305. DOI: 10.3201/eid2202.150882.
- Mizroch SA, Rice DW, Zwiefelhofer D, Waite J, Perryman WL. 2009. Distribution and movements of fin whales in the North Pacific Ocean. Mammal Review 39(3):193–227. DOI: 10.1111/j.1365-2907.2009.00147.x.
- Mussi B, Miragliuolo A, Monzini E, Diaz Lopez B, Battaglia M. 1999. Fin whale (Balaenoptera physalus) feeding ground in the coastal waters of Ischia (Archipelago Campano). European Research on Cetaceans 13:330–335.
- Nichol L, Heise K. 1992. The historical occurrence of large whales off the Queen Charlotte Islands. Research Paper Report for South Moresby/Gwaii Haanas National Park Reserve, Canadian Parks Service. Queen Charlotte City, British Columbia. pp. 68.
- North Atlantic Marine Mammal Commission. 2020. Estimates of Cetacean abundance in the North Atlantic of relevance to NAMMCO. NAMMCO Scientific Publications. p. 11. DOI: 10.7557/3.5732.
- Notarbartolo di Sciara G, Agardy T, Hyrenbach D, Scovazzi T, Van Klaveren P. 2008. The Pelagos sanctuary for Mediterranean marine mammals. Aquatic Conservation: Marine and Freshwater Ecosystems 18(4):367–391. DOI: 10.1002/aqc.855.
- Notarbartolo di Sciara G, Castellote M, Druon JN, Panigada S. 2016. Fin whales, Balaenoptera physalus: At home in a changing Mediterranean Sea? Advances in Marine Biology 75:75–101. DOI: 10.1016/bs.amb.2016.08.002.
- Notarbartolo di Sciara G, Zanardelli M, Jahoda M, Panigada S, Airoldi S. 2003. The fin whale Balaenoptera physalus (L. 1758) in the Mediterranean Sea. Mammal Review 33(2):105–150. DOI: 10.1046/j.1365-2907.2003.00005.x.
- Palsbøll PJ, Bérubé M, Aguilar A, Notarbartolo di Sciara G, Nielsen R. 2004. Discerning between recurrent gene flow and recent divergence under a finite‐site mutation model applied to North Atlantic and Mediterranean Sea fin whale (Balaenoptera physalus) populations. Evolution 58(3):670–675. DOI: 10.1111/j.0014-3820.2004.tb01691.x.
- Palsbøll PJ, Clapham PJ, Mattila DK, Larsen F, Sears R, Siegismund HR, Sigurjónsson J, Vasquez O, Arctander P. 1995. Distribution of mtDNA haplotypes in North Atlantic humpback whales: The influence of behaviour on population structure. Marine Ecology Progress Series 116:1–10. Available: http://www.jstor.org/stable/44634989.
- Pampoulie C, Gíslason D, Ólafsdóttir G, Chosson V, Halldórsson SD, Mariani S, Elvarsson BÞ, Rasmussen MH, Iversen MR, Daníelsdóttir AK, Víkingsson GA. 2021. Evidence of unidirectional hybridization and second‐generation adult hybrid between the two largest animals on Earth, the fin and blue whales. Evolutionary Applications 14(2):314–321. DOI: 10.1111/eva.13091.
- Panigada S, Notarbartolo di Sciara G. 2012. Balaenoptera physalus. The IUCN Red List of Threatened Species 2012: E.T2478A2787161. Accessed February 2022 20.
- Panigada S, Pesante G, Zanardelli M, Capoulade F, Gannier A, Weinrich MT. 2006. Mediterranean fin whales at risk from fatal ship strikes. Marine Pollution Bulletin 52(10):1287–1298. DOI: 10.1016/j.marpolbul.2006.03.014.
- Pérez-Alvarez MJ, Kraft S, Segovia NI, Olavarría C, Nigenda-Morales S, Urbán RJ, Viloria-Gómora L, Archer F, Moraga R, Sepúlveda M, Santos-Carvallo M, Pavez G, Poulin E. 2021. Contrasting phylogeographic patterns among Northern and Southern Hemisphere fin whale populations with new data from the Southern Pacific. Frontiers in Marine Science 8:630233. DOI: 10.3389/fmars.2021.630233.
- Robineau D. 2005. Cétacés de France. Faune de France vol. 89. Paris: Fédération Française des Sociétés de Sciences Naturelles. pp. 644.
- Rychel AL, Reeder TW, Berta A. 2004. Phylogeny of mysticete whales based on mitochondrial and nuclear data. Molecular Phylogenetics and Evolution 32(3):892–901. DOI: 10.1016/j.ympev.2004.02.020.
- Sambrook J. 1989. Molecular cloning: A laboratory manual. New York: Cold Spring Harbor Laboratory Press.
- Sears R, Perrin WF. 2009. Blue whale: Balaenoptera musculus. In: Perrin WF, Würsig B, Thewissen JGM, editors. Encyclopedia of marine mammals. 2nd ed. San Diego: Academic Press. pp. 120–124. DOI: 10.1016/B978-0-12-373553-9.00033-X.
- Silva MA, Prieto R, Jonsen I, Baumgartner MF, Santos RS. 2013. North Atlantic blue and fin whales suspend their spring migration to forage in middle latitudes: Building up energy reserves for the journey? PLoS One 8(10):e76507. DOI: 10.1371/journal.pone.0076507.
- Spilliaert R, Vikingsson G, Arnason U, Palsdottir A, Sigurjonsson J, Arnason A. 1991. Species hybridization between a female blue whale (Balaenoptera musculus) and a male fin whale (B. physalus): Molecular and morphological documentation. Journal of Heredity 82(4):269–274. DOI: 10.1093/oxfordjournals.jhered.a111085.
- Tershy BR, Urban-R J, Breese D, Rojas-B L, Findley LY. 1993. Are fin whales resident to the Gulf of California? Revista de Investigación Científica 1:69–72.
- Tethys Research Institute. 2021. Photo uploaded to Facebook. Available: https://www.facebook.com/baleneedelfini/photos/a.121544236057/10157396889441058/. Accessed February 2022 21.
- Tomilin AG. 1956. Thermoregulation and the geographical races of cetaceans (Termoregulyatsiya I geograficheskie racy kitoobraznykh). Doklady Akademii Nauk CCP 54 (5):465–472.
- Tomilin AG. 1957. Kitoobraznye [Cetaceans]. Vol. 9. In: Heptner VG, editor. Zveri SSSR I prilezhashchickh stran. Zveri vostochnoi Evropy I severnoi Azii [Mammals of U.S.S.R. and adjacent countries. Mammals of eastern Europe and northern Asia]. Izdatel’stvo Akademi Nauk, SSSR, Moskva. pp. 756. Translation by Ronen O. 1967. Mammals of the U.S.S.R. and adjacent countries. Mammals of eastern Europe and adjacent countries [sic]. Israel Program for Scientific Translations, Jerusalem. pp. 717.
- Wirtz P. 1999. Mother species-father species: Unidirectional hybridization in animals with female choice. Animal Behaviour 58(1):1–12. DOI: 10.1006/anbe.1999.1144.
- Yablokov AV, Bel’kovich VM, Borisov VI. 1972. Whales and dolphins. Izd-vo Nauka [Acad. Sci. USSR], Moscow. pp. 472 (in Russian). English translation, Joint Publications Research Service, Virginia, 1974 (Part I and II).