Abstract
Two mitochondrial genes (cytochrome oxidase I and cytochrome b) were examined to compare an isolated population of the Italian minnow in the central Apennines to other populations in the species’ range (Po plain) and other European minnow species. Both mitochondrial markers showed a new haplotype, fixed in a sample of 30 specimens, never observed in the main species distribution range, and ostensibly divergent from other Italian minnow haplotypes. The result suggests a long history of isolation, probably preceding the Holocene retreat northwards of the Po River. This relict population is now at serious extinction risk owing to habitat loss and predation by alien trout restocked into the wild for recreational fishing purposes.
Introduction
Freshwater vertebrates (lampreys and bony fishes) inhabiting peninsular Italy appear to be of recent origin and to have been influenced by events beginning c. five million years ago (mya) during the Messinian salinity crisis and continuing until recently. Paleontological data indicate that the Adriatic side of the Italian peninsula was inhabited by only marine fish until 2 mya (e.g., Landini & Sorbini Citation1989). Subsequent tectonic and sedimentary events led to the formation of the Po Plain and Apennine (Ambrosetti et al. Citation1981; Dondi & D’Andrea Citation1986), producing the present river basins. The distribution of freshwater fish in this area appears to have been strongly influenced by river confluences during the last (Würmian) marine regression c. 15,000–10,000 years ago. As a result of this regression, the Po basin extended approximately up to the margin of the meso-Adriatic depression (Waelbroeck et al. Citation2002; Amorosi et al. Citation2016), allowing fishes to disperse into the basin (). This explains why many species of fish currently living in the Po basin have the limit of their distribution range in central Italy, therefore included in the Padano-Venetian biogeographic district (according to Bianco et al. Citation1987; Bianco Citation1995a). For instance, the freshwater goby, Padogobius martensi, and several cyprinid species, including Protochondrostoma genei, Rutilus aula and Romanogobio benacensis, shared this distribution range whose southern limit is represented approximately by the basin of the Vomano river (Bianco Citation1995a; Kottelat & Freyhof Citation2007). In addition, a relict population of the Adriatic brook lamprey (Lampetra zanandreai) was discovered in the river Potenza (Bianco Citation1992; Caputo et al. Citation2009a) and genetic data suggest a past presence of Marble trout (Salmo marmoratus) in this and other central Apennine rivers once flowed in the paleo-Po River basin (Splendiani et al. Citation2006). Thus, it was not surprising to have recently found in Potenza (Marconi Citation2009) and Esino (ARPAM Citation2014 and this study) river basins (Adriatic slope of central Italy) isolated populations of Italian minnow, approximately 400 km south of its previously known range, so far gone unnoticed (). Unfortunately, mismanagement of freshwater fishes, especially in the twentieth century, has dramatically altered the original distribution of native populations, thus making very difficult to track the original ranges of many primary freshwater fishes in Italy (e.g., Bianco Citation1990, Citation1995a; Caputo et al. Citation2009b; Splendiani et al. Citation2020). Despite having little direct interest for sport fishing, populations of Italian minnow have been affected by human mediated translocation for their use as (i) live bait for angling activities on trout like species, (ii) forage fish to sustain introduced salmonids populations into Alpine lakes (De Santis et al. Citation2021) and (iii) part of a mixture of fish species (including minnow, chub, barbel, cobitids, gobies, etc.) object during the twentieth century of deliberated translocations in Italy (i.e., “pesce bianco” sensu Bianco Citation1990).
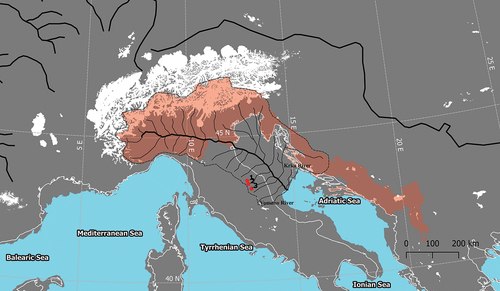
Figure 1. Location of Phoxinus lumaireul observed in the Marche Region: (1) Rio Bono (Esino River), source: ARPAM, Relazione sullo stato di qualità dei corpi idrici fluviali per l’anno 2014; (2) Torrente Sanguirone (Esino River), this study; (3) Potenza River (Mario Marconi, personal communication). The LGM Po River drainage area (dashed line) was desumed from Maselli et al. (Citation2011). The Mediterranean basin coast line during LGM was downloaded from Zickel et al. (Citation2016) GIS dataset. The LGM ice cover (white areas) was downloaded from Ehlers et al. (Citation2011). The IUCN range map of P. lemaireul is represented by the red shaded area.
The aim of the present paper was to examine mitochondrial DNA (mtDNA) variability of this biogeographic isolate to estimate relatedness with populations in the main part of the species’ range and with other species of minnows, in order to evaluate if its presence in central Italy is natural or due to human mediated translocations. The mtDNA cytochrome oxidase I (COI) and cytochrome b (cytb) markers were chosen because their wide use in scientific literature to describe genetic variability in Phoxinus genus make them the best candidates for resolving phylogeographic issues concerning this cyprinid fish (e.g., Palandačić et al. Citation2015; Vucić et al. Citation2018).
Materials and methods
Thirty individuals of P. lumaireul were collected from the Sanguirone stream, within the Esino River basin (Marche region, central Italy) (). Species identification was made using colour pattern, body proportion and scalation according to Kottelat and Freyhof (Citation2007) (). Total genomic DNA was extracted and purified from 95% ethanol preserved fin clips using the Mag-Bind Blood & Tissue DNA HDQ 96 Kit (Omega Bio-tek Inc., Norcross, GA, USA) on a KingFisher sample preparation instrument (Thermo Fisher Scientific Inc., Fremont, CA, USA), according to manufacturer protocols. Mitochondrial DNA cytochrome oxidase I (COI) was amplified using the universal primers LCO1490 (5'-GGTCAACAAATCATAAAGATATTGG-3') and HCO2198 (5'-TAAACTTCAGGGTGACCAAAAAATCA-3') (Folmer et al. Citation1994), with PCR conditions as follows: initial DNA denaturation (95°C, 2 min) and 35 successive cycles of strand denaturation (95°C, 30 s), primer annealing (50°C, 30 s), DNA extension (72°C, 60 s) and final DNA extension (72°C, 5 min). All DNA amplifications were performed in a Veriti 96-Well Fast Thermal Cycler (AB Applied Biosystems). A total volume of 50 µL of reaction mix contained 2.00 µL of genomic DNA extract, 0.20 µM of each primer, 0.20 mM each dNTP, 2.00 mM MgCl2, 1× Flexi buffer, 1.25 U of GoTaq G2 Hot Start Polymerase (Promega, Madison, WI, USA). Mitochondrial DNA cytochrome b locus (cytb) was amplified using the primers Glu-F (5'-GAAGAACCACCGTTGTTATTCAA-3') and Thr-R (5'-ACCTCCRATCTYCGGATTACA-3') (Zardoya & Doadrio Citation1998) using the same PCR concentrations and thermal profile as for COI locus, except for annealing temperature set at 56°C.
Figure 2. A scientific illustration of a specimen of Italian minnow sampled in the Sanguirone stream (Esino River basin, central Apennines, Italy). Scale bar = 10 mm. Illustration by Paul Vecsei.

PCR products for the two loci were then sequenced in forward and reverse directions with the same primers, using the BigDye Terminator Cycle Sequencing kit (Applied Biosystems, Foster City, CA, USA) and run on a 3130XL sequencer (Applied Biosystems).
All forward and reverse partially overlapping sequences were aligned - per individual and per locus - and visually checked using Sequencher (version 5.0.1, Gene Codes) to obtain a consensus sequence. The assembled sequences were then aligned in the GenBank database by BLAST (Altschull et al. Citation1990) to obtain preliminary taxonomic confirmation. Subsequently, cytb and COI sequences were respectively aligned using CLUSTALW with default settings (Thompson et al. Citation1994; Larkin et al. Citation2007) with two reference datasets retrieved from GenBank and composed as follows:
COI: all sequences available in GenBank from Geiger et al. (Citation2014), Palandačić et al. (Citation2017), (Citation2020), Ramler et al. (Citation2017) and Schönhuth et al. (Citation2018), assigned to genetic lineage 1 (1a to 1 f) according to Palandačić et al. (Citation2020), and sequences from De Santis et al. (Citation2021) assigned by these authors to P. lumaireul, having a sequence length of at least 633 bp (N = 260, including sequences from present work);
cytb: a preliminary screening included all sequences available in GenBank from Imoto et al. (Citation2013) Palandačić et al. (Citation2015), (Citation2017), Citation2020), Ramler et al. (Citation2017), Vucić et al. (Citation2018), assigned to genetic lineage 1 (1a to 1 f) according to Palandačić et al. (Citation2020), and having a sequence length of at least 1091 bp (N = 479, including sequences from present work); in a second analysis the dataset was reduced to sequences assigned to genetic sublineage 1a (Palandačić et al. Citation2020), highlighting the further geographic subdivisions in lineages as proposed by Vucić et al. (Citation2018) (N = 160, including sequences from present work).
The aligned datasets, including sequences from the Esino sample, were used to build two median-joining networks (Bandelt et al. Citation1999) with PopART (Leigh & Bryant Citation2015), with default settings.
Results
For each individual included in the analysis a total of 633 bp and 1134 bp long sequences were obtained for COI and cytb, respectively. All thirty individuals shared the same haplotype at both loci. The COI haplotype (named ESNCOI-1, GenBank Acc. no. ON459639 – ON459668) and the cytb haplotype (named ESNcytb-1, GenBank Acc. no. ON494524 – ON494553) resulted to be unique and undescribed before, having no complete identity with any sequence deposited in the GenBank database.
Maximum observed similarity for ESNCOI-1 was 99.84%, over 633 bp, with the sequence observed in 14 specimens (see GenBank Acc. Ns in ), due to a single base mutation; maximum observed similarity for ESNcytb-1 was 99.18%, over a length of 1091 bp, with the sequence observed in six isolates (see GenBank Acc. Ns in ) differing for nine single base mutations
Figure 3. Median-Joining haplotype network built on 633 bp of the mtDNA cytochrome oxidase I locus of Phoxinus lumaireul. Each circle represents a haplotype with size proportional to its frequency in the considered dataset; GenBank Acc. Ns are shown besides each haplotype; different colours are used for subclades 1a–f (Palandačić et al. Citation2020) and for the new haplotype described for the Esino River sample (ESNCOI-1), with black circles indicating haplotypes never sampled or extinct; single-mutational steps are indicated as dashes along connections.
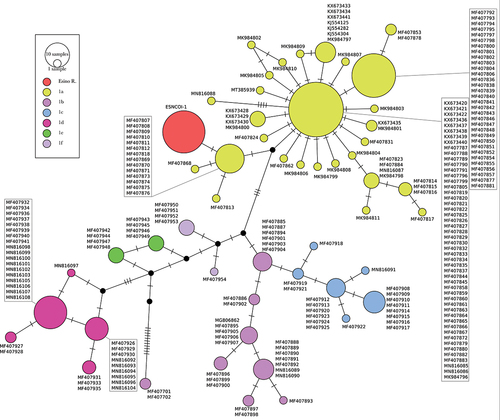
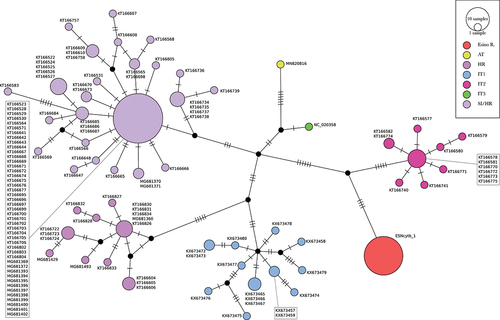
Figure 4. Median-Joining haplotype network built on 1091 bp of the mtDNA cytochrome-b locus of Phoxinus lumaireul. Each circle represents a haplotype with size proportional to its frequency in the considered dataset; GenBank Acc. Ns are shown besides each haplotype; different colours are used for lineages (AT: Austria, HR: Croatia, IT1-3: Italy, SI/HR:Slovenia and Croatia; Vucić et al. Citation2018) geographically identified within subclade 1a (Palandačić et al. Citation2020) and for the new haplotype described for Esino River sample (ESNcytb-1), with black circles indicating haplotypes never sampled or extinct; single-mutational steps are indicated as dashes along connections.
All the polymorphisms observed at the two loci were synonymous mutations.
The closest sequences found in GenBank for the two loci were from fish sampled in Ticino River, North of Italy.
The median joining network built on COI dataset () confirms the newly described haplotype ESNCOI-1 as belonging to the P. lumaireul clade 1a previously described by Palandačić et al. (Citation2020) in North Italian, Slovenian and Croatian samples. In detail, ESNCOI-1 haplotype differs from the central haplotype of the classically starlike-shaped clade 1a by three mutational steps, and from the center of a Ticino River subclade by a single mutational step.
The median joining network built on cytb dataset () reveals the relationships of newly found and unique ESNcytb-1 haplotype in respect to the known variability described so far for the clade 1a, having a peri-Adriatic distribution (Palandačić et al. Citation2020). The genetic distance between the ESNcytb-1 haplotype and the rest of cytb haplotypes of the P. lumaireul clade 1a ranged from a minimum of nine mutational steps, separating it from the central cytb haplotype observed in the classical starlike-shaped subclade formed by the sequences detected in the Ticino River basin (IT2 in ) to a maximum of 19 mutational steps, separating it from the central cytb haplotype of the Croatian subclade, observed in the Zrmanja River basin (HR in ).
Discussion
Although the Italian peninsula has numerous endemic species, whose origin can be traced to dispersion events of the Lago-Mare phase of Mediterranean Basin (about 5 mya), their distribution pattern appears to be strongly influenced by more recent Quaternary events. Considering the biogeographic scenario depicted by Bianco (Citation1995a), Alpine and central Apennine Adriatic rivers coincide with the Padano-Venetian ichthyological district, reaching the Vomano River in Italy and the Krka River in Croatia, southward (). The geographic extent of this district reflects the expansion of the Po River basin down to the meso-Adriatic depression, which occurred during the last glacial maximum (about 15,000–18,000 years ago) in concomitance with the eustatic sea level regression of 120 m). The presence of isolated populations of P. lumaireul in central Italy (Esino and Potenza river basins), about 400 km south of the previously known range for the species, could be attributed to a dispersion event following habitat and river connectivity expansion. The detection of a single private haplotype in both COI and cytb loci in a sample of 30 individuals from the Esino River strongly supports this hypothesis. In addition, the absence of intra-population diversity may be due to historical population demographic fluctuations linked to the small and isolated characteristics of the central Apennine streams, leading to the loss of genetic variability. On the other hand, the marked nucleotide divergence observed between central Italy and North Adriatic haplotypes suggests a long history of isolation, probably preceding the Holocene retreat northwards of the Po River. The detection of three mutational steps between the haplotype ESNCOI-1 (Esino River) and the central haplotype of the clade 1a (North Adriatic) () is coherent with an allopatric evolutionary process as suggested above. A similar scenario of interrupted gene-flow could also explain the high genetic divergence observed between the endemic cytb haplotype (ESNcytb-1) and the North Adriatic haplotypes (). Therefore, the uniqueness of both COI and cytb haplotypes described in this study leads us to exclude the presence of Italian minnow in central Italy as a consequence of translocation practices. Although introduction by human activity is known for minnow species in Europe and Italy (see Palandačić et al. Citation2020; De Santis et al. Citation2021, respectively), the possibility that the Esino River population could have originated from stocking activities seems unlikely because Italian minnow is not used as live bait and/or forage fish in central Italy. Further, Bianco (Citation1991) recognised the occurrence of stocking practices with “pesce bianco” in Esino River waters, listing all the nominal species introduced and thus recorded as exotic. The Italian minnow is not included by Bianco (Citation1991) among stocked fish, but it is not recorded among native ones as well. However, the study by Bianco was focussing on the lower part of the Esino Basin – where stocking practices were concentrated – not including the sampling area of the present study, thus explaining the cryptic occurrence of the Italian minnow in the Esino basin.
The natural presence of the Italian minnow in the Esino River could also be supported by historical records. In fact, small cyprinids of the Apennine streams were well known in the past as a source of food for the poor local human populations. Indeed, among other freshwater fish utilized as food from the peasants, a local author of the second half of the nineteenth century (Marcoaldi Citation1873) remembers a small freshwater fish locally known as “rosciolo” (that is, “small red fish” in the local dialect) which “is fished in May”. In fact, the Italian minnow spawns precisely in late spring (from April to July), with a peak in May (Gandolfi et al. Citation1991), suggesting that the “rosciolo” is indeed the Phoxinus lumaireul, more easily catchable in that season when males have an aggregate behavior and show a nuptial livery with a characteristic red ornamentation. The fact that until now the species has gone overlooked by Italian ichthyologists (e.g., Bianco Citation1991, Citation1995b) in this region, could be a direct consequence of sporadic and occasional freshwater fish faunal surveys on the national territory, while predominant effort is dedicated to field activities strictly related to management of “charismatic species” for sport fishing, first salmonids. It is furthermore the fishery management focussed on and linked to angling activities that seriously risks compromising the survival of these “minor” populations. Local, small and even endemic fish and amphibian populations represent potential prey of allochthonous salmonids yearly illegally released into rivers in huge quantities to meet the expectations of anglers. The recent modifications to the Italian national legislation, opening to the introduction of allochthonous fish in nature (DPR 102/2019, at “https://www.gazzettaufficiale.it/eli/id/2019/09/05/19G00108/sg”, and following Legislative decree of 2 April 2020, at “https://www.gazzettaufficiale.it/eli/id/2020/04/14/20A02112/sg”) probably represent the tombstone to what remains of the vulnerable biodiversity of the inland waters of the Peninsula, still little known. In the last two decades, the description of a native species of pike in Italy (Esox cisalpinus, Bianco & Delmastro Citation2011) and the recent description and revalidation of two barbel species in southern Italy, Barbus samniticus and B. fucini (Lorenzoni et al. Citation2021; Costa Citation1853, respectively), highlight how still scarce is our knowledge on the Italian freshwater fish fauna. Further, the current disjunct and fragmentary distribution of various freshwater fish belonging to the ancient paleo-Po River basin appears to reflect a recent history of habitat loss due to climate change, industrial and agricultural development and restocking with allochthonous fish species that caused the disappearance of intermediate populations (e.g., Bianco Citation1990). A fraction of this biodiversity thus risks disappearing forever before it is even known to science, as the case of the Italian minnow in central Apennine here described well testifies.
Acknowledgements
This work was supported by local funds of the Polytechnic University of Marche. We thank the Wildlife Administration of Marche Region for permitting specimen collection (Authorization decree no. 185, March 24, 2021). We also thank Lorenzo Tancioni for his helpful suggestions during the preparation of the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Altschull SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. Journal of Molecular Biology 215:403–410. DOI: 10.1016/S0022-2836(05)80360-2.
- Ambrosetti P, Centamore E, Deiana G, Dramis F, Pierucci U. 1981. Schema di evoluzione neotettonica dell’area Umbro-Marchigiana tra il Tronto e il Metauro. Rendiconti della Società Geologica Italiana 4:471–475.
- Amorosi A, Maselli V, Trincardi F. 2016. Onshore to offshore anatomy of a late Quaternary source-to-sink system (Po Plain–Adriatic Sea, Italy). Earth-Science Reviews 153:212–237. DOI: 10.1016/j.earscirev.2015.10.010.
- ARPAM (2014). Relazione sullo stato di qualità dei corpi idrici fluviali per l’anno 2014. Agenzia Regionale per la Protezione Ambientale Marche, Ancona.
- Bandelt HJ, Forster P, Röhl A. 1999. Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution 16(1):37–48. DOI: 10.1093/oxfordjournals.molbev.a026036.
- Bianco PG, Bullock AM, Miller PJ, Roubal FR. 1987. A unique teleost dermal organ in a new European genus of fishes (Teleostei, Gobioidei). Journal of Fish Biology 31:797–803. DOI: 10.1111/j.1095-8649.1987.tb05280.x.
- Bianco PG. 1990. Vanishing freshwater fishes in Italy. Journal of Fish Biology 37(Suppl.A):235–237. DOI: 10.1111/j.1095-8649.1990.tb05050.x.
- Bianco PG. 1991. Sui pesci d’acqua dolce del fiume Esino (Marche, Italia centrale). Atti della Società Italiana di Scienze Naturali e del Museo Civico di Storia Naturale in Milano 132:49–60.
- Bianco PG. 1992. Zoogeographical implications of a first record of Lethenteron zanandreai on the Adriatic slope of central Italy (Cyclostomata: Petromyzontidae). Ichthyological Exploration of Freshwaters 3:183–186.
- Bianco PG. 1995a. Factors affecting the distribution of freshwater fishes especially in Italy. Cybium 19:241–259.
- Bianco PG. 1995b. I pesci d’acqua dolce delle Marche: Origini, problemi di conservazione e nuove prospettive di gestione. In: Salvaguardia e gestione dei beni ambientali nelle Marche, Ancona: pp. 229–257. Accademia marchigiana di Scienze, lettere e Arti.
- Bianco PG, Delmastro GB. 2011. Recenti novità tassonomiche riguardanti i pesci d’acqua dolce autoctoni in Italia e descrizione di una nuova specie di luccio. Researches on Wildlife Conservation 2(suppl.):1–13. IGF Publishing, USA.
- Caputo V, Giovannotti M, Nisi Cerioni P, Splendiani A, Marconi M, Tagliavini J. 2009a. Mitochondrial DNA variation of an isolated population of the Adriatic brook lamprey (Lampetra zanandreai Vladykov, 1955): Phylogeographic and phylogenetic inferences (Agnatha: Petromyzontidae). Journal of Fish Biology 75:2344–2351. DOI: 10.1111/j.1095-8649.2009.02413.x.
- Caputo V, Giovannotti M, Nisi Cerioni P, Splendiani A, Olmo E. 2009b. Chromosomal study of native and hatchery trout from Italy (Salmo trutta complex, Salmonidae): Conventional and FISH analysis. Cytogenetics and Genome Research 124:51–62. DOI: 10.1159/000200088.
- Costa OG. 1853. La fauna del regno di Napoli ossia enumerazione di tutti gli animali che abitano le diverse regioni di questo regno e le acque che le bagnano. Pesci. Parte I. Napoli, Italia: Stabilimento Tipografico di Filippo Azzolino.
- De Santis V, Delmastro GB, Vanetti I, Britton JR, Zaccara S. 2021. Species composition of introduced and natural minnow populations of the Phoxinus cryptic complex in the westernmost part of the Po River Basin (north Italy). Biological Invasions 23:657–668. DOI: 10.1007/s10530-020-02406-.
- Dondi L, D’Andrea MG. 1986. La Pianura Padana e Veneta dall’Oligocene superiore al Pleistocene. Giornale di Geologia 48:197–225.
- Ehlers J, Gibbard PL, Hughes PD, eds. 2011. Quaternary Glaciations – Extent and Chronology: A Closer Look. Oxford, UK: Elsevier. Online supplementary information. http://booksite.elsevier.com/9780444534477/index.php. Accessed Feb 2014 3.
- Folmer O, Black M, Hoen W, Lutz R, Vrijenhoek R. 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology 3:294–299.
- Gandolfi G, Torricelli P, Zerunian S, Marconato A. 1991. I pesci delle acque interne italiane. Rome: Unione Zoologica Italiana, Istituto Poligrafico e Zecca dello Stato. pp. 616.
- Geiger MF, Herder F, Monaghan MT, Almada V, Barbieri R, Bariche M et al. 2014. Spatial heterogeneity in the Mediterranean Biodiversity Hotspot affects barcoding accuracy of its freshwater fishes. Molecular Ecology Resources 14:1210–1221. DOI: 10.1111/1755-0998.12257.
- Imoto JM, Saitoh K, Sasaki T, Yonezawa T, Adachi J, Kartavtsev YP et al. 2013. Phylogeny and biogeography of highly diverged freshwater fish species (Leusciscinae, Cyprinidae, Teleostei) inferred from mitochondrial genome analysis. Gene 514:112–124. DOI: 10.1016/j.gene.2012.10.019.
- Kottelat M, Freyhof J. 2007. Handbook of European freshwater fishes. Cornol, Switzerland: Publications Kottelat.
- Landini W, Sorbini L. 1989. Ichthyofauna of the evaporitic Messinian in the Romagna and Marche regions. Bollettino della Società Paleontologica Italiana 28:287–293.
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23(21):2947–2948. DOI: 10.1093/bioinformatics/btm404.
- Leigh JW, Bryant D. 2015. PopART: Full-feature software for haplotype network construction. Methods in Ecology and Evolution 6(9):1110–1116. DOI: 10.1111/2041-210X.12410.
- Lorenzoni M, Carosi A, Quadroni S, De Santis V, Vanetti I, Delmastro GB, Zaccara S. 2021. Cryptic diversity within endemic Italian barbels: Revalidation and description of new Barbus species (Teleostei: Cyprinidae). Journal of Fish Biology 98(5):1433–1449. DOI: 10.1111/jfb.14688.
- Marcoaldi O. 1873. Guida e statistica della città e comune di Fabriano. Fabriano: Tipografia Crocetti.
- Marconi M. 2009. Carta Ittica della Provincia di Macerata: Acque di categoria B e C. Macerata: Provincia di Macerata.
- Maselli V, Hutton EW, Kettner AJ, Syvitski JPM, Trincardi F. 2011. High-frequency sea level and sediment supply fluctuations during Termination I: An integrated sequence-stratigraphy and modeling approach from the Adriatic Sea (Central Mediterranean). Marine Geology 287:54–70. DOI: 10.1016/j.margeo.2011.06.012.
- Palandačić A, Bravničar J, Zupančič P, Šanda R, Snoj A. 2015. Molecular data suggest a multispecies complex of Phoxinus (Cyprinidae) in the Western Balkan Peninsula. Molecular Phylogenetics and Evolution 92:118–123. DOI: 10.1016/j.ympev.2015.05.024.
- Palandačić A, Naseka A, Ramler D, Ahnelt H. 2017. Contrasting morphology with molecular data: An approach to revision of species complexes based on the example of European Phoxinus (Cyprinidae). BMC Evolutionary Biology 17:184. DOI: 10.1186/s12862-017-1032-x.
- Palandačić A, Kruckenhauser L, Ahnelt H, Mikschi E. 2020. European minnows through time: Museum collections aid genetic assessment of species introductions in freshwater fishes (Cyprinidae: Phoxinus species complex). Heredity 124:410–422. DOI: 10.1038/s41437-019-0292-1.
- Ramler D, Palandačić A, Delmastro GB, Wanzenböck J, Ahnelt H. 2017. Morphological divergence of lake and stream Phoxinus of Northern Italy and the Danube basin based on geometric morphometric analysis. Ecology and Evolution 7(2):572–584. DOI: 10.1002/ece3.2648.
- Schönhuth S, Vukić J, Šanda R, Yang L, Mayden RL. 2018. Phylogenetic relationships and classification of the Holarctic family Leuciscidae (Cypriniformes: Cyprinoidei). Molecular Phylogenetics and Evolution 127:781–799. DOI: 10.1016/j.ympev.2018.06.026.
- Splendiani A, Giovannotti M, Nisi Cerioni P, Caniglia ML, Caputo V. 2006. Phylogeographic inference on the native brown trout mtDNA variation in central Italy. Italian Journal of Zoology 73:179–189. DOI: 10.1080/11250000600679751.
- Splendiani A, Berrebi P, Tougard C, Righi T, Reynaud N, Fioravanti T, Lo Conte P, Delmastro GB, Baltieri M, Ciuffardi L, Candiotto A, Sabatini A, Caputo Barucchi V. 2020. The role of the south-western Alps as a unidirectional corridor for Mediterranean brown trout (Salmo trutta complex) lineages. Biological Journal of the Linnean Society 131(4):909–926. DOI: 10.1093/biolinnean/blaa125.
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research 22:4673–4680. DOI: 10.1093/nar/22.22.4673.
- Vucić M, Jelić D, Žutinić P, Grandjean F, Jelić M. 2018. Distribution of Eurasian minnows (Phoxinus: Cypriniformes) in the Western Balkans. Knowledge & Management of Aquatic Ecosystems 419(419):11. DOI: 10.1051/kmae/2017051.
- Waelbroeck C, Labeyrie L, Michel E, Duplessy JC, McManus JF, Lambeck K, Balbon E, Lambracherie M. 2002. Sea-level and deep water temperature changes derived from benthic foraminifera isotopic records. Quaternary Science Reviews 21:295–305. DOI: 10.1016/S0277-3791(01)00101-9.
- Zardoya R, Doadrio I. 1998. Phylogenetic relationships of Iberian cyprinids: Systematic and biogeographical implications. Proceedings of the Royal Society B: Biological Sciences 265(1403):1365–1372. DOI: 10.1098/rspb.1998.0443.
- Zickel M, Becker D, Verheul J, Yener Y, Willmes C. 2016. Paleocoastlines GIS dataset. CRC806-Database. DOI: 10.5880/SFB806.20.
