Abstract
Individuals identified as Aricidea assimilis Tebble, 1959 were collected from ten localities across the Mediterranean Sea from 0.5 to 225 m depth in order to have a wide coverage of the species habitats and geographic range and to assess the effects of environmental factors and biogeographical barriers on molecular and morphological diversity. Two mitochondrial and one nuclear markers were used to reconstruct phylogenetic relationships and test the occurrence of cryptic species. We observed two highly divergent lineages, one including all individuals from shallow, sandy environments (<10 m depth) and the other with the individuals from deeper muddy bottoms (30–225 m depth). Less pronounced divergence was detected between morphologically distinct brackish-water individuals and the remaining shallow-water individuals. The divergence observed between deep-water and shallow-water lineages is consistent with the hypothesis of distinct species. The ambiguous results of species delimitation tests applied to the two shallow-water sub-lineages might instead suggest a process of incipient speciation, even if this hypothesis needs additional evidence. These results suggest that sediment represents the main factor driving genetic divergence and ultimately cryptic speciation in A. assimilis, while other depth-associated factors and geographical barriers do not seem to significantly contribute to the genetic architecture of this species, suggesting the occurrence of wide-range larval dispersal.
Introduction
Among polychaetes, the family Paraonidae is a rather diverse group with more than 120 described species occurring on soft bottoms from the tidal zone to the abyssal depths (Grosse et al. Citation2021). In several marine environments, Paraonidae represent the dominant taxon in terms of abundance and biomass, and contribute to sediment dynamics, food webs and many other ecological processes (Gibbs Citation1965; Blake Citation1996; Quiroz-Martinez et al. Citation2012). Despite their importance, many aspects of the biology of Paraonidae, such as their reproductive features and population connectivity, are still scarcely known (Grosse et al. Citation2021). Moreover, many nominal species show a very wide distribution and some of them are considered cosmopolitan (Strelzov Citation1973). This characteristic, together with the high level of intraspecific morphological variability, was interpreted as a possible clue of cryptic speciation (Katzmann & Laubier Citation1975). In fact, molecular data reported by Langeneck et al. (Citation2019) showed the occurrence of several cases of divergent lineages within single nominal species, suggesting that the number of species within Paraonidae is currently underestimated. Nonetheless, cryptic diversity and speciation processes in Paraonidae are largely unknown, in particular as regards the role played by environmental factors and biogeographical breaks and boundaries in the diversification of this family.
According to scientific literature, Aricidea assimilis Tebble Citation1959, is a species that typically occurs in sandy and muddy bottoms from 2 to 300 meters depth (Castelli Citation1987). As for several paraonid species, its actual distribution is somewhat unclear. Even though Strelzov (Citation1973) reported A. assimilis from the Northern Pacific Ocean, Red Sea and Southern Atlantic Ocean, the majority of reliable records are referred to the Mediterranean Sea and adjacent Atlantic waters (Tebble Citation1959; Laubier & Ramos Citation1974; Katzmann & Laubier Citation1975; Castelli Citation1985, Citation1987; Çinar et al. Citation2014; Erdoğan-Dereli & Çinar Citation2020). Blake (Citation1996) argued that records from the Pacific Ocean by Strelzov (Citation1973) and Hobson (Citation1976) should probably be referred to different species, and this most likely accounts also for Lovell’s (Citation2002) report for the Andaman Sea (Indian Ocean). In addition, the high degree of morphological variability within A. assimilis accounts for taxonomic uncertainties surrounding this taxon. In fact, the species was misidentified as Aricidea fauveli Hartman, 1957 (= A. lopezi) (Bellan Citation1965), Aricidea fragilis Webster, 1879 (Amoureux Citation1970), and Aricidea lopezi Berkeley & Berkeley, 1956 (Strelzov Citation1973). Moreover, A. assimilis was redescribed as Aricidea mutabilis by Laubier and Ramos (Citation1974), who highlighted the high degree of intraspecific morphological variability. These authors observed a high degree of variability in the size and shape of prostomial antenna, which may vary from very long to relatively short. Even though Katzmann and Laubier (Citation1975) raised the doubt that short-antenna and long-antenna forms of A. assimilis could actually represent separate species, they provisionally considered them conspecific. It is noteworthy that the short-antenna form was erroneously interpreted as conspecific with the Pacific A. lopezi (Strelzov Citation1973; Castelli Citation1987). More recently, Erdoğan-Dereli and Çinar (Citation2020) described Aricidea pseudoassimilis for the Sea of Marmara, suggesting that A. assimilis specimens with short prostomial antenna might belong to this species.
The exclusive use of morphological data in several cases revealed itself misleading when describing the actual diversity of a group of organisms. This is particularly true for annelids, where cryptic and pseudo-cryptic species are continuously discovered (Nygren Citation2014) and morphological variability and phenotypic plasticity might lead to incorrect conclusions on boundaries between species (Meyer et al. Citation2008; Syomin et al. Citation2017; Righi et al. Citation2019). An integrated approach is therefore necessary to disentangle the actual diversity of polychaete taxa. Moreover, the vast majority of studies on cryptic speciation in annelids focused on genetic differences and geographical distribution of lineages. While the role of biogeography is obvious in the separation of lineages occurring in different biogeographical sectors, or with a strongly skewed geographical distribution (Iannotta et al. Citation2009; Cossu et al. Citation2015), in several cases cryptic species occur in sympatry or even in syntopy (Carr et al. Citation2011; Nygren & Pleijel Citation2011; Langeneck et al. Citation2020). In this case, the evolutionary reasons underlying lineage separation are often unclear, and might involve adaptation to different environmental conditions, such as depth, sediment grain (Bleidorn et al. Citation2006; Luttikhuizen & Dekker Citation2010), or even finer adaptations, such as a shift in the reproductive period (Boidin-Wichlachz et al. Citation2021).
In this study, we employed an integrative approach to assess the occurrence of cryptic species within A. assimilis in the Mediterranean Sea, in order to understand if different lineages are morphologically distinguishable, and to identify possible environmental drivers leading to genetic divergence and ultimately speciation.
Materials and methods
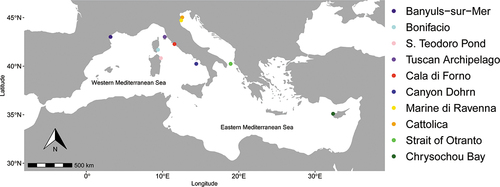
A total of 88 individuals of Aricidea assimilis were collected from ten Mediterranean localities, six in the Western and four in the Eastern Mediterranean Sea. The sampling depth varied from 0.5 to 225 m and the local sample size ranged from 2 to 28 individuals; small sample size was due to the low density reached by the species, and the impossibility to obtain additional samples in some localities. (; ).
Table I. Sampling localities of the Aricidea assimilis individuals employed in this study. Legend: BW: Brackish–water; SM: Shallow marine; IM: Intermediate marine; DM: Deep marine; S: Silty sand; C: Silt or clayish silt; N: number of specimens.
Sediment samples were collected with a Van Veen grab or with a hand corer by free-diving, and subsequently sieved with a 0.5 mm mesh. When possible, individuals of A. assimilis were sorted alive, otherwise sorting was carried out after fixation of the whole sample in 96% ethanol. Prior to DNA extraction, morphological traits, and especially the length of the antenna, were observed under microscope; it should be noted, however, that in most specimens the prostomial antenna was often missing or damaged. Specimens were stored in 96% ethanol at 4 or −20°C until DNA extraction. DNA was extracted whenever possible from the posterior part of the individual, while the anterior part was kept as morphological voucher and deposited in the polychaete collection of the Natural History Museum of the University of Pisa (MSNP); for some specimens, however, the extraction of DNA from the whole individual was necessary.
DNA extraction was carried out using the GenElute™ Mammalian Genomic DNA Miniprep Kit distributed by Sigma-Aldrich, following the manufacturer’s instructions. Individuals fixed with 4% neutralised formaldehyde in seawater (a part of the Tuscan Archipelago material) were first de-contaminated with diluted sodium hypochlorite, and then washed ten times in phosphate buffered saline solution before extraction, following the protocol by Forcina et al. (Citation2015). The mitochondrial regions coding for 16S rRNA and cytochrome c oxidase subunit I (COI) and a nuclear region including the ITS regions with a small portion of the 28S rRNA (henceforth ITS) were amplified. 16S rDNA amplification was obtained using the primer pair 16S ANNNF (5’-GCGGTATCCTGACCGTRCWAAGGTA-3’) and 16S_ANNR (5’-TCCTAAGCCAACATCGAGGTGCCAA-3’) (Sjölin et al. Citation2005), whereas for COI amplification the annelid-specific primers POLYLCO (5’-GAYTATWTTCAACAAATCATAAAGATATTGG-3’) and POLYHCO (5’-TAMACTTCWGGGTGACCAAARAATCA-3’) (Carr et al. Citation2011) were employed. In some cases semi-nested PCRs were performed using the combination of POLYLCO and the custom-designed Par-R-2 (5’- GGRTCAWAGAAWGT-3’) and of the custom-designed Par-F-1 (5’- CACGCCTTCCTAATAAT-3’) and POLYHCO. ITS amplification was carried out using the primers ITS-F (5’-TCGTAACAAGGTTTCCGTAGG-3’) and ITS-R (5’- GGTCCGTGTTTCAAGACGGG-3’) (Di Giuseppe et al. Citation2013). Polymerase chain reaction (PCR) amplifications were carried out in 20 μL solutions using 1.5 mM of MgCl2, 0.2 mM of each dNTP, 0.1 μM of each primer, 1 U of DreamTaq DNA polymerase (Thermo Scientific), and ~2.5 ng of template DNA. For 16S rDNA and ITS the PCR profile was set as follows: initial denaturing step at 94°C for 3 min, 34 cycles of denaturing at 94°C for 45s, annealing at 50°C for 1 min, and extending at 72°C for 1 min, with a final extending step at 72°C for 7 min. For COI, annealing temperature was set at 45°C. A negative control was included in each reaction. PCR products were precipitated with sodium acetate and absolute ethanol and sent to Macrogen Europe for sequencing.
Sequences were aligned with ClustalX v. 2.1 (Larkin et al. Citation2007) and alignments were checked and edited with BIOEDIT v. 7.2.5 (Hall Citation1999). To ensure the reliability of the alignments for phylogenetic inference, automatic alignment trimming was performed by the program trimAl v. 1.4 (Capella-Gutierrez et al. Citation2009) with the method option –automated1. Sequence evolution models were assessed using MEGA X v. 10.2.5 (Kumar et al. Citation2016) and under the Akaike Information Criterion (AIC) (Akaike Citation1974). Alignments of the three different genes were concatenated in Sequence Matrix v. 1.8 (Vaidya et al. Citation2011). Sequences of Cirrophorus branchiatus Ehlers, 1908, Cirrophorus nikebianchii Langeneck, Barbieri, Maltagliati & Castelli, 2017, Levinsenia demiri Çinar, Dağli & Açik, 2011, Paradoneis armata Glémarec, 1966, Paradoneis cf. ilvana Castelli Citation1985 and Paradoneis lyra (Southern, 1914), were used as outgroup (Genbank accession numbers: see Table SM1). A Bayesian consensus phylogenetic tree based on the three concatenated markers was constructed using MrBayes v. 3.2.7 (Ronquist et al. Citation2011). Two replicate runs were carried out with three Markov chains per run for 2 × 106 generations. Each chain was sampled every 100 generations to obtain 20000 sampled trees. The first 5000 sampled trees (25%) were discarded as burn-in, with the remaining 15000 trees used to estimate the Bayesian posterior probability (PP) of tree nodes. The convergence of Bayesian analyses was checked through the standard deviation of split frequencies that should reach a value <0.01 at the end of the analysis (Ronquist et al. Citation2011). A maximum-likelihood tree of the concatenated alignment was produced with MEGA X v. 10.2.5 (Kumar et al. Citation2016) with the computation of 1000 bootstrap replications. Additionally, phylogenetic trees were inferred independently for each gene and each method according to parameters described above.
Pairwise K2P distances (Kimura Citation1980) were calculated in R using the ape package (Paradis et al. Citation2004). The separation at species level of the identified lineages was tested using two different single-locus species delimitation tests. The Automatic Barcoding Gap Discovery approach (ABGD) uses a range of prior intraspecific divergences to infer from sequence data a model-based one-sided confidence limit for intraspecific divergence. Thereafter, the algorithm detects the barcoding gap as the first significant gap beyond this limit and uses it to partition data, automatically sorting sequences into hypothetical species (Puillandre et al. Citation2012). The Poisson Tree Processes approach (PTP), on the other hand, uses phylogenetic trees, and in particular branch length (as proxy of number of substitutions), based on the principle that the number of substitutions between species is significantly higher than the number of substitutions within species (Zhang et al. Citation2013).
Haplotype networks were constructed by using the R package pegas (Paradis Citation2010), following the statistical parsimony method of Templeton et al. (Citation1992). This method estimates the maximum number of differences among haplotypes as a result of single mutation. It groups haplotypes differing from one substitution together, then from two, three, etc., and computes a cladogram displaying linkages that have a probability >0.95 of being true.
Results
Phylogenetic reconstruction and species delimitation
We obtained 46 sequences of a 606 bp portion of COI (GenBank accession numbers OM416165 to OM416210), 84 sequences of a 285 bp portion of 16S (GenBank accession numbers OM416044 to OM416129) and 17 sequences of a 643 bp portion of ITS (GenBank accession numbers OM419196 to OM419212). The best fitting nucleotide substitution model was GTR+G + I (Tavaré Citation1986) for all markers. The concatenated alignment, including the outgroup sequences, was 1595 bp long.
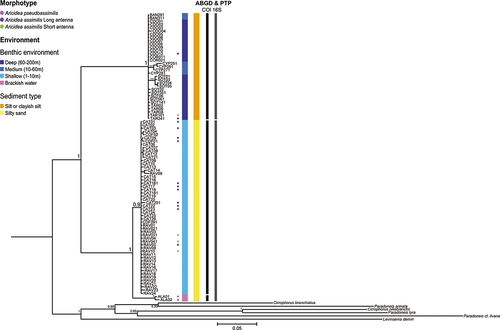
The Bayesian tree () showed a high degree of molecular divergence between deep- and shallow-water individuals. Less pronounced divergence can be observed between all marine shallow-water individuals and specimens from the brackish-water San Teodoro pond. The above-described clusters are supported by high values of posterior probability (PP > 0.95) (). These groups are further supported by the maximum-likelihood concatenated tree (Fig. SF1) with a bootstrap support of 100 for the deep-water and shallow-water clade and 100 for the shallow subgroup. All single-gene phylogenies supported the first clade regardless of the phylogenetic method, although sometimes with lower node support (Fig. SF2-SF7).
Figure 2. Bayesian inference phylogenetic tree of Aricidea assimilis based on the concatenated alignments of COI, 16S and ITS markers. Node values indicate Bayesian posterior probabilities. Each label prefix corresponds to the specimen sampling locality with BAN: Banyuls-sur-Mer; COR: Bonifacio; ALA: San Teodoro Pond; TAR: Tuscan Archipelago; CDF: Cala di Forno; CDO: Canyon Dohrn; RAV: Marina di Ravenna; CAT: Cattolica; SOT: Strait of Otranto; CYP: Chrysochou Bay. Coloured dots next to tip labels represent the individual morphotypes when they could be determined. Benthic environment and sediment type data associated with the specimen are represented by colour bars. Species delimitations proposed by ABGD and PTP tests are represented by interrupted lines on the right side of the tree, in black for COI and in grey for 16S.
For the COI dataset, K2P distances within groups ranged between 0% and 4%. Sequences of shallow-water marine and brackish-water individuals were separated by 6–7% genetic distances. Distances between deep-water and shallow-water individuals ranged from 19% to 28% (); however, those calculated for the 16S and ITS markers were not as wide. For 16S, within-group distances were 0% to 1.5%, while slightly higher distances were retrieved between marine and brackish shallow-water individuals (2–3%). The distance between deep-water and shallow-water individuals ranged from 14% to 17% (). Pairwise K2P distances calculated for ITS reached 2% of divergence within groups and 12–16% between the shallow-marine and deep-marine group (); unfortunately, we were not able to obtain ITS sequences for the San Teodoro Pond specimens.
Figure 3. K2P distance ranges observed in the three markers examined for A. assimilis. White: distances between individuals sampled in the same habitat type; Gray: distances between brackish–water and marine shallow individuals; Black: distances between deep and shallow–water individuals.
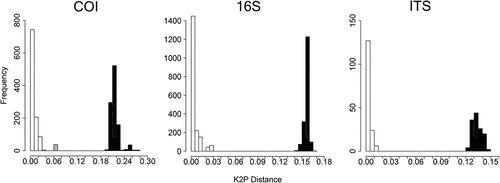
ABGD and PTP species delimitation tests were consistent in supporting the separation at species level of the shallow-water and the deep-water lineages, according to COI and 16S markers (). Nevertheless, the two tests provided conflicting results regarding the brackish-water and marine shallow groups (). In fact, the COI dataset suggested the separation of these lineages into different species, while the 16S dataset did not.
Haplotype networks
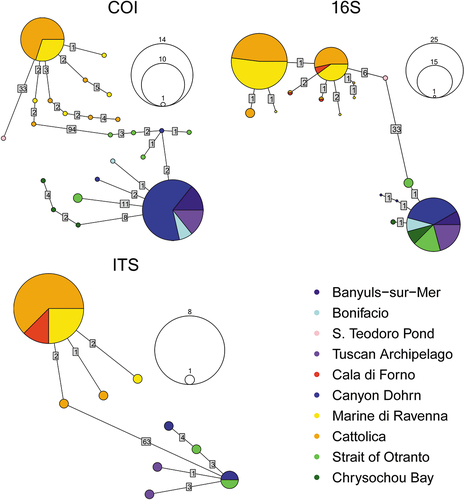
All haplotype networks showed a reciprocally monophyletic structure (Jenkins et al. Citation2018), and the shallow and deep lineages were separated by a high number of mutational steps: 99 for COI (), 33 for 16S () and 63 for the ITS network (). Sequences of the brackish-water individuals from San Teodoro Pond were separated from shallow-water marine ones by 33 steps in the COI haplotype network and 6 steps in the 16S network. In each of the shallow-water and deep-water groups, all networks contained a highly frequent haplotype, shared by a large number of individuals; up to 8, 14, 25 individuals in ITS, COI, 16S networks, respectively. The 16S network also displayed an additional highly frequent haplotype (15 sequences) corresponding to sequences from shallow-water marine individuals. Except for the shallow-water group in the COI network, specimens from populations of the Western and Eastern Mediterranean basins shared the most frequent haplotypes. This pattern was also found in two less frequent haplotypes of the 16S representation (), which included sequences from Cala di Forno (Western Mediterranean) and Cattolica (Eastern Mediterranean). Many unique haplotypes were separated by one or two mutational steps from the most frequent haplotypes in the 16S and ITS networks. Conversely, in the COI network, unique haplotypes were often separated from the most frequent haplotype by a higher number of mutational steps. This is especially true for sequences from Chrysochou Bay (Cyprus, Eastern Mediterranean), connected to the most frequent haplotype by eight mutational steps, and a part of the sequences from the Strait of Otranto, connected to the most frequent haplotype by 11 mutational steps (however, the majority of sequences from the Strait of Otranto were closer to the shared haplotype and intermixed with a Tyrrhenian sequence from Canyon Dohrn).
Figure 4. Statistical parsimony haplotype networks of COI, 16S and ITS markers in the A. assimilis complex. In the networks, each haplotype is represented by a circle. The size of the circle corresponds to the number of individuals displaying that haplotype. The haplotype scales are drawn on the right of the haplotype networks. Numbers on links between haplotypes indicate the number of mutational steps separating them. Haplotypes are colored according to the origin of individuals.
Discussion
Although several clues of cryptic speciation in Paraonidae were found in Langeneck et al.’s (Citation2019) molecular phylogenetic reconstruction, the present study gave for the first time a deeper insight into the distribution of cryptic lineages across the Mediterranean Sea, allowing to infer on possible reasons for the observed diversification. Katzmann and Laubier’s (Citation1975) hypothesis about the occurrence of a species complex within the morphospecies Aricidea assimilis is validated by the current study. Nevertheless, results did not support the hypothesis on taxonomic separation between short- and long-antenna individuals (Strelzov Citation1973; Laubier & Ramos Citation1974), suggesting that a correct interpretation of morphological characters is more complex than previously considered. In fact, both marine clades included specimens with short and long antenna. The holotypes of A. assimilis and Aricidea mutabilis, which are considered synonymous (Katzmann & Laubier Citation1975), were sampled on low infralittoral/high circalittoral bottoms sensu Pérès and Picard (Citation1964) (50 to 60 m depth) (Tebble Citation1959; Laubier & Ramos Citation1974), suggesting that the two taxa are actually synonymous, and the name Aricidea assimilis should be employed for the deep-water lineage (30–120 m). In fact, the specimens from Banyuls-sur-Mer, sampled rather close to the type locality of A. mutabilis, and the Levantine specimens examined are likely to correspond to A. assimilis s.s. However, topotypic material would be needed to clarify this point. Specimens from San Teodoro pond showed a good morphological correspondence with the recently described Aricidea pseudoassimilis, which is characterised by a shorter, blunt antenna and branchiae with blunt tips, but the genetic distance towards the remaining marine individuals did not allow to consider them as univocally separated at species level. The holotype and the majority of paratypes of A. pseudoassimilis were sampled at around 10 m depth, although the examined material included specimens collected down to 100 m (Erdoğan-Dereli & Çinar Citation2020). Even though we did not have the opportunity to examine specimens from the Sea of Marmara (type locality of A. pseudoassimilis), it is possible that this name could be applied to the shallow-water lineage. However, the morphological differences highlighted by Erdoğan-Dereli and Çinar (Citation2020) between the two taxa do not seem to be relevant to separate the two clades. The inconsistency between morphotypes and genetic lineages suggests that the aforementioned morphological features may depend on factors other than phylogenetic relationships, such as ontogeny or phenotypic plasticity, and caution should be taken when using such features to diagnose Paraonidae species, as already observed for other polychaete families (Meyer et al. Citation2008; Iannotta et al. Citation2009; Langeneck et al. Citation2020).
Results of the present study highlighted that the individuals of A. assimilis analysed are separated in three mitochondrial lineages. The deep-water lineage is clearly distinguished from the shallow-water counterpart, which in turn is composed by one widespread marine sub-lineage, and another sub-lineage detected only in the brackish-water San Teodoro Pond, as depicted in the phylogenetic tree (). Even if all nodes at the basis of these clades showed high statistical support, the divergence between the deep-water and the shallow-water groups is remarkably higher than that between the two shallow-water sub-lineages. Genetic distance values between deep-water and shallow-water individuals () are clearly in the range of interspecific distances detected by other studies on polychaetes (Pleijel et al. Citation2009; Nygren & Pleijel Citation2011; Neal et al. Citation2014). This outcome is confirmed by the pattern retrieved with the nuclear ITS fragment and, along with the consistent result of the two species delimitation tests on the three genes, allowed to consider the deep- and shallow-water lineages as separated at species level. On the other hand, the distances observed between the two shallow-water sub-lineages are approximately four- to five-fold lower, even though the distance calculated through COI sequences is remarkably higher than the 3% value proposed as species’ boundary by Hebert et al. (Citation2003). It is worth noting, however, that recent studies stressed that the identification of a barcoding gap is more important than the setting of a fixed threshold (Čandek & Kuntner Citation2015; Kvist Citation2016). Moreover, intraspecific distances of several annelid taxa turned out to be higher than 3%, being closer to the values of 6.2–6.4% identified in this study (Kvist Citation2016; Lobo et al. Citation2016). Therefore, the divergence observed between the two shallow-water sub-lineages is consistent with the hypothesis of conspecific individuals, which is also in agreement with the results of the species delimitation tests performed on the 16S dataset. However, this interpretation is poorly satisfying for two reasons. The first clue towards a different interpretation of these results is represented by the absence of geographical segregation between the two sub-lineages. In fact, Tyrrhenian shallow marine individuals are genetically closer to Adriatic shallow marine ones than to brackish-water individuals from the Tyrrhenian Sea. This suggests that, even if the separation between the brackish-water and the shallow marine lineages is more recent than the separation between the deep and the shallow clades, it is nevertheless old enough to be detected over the geographical separation. A more formal clue was provided by the ambiguous results of species delimitation tests, that in the case of COI separated the two groups at species level, but failed in doing so with 16S rDNA (). A similar ambiguous situation has been retrieved between the closely related species Diopatra neapolitana (Delle Chiaje, 1828) and Diopatra aciculata Knox & Cameron, 1971. Also in this case, COI sequences allowed to readily separate the two species, while the distinction was distinctly lower when considering 16S rDNA sequences, and nuclear markers did not show any separation between the two alleged species, suggesting that the speciation process is still ongoing (Elgetany et al. Citation2020). The relationship between the shallow-water marine lineage and the brackish-water lineage is very similar to that retrieved between D. neapolitana and D. aciculata by Elgetany et al. (Citation2020) and suggests that these lineages should be regarded as two incipient species (Mallet Citation2007), within the so-called “grey zone” of speciation. Hausdorf (Citation2011) underlined that randomly sampled molecular markers do not always allow to distinguish between incipient species. This is particularly true when considering recombinant nuclear markers with lower mutation rates, but it can also be retrieved in case of mitochondrial markers with different mutation rates (Elgetany et al. Citation2020). Nonetheless, despite having been historically considered as neutral, and as such widely employed for phylogeographical reconstructions, mitochondrial markers may be subject to a certain degree of selection, which may impair their informativeness (Ballard & Whitlock, 2004). In fact, inconsistent diversity patterns between COI and other markers have been retrieved in marine invertebrates; for instance, Casu et al. (Citation2011) found in the ribbed limpet Patella ferruginea Gmelin, 1791 an almost monomorphic COI, against a clear spatial genetic structure retrieved with ISSR markers. In this case, COI seemed instead to show a higher variability with respect to 16S rDNA, but in both cases a selective process (stabilising selection in the case of P. ferruginea, divergent selection in the case of brackish-water A. assimilis) might be responsible for this discordance in genetic patterns. Moreover, although interesting, these data are based on a very limited number of individuals from a single brackish-water environment, and therefore any conclusion drawn should be taken with considerable caution.
While the shallow-water clades occurred from the surface to 10 m depth, the deep-water clade was widespread from around 30 to more than 200 m depth, ranging from the mid-infralittoral stage to the upper bathyal stage (Pérès & Picard Citation1964). Therefore, A. assimilis does not seem to be significantly affected by some of the environmental factors associated with depth, such as pressure, or temperature, as these factors show significant variations in the bathymetric range where the deep-water clade occurs. Also a direct influence of the seasonal thermocline can be excluded, since in the Mediterranean Sea seasonal variations in temperature around 10°C are detectable down to 60–70 m (Houpert et al. Citation2015), well below the 30 m isobath where the deeper clade was retrieved, both in the Western and in the Eastern Mediterranean Sea. It is worth noting that the shallower sampling sites in the Western Mediterranean (mid-infralittoral to high-circalittoral, 30–100 m) are located in the superficial Atlantic Water mass, the deeper Tyrrhenian ones (100–225 m) are included into the Tyrrhenian Intermediate Water mass (Napolitano et al. Citation2019), the deep Adriatic sample has been obtained in an area characterised by mixing between Levantine Intermediate Water and South Adriatic Deep Water (Modified Levantine Intermediate Water – Orlić et al. Citation1992), and lastly, the sample from Cyprus is included into the Levantine Surface Water mass (Sur et al. Citation1993). The absence of genetic diversification between populations of A. assimilis associated to different water masses supports the scarce effect of physical-chemical variables typically associated to the distinction of water masses, as temperature and salinity. Instead, it is likely that the most relevant ecological factor distinguishing the two main clades of A. assimilis is represented by sediment granulometry, which is related to depth, but shows a major change within the first tens of meters. In fact, samples in the shallow-water clade have been collected on sand, with a limited amount of silt, whereas samples in the deep-water clade have been obtained from silt or silty clay. Interestingly, despite the pronounced molecular divergence, individuals from shallow-water marine and deep-water marine environments could not be morphologically distinguished. It has been argued that early divergent species accumulate first differences in physiological, behavioral or reproductive traits rather than morphological ones (Struck et al. Citation2018), thus, they can remain morphologically undistinguishable. However, the divergence between these lineages does not suggest the speciation event to be more recent than for other morphologically distinct species of Aricidea, according to phylogenetic reconstructions carried out on Paraonidae (Langeneck et al. Citation2019). The inconsistency between morphological and genetic characters might be the result of drift or stabilizing selection, and may represent a case of morphological stasis (Struck et al. Citation2018). On the other hand, despite a less pronounced molecular divergence, the brackish lineage and shallow marine lineages can be separated by the size of the antenna, which is always short and with blunt tip in brackish individuals, and the length of branchiae, which is constant in individuals from San Teodoro pond but gradually increases in all marine specimens. This observation could suggest a limited effect of brackish-water environments on molecular evolution, probably due to the connectivity between brackish-water and marine environments that allows a certain degree of gene flow (Cognetti & Maltagliati Citation2000). The occurrence of speciation processes in brackish-water environments has been confirmed in recent years by molecular studies (Maltagliati et al. Citation2000, Citation2001; Beheregaray & Sunnucks Citation2001; Trabelsi et al. Citation2002; Sanna et al. Citation2013) and, at some extent, present results are consistent with the hypothesis that these environments may play an important role in lineage diversification. The selective pressure of brackish-water environments is often considered a strong driver for morphological diversification; in fact, brackish-water environments are often characterised by clearly differentiated morphotypes (Cognetti Citation1954; Maltagliati et al. Citation2001), even if these differences often do not reflect patterns of genetic diversity (Heras & Roldán Citation2011; Jimoh et al. Citation2013), or appear to be distinctly wider than molecular data would suggest (Maltagliati et al. Citation2001). On the other hand, the frequency of unfavourable events and stressful conditions in brackish-water environments might cause local extinctions and thus hamper diversification processes. In particular, the divergent morphotype of A. assimilis known for San Teodoro pond since the 1990s (Martinelli et al. Citation1997) was not found in a subsequent sampling of 2019, due to an extensive dystrophic crisis that significantly affected environmental quality of this brackish-water ecosystem (J. Langeneck, pers. obs.). The distribution of the brackish-water lineage of A. assimilis is currently unknown, but it is likely that it is vulnerable to the current increase of extreme weather events associated to climate change affecting brackish-water environments (Vignes et al. Citation2009). The comparison of the studied specimens with Atlantic sequences of A. laubieri deposited in GenBank showed that this species is clearly distinct from Mediterranean specimens and does not belong to the A. assimilis species complex.
Geographical boundaries seem to have a distinctly lower effect on diversification within the Aricidea assimilis complex. Our work showed that Adriatic and Tyrrhenian individuals were not separated by phylogenetic reconstruction (). A slight diversification of Eastern Mediterranean haplotypes from Chrysochou Bay was retrieved in the COI network; nonetheless, a similar pattern was found in a part of the specimens from the Strait of Otranto, while other specimens from the same population were closer to the most frequent haplotype. The co-occurrence of separate haplotype clusters within the same population was retrieved in both vertebrates (Angiulli et al. Citation2016) and invertebrates (Langeneck et al. Citation2020) characterised by wide-range dispersal of larval stages and might depend on past events of vicariance driven by biogeographical barriers that subsequently disappeared (Avise Citation2000). While most frequent haplotypes that are shared by individuals from different Mediterranean sites may alternatively be explained by high population size or haplotype ancestry (Posada & Crandall Citation2001), the fact that much rarer haplotypes are shared by shallow-water specimens of the Tyrrhenian and Adriatic Seas strongly indicates a high degree of connectivity. Indeed, these seas are separated by two phylogeographic breaks (Villamor et al. Citation2014), namely the Sicilian Strait and the Strait of Otranto. Usually, polychaetes and other marine invertebrates with direct development are geographically structured between the Adriatic and the Tyrrhenian Sea (Abbiati & Maltagliati Citation1996; Virgilio & Abbiati Citation2004; Cossu et al. Citation2015), whereas for species with dispersal phases genetic divergence between these two basins is lower or absent (Abbiati & Maltagliati Citation1992; Iannotta et al. Citation2007; Weber et al. Citation2015; Modica et al. Citation2017). In the present study, both the deep- and shallow-water lineages showed the absence of geographical structuring. Thus, the observed phylogeographic pattern in the two species identified in the A. assimilis complex suggests that their development comprises a relatively long-lived pelagic larval phases with high potential for dispersal. However, there are no reliable reports of planktonic larvae that can be assigned to this family (Blake Citation1996), and several species show epitoke modifications, large-sized eggs and sometimes dorsal brooding of juveniles (Grosse et al. Citation2021). These features led to the hypothesis that this family is characterised by direct development (Giangrande Citation1997), which is however not supported by the phylogeographic pattern detected in this study. The reason for the scarcity of Paraonidae larvae in planktonic samples is unclear; it is possible that reproductive events are sporadic and rather limited throughout the year, and that this feature makes their detection difficult, but the hypothesis of direct development for a part of the known species cannot be discarded based on current data. According to the molecular data presented by Langeneck et al. (Citation2019), some nominal taxa (e.g., Aricidea cerrutii Laubier, 1966) show the occurrence of several cryptic lineages, while others (e.g., Aricidea claudiae Laubier, 1967) are genetically homogeneous across the Mediterranean Sea. Further detailed studies on a wider array of Paraonidae species are needed to clarify if the phylogeographic pattern detected in the A. assimilis species complex can be generalised to all representatives of this family, or if different species and groups are characterised by different reproductive and developmental features.
Geolocation information
Banyuls-sur-Mer, France (42.485°N, 3.1618°E); Bonifacio, Corsica, France (41.4166°N, 9.3000°E); San Teodoro Pond, Sardinia, Italy (40.8096°N, 9.6766°E); Tuscan Archipelago, Italy (43.5347°N, 9.9955°E); Cala di Forno, Italy (42.6193°N, 11.0840°E); Canyon Dohrn, Italy (40.7382°N, 14.2040° E); Marina di Ravenna, Italy (44.4699°N, 12.3149°E); Cattolica, Italy (43.9822°N, 12.7525°E); Strait of Otranto, Italy (41.1757°N, 16.9021°E); Latsi, Chrysochou Bay, Cyprus (35.0659°N, 32.3943°E).
Supplemental Material
Download MS Excel (20.2 KB)Acknowledgements
We would like to thank S. Aliani, P. Bouchet, O. Bresciani, M. Grosse, L. Le Gall, C. Mazziotti, A. Pavia, L. Romani for providing ethanol-fixed individuals of Aricidea assimilis for this work; M. Casu, F. Crocetta, M. Oliva, L. Pacciardi, M. Pertusati, M. Ponti, C. Pretti, S. Stefanni and A. Vannucci for their invaluable help in field sampling; T. Ravaglia and F. Squarcia for their help in molecular laboratory work. Specimens from Bonifacio were sampled in the frame of the expedition Corsicabenthos 2, organised by the Muséum National d’Histoire Naturelle, Paris, in partnership with Université de Corse Pasquale Paoli and Office de l’Environnement de la Corse (OEC), with the support of Office Français de la Biodiversité (OFB) and Collectivité Territoriale de Corse (CTC) and in collaboration with Réserve Naturelle des Bouches de Bonifacio. Specimens from Canyon Dohrn were collected in the frame of the EARTH CRUISERS project (EARTH’s CRUst Imagery for investigating SEismicity, volcanism and marine natural Resources in the Sicilian offshore) funded by the Italian Ministry of University and Research. The material from Cyprus was collected by the Department of Fishery and Marine Research (DFMR) of the Ministry of Agriculture, Rural Development and Environment of the Republic of Cyprus as part of the implementation of: (i) the Water Framework Directive (2000/60/EC) and the Marine Strategy Framework Directive (2008/56/EC) and (ii) the project “Investigation and evaluation of sensitive benthic marine ecosystems in the territorial waters and the Exclusive Economic Zone of the Republic of Cyprus” which is co-financed by the Operational Program Thalassa 2014–2020 (European Maritime and Fisheries Fund (EMFF) 2014–2020 and national resources). C.J.L. Fourreau thanks Erasmus+ programme for travel funding and an internship scholarship. We are grateful to two anonymous reviewers whose comments improved the manuscript.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/24750263.2022.2138588
Disclosure statement
The authors do not have any conflict of interest to declare.
Data availability statement
Sequences were deposited in GenBank and examined specimens in the polychaete collection of the Natural History Museum of University of Pisa and (for specimens from Bonifacio) the Muséum National d’Histoire Naturelle of Paris. GenBank accession numbers for each specimen are provided in the electronic supplementary files (https://www.ncbi.nlm.nih.gov/genbank).
Additional information
Funding
References
- Abbiati M, Maltagliati F. 1992. Genetic population structure of Neanthes succinea (Polychaeta: Nereididae). Journal of the Marine Biological Association UK 72(3):511–517. DOI:10.1017/S0025315400059300.
- Abbiati M, Maltagliati F. 1996. Allozyme evidence of genetic differentiation between populations of Hediste diversicolor (Polychaeta: Nereididae) from the Western Mediterranean. Journal of the Marine Biological Association UK 76(3):637–647. DOI:10.1017/S0025315400031349.
- Akaike H. 1974. A new look at the statistical model identification. IEEE Transaction on Automatic Control 19(6):716–723. DOI:10.1109/TAC.1974.1100705.
- Amoureux L. 1970. Annélides Polychètes du Golfe de Tarente. Resultats des nouvelles croisieres de l’Albatros (1968, 1969). Annali del Museo Civico di Storia Naturale di Genova 78:1–11.
- Angiulli E, Sola L, Ardizzone G, Fassatoui C, Rossi AR. 2016. Phylogeography of the common pandora Pagellus erythrinus in the central Mediterranean Sea: Sympatric mitochondrial lineages and genetic homogeneity. Marine Biology Research 12(1):4–15. DOI:10.1080/17451000.2015.1069355.
- Avise JC. 2000. Phylogeography: The history and formation of species. Cambridge: Harvard University Press. pp. 447.
- Beheregaray LB, Sunnucks P. 2001. Fine-scale genetic structure, estuarine colonization and incipient speciation in the marine silverside fish Odontesthes argentinensis. Molecular Ecology 10(12):2849–2866. DOI:10.1046/j.1365-294X.2001.t01-1-01406.x.
- Bellan G. 1965. Contribution à l’étude des Polychètes profondes des parages de Monaco et des cotes de la Corse. Bulletin de l’Institut Océanographique de Monaco 65:1–24.
- Blake JA. 1996. Family Paraonidae Cerruti, 1909. In: Blake JA, Hilbig B, Scott PH, editors. Taxonomic atlas of the Benthic Fauna of the Santa Maria Basin and Western Santa Barbara Channel. 6 – The Annelida Part 3. Polychaeta: Orbiniidae to Cossuridae. Santa Barbara Museum of Natural History, Santa Barbara, California, the U.S.A. pp. 27–70.
- Bleidorn C, Kruse I, Albrecht S, Bartolomaeus T. 2006. Mitochondrial sequence data expose the putative cosmopolitan polychaete Scoloplos armiger (Annelida, Orbiniidae) as a species complex. BMC Evolutionary Biology 6(1):47. DOI:10.1186/1471-2148-6-47.
- Boidin-Wichlachz C, Jollivet D, Papot C, Roisin L, Massol F, Tasiemski A. 2021. Genetic diversification and life-cycle of the polychaete Capitella spp. from the English Channel: Evidence for sympatric cryptic species and alternative reproductive strategies. Marine Biology 168(12):176. DOI:10.1007/s00227-021-03972-2.
- Čandek K, Kuntner M. 2015. DNA barcoding gap: Reliable species identification over morphological and geographical scales. Molecular Ecology Resources 15(2):268–277. DOI:10.1111/1755-0998.12304.
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. 2009. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25(15):1972–1973. DOI:10.1093/bioinformatics/btp348.
- Carr CM, Hard SM, Brown TM, Macdonald TA, Hebert PDN. 2011. A tri-oceanic perspective: DNA barcoding reveals geographic structure and cryptic diversity in Canadian polychaetes. Plos One 6(7):e22232. DOI:10.1371/journal.pone.0022232.
- Castelli A. 1985. Paraonidae (Annelida, Polychaeta) des fonds meubles infralittoraux des côtes toscanes. Cahiers de Biologie Marine 26:267–279.
- Castelli A. 1987. Censimento dei policheti dei mari italiani: Paraonidae Cerruti, 1909. Atti della Società Toscana di Scienze Naturali. Memorie, Serie B 94:319–340.
- Casu M, Rivera-Ingraham GA, Cossu P, Lai T, Sanna D, Dedola GL, Sussarellu R, Sella G, Cristo B, Curini-Galletti M, García-Gómez JC, and Espinosa F. 2011. Patterns of spatial genetic structuring in the endangered limpet Patella ferruginea: Implications for the conservation of a Mediterranean endemic. Genetica 139:1293–1308. DOI:10.1007/s10709-012-9631-3.
- Çinar ME, Dağli E, Kurt–Şahin G. 2014. Checklist of Annelida from the coasts of Turkey. Turkish Journal of Zoology 38:734–764. DOI:10.3906/zoo-1405-72.
- Cognetti G. 1954. Forme della Mercierella enigmatica nella nuova stazione del Lago di Patria. Bollettino di Zoologia 21:41–45.
- Cognetti G, Maltagliati F. 2000. Biodiversity and adaptive mechanisms in brackish water fauna. Marine Pollution Bulletin 40:7–14.
- Cossu P, Maltagliati F, Pannacciulli FG, Simonini R, Massamba-N’Siala G, Casu M, Lardicci C, Prevedelli D, Castelli A. 2015. Phylogeography of Ophryotrocha labronica (Polychaeta, Dorvilleidae) along the Italian coasts. Marine Ecology 36:1088–1097.
- Di Giuseppe G, Barbieri M, Vallesi A, Luporini P, Dini F. 2013. Phylogeographical pattern of Euplotes nobilii, a protist ciliate with a bipolar biogeographical distribution. Molecular Ecology 22:4029–4037.
- Elgetany AH, van Rensburg H, Hektoen M, Matthee C, Budaeva N, Simon CA CA, Struck TH. 2020. Species delineation in the speciation grey zone – The case of Diopatra (Annelida, Onuphidae). Zoologica Scripta 49:516–534.
- Erdoğan-Dereli D, Çinar ME. 2020. The diversity of the genus Aricidea (Polychaeta: Paraonidae) from the Sea of Marmara, with descriptions of two new species and two new records for the Mediterranean fauna. Zootaxa 4844:1–73.
- Forcina G, Guerrini M, van Grouw H, Gupta BK, Panayides P, Hadjigerou P, Al–Sheikhly OF, Awan MN, Khan AA, Zeder MA, Barbanera F, 2015. Impacts of biological globalization in the Mediterranean: Unveiling the deep history of human–mediated gamebird dispersal. Proceedings of the National Academy of Sciences 112:3296–3301.
- Giangrande A. 1997. Polychaete reproductive patterns, life cycles and life histories: An overview. Oceanography and Marine Biology 35:323–386.
- Gibbs PA. 1965. Recent additions to the marine fauna of Whitstable, with a description of Aricidea minuta Southward (Annelida: Polychaeta). Annals and Magazines of Natural History 7:33–36.
- Grosse M, Zhadan A, Langeneck J, Fiege D, Martínez A. 2021. Still digging: Advances and perspectives in the study of the diversity of several sedentarian annelid families. Diversity 13:132.
- Hall TA. 1999. BioEdit: A user–friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41:95–98.
- Hausdorf B. 2011. Progress towards a general species concept. Evolution 65:923–931.
- Hebert PDN, Cywinska A, Ball SL, deWaard JR, 2003. Biological identification through DNA barcode. Proceedings of the Royal Society of London Series B 270:313–321.
- Heras S, Roldán MI. 2011. Phylogenetic inference in Odontesthes and Atherina (Teleostei: Atheriniformes) with insights into ecological adaptation. Comptes Rendus Biologies 334:273–281.
- Hobson KD. 1976. Protoariciella oligobranchia new species (Orbiniidae) and six new records of Orbiniidae, Questidae, and Paraonidae (Annelida, Polychaeta) from British Columbia. Canadian Journal of Zoology 54:591–596.
- Houpert L, Testor P, Durrieu de Madron X, Somot S, D'Ortenzio F, Estournel C, and Lavigne H. 2015. Seasonal cycle of the mixed layer, the seasonal thermocline and the upper-ocean heat storage rate in the Mediterranean Sea derived from observations. Progress in Oceanography 132:333–352. DOI:10.1016/j.pocean.2014.11.004.
- Iannotta MA, Gambi MC, Patti FP. 2009. Molecular evidence of intraspecific variability in Lysidice ninetta (Polychaeta: Eunicidae) in the Mediterranean Sea. Aquatic Biology 6:121–132.
- Iannotta MA, Patti FP, Ambrosino M, Procaccini G, Gambi MC. 2007. Phylogeography of two species of Lysidice (Polychaeta, Eunicidae) associated to the seagrass Posidonia oceanica in the Mediterranean Sea. Marine Biology 150:1115–1126.
- Jenkins TL, Castilho R, Stevens JR. 2018. Meta-analysis of northeast Atlantic marine taxa shows contrasting phylogeographic patterns following post-LGM expansions. PeerJ 6:e5684.
- Jimoh AA, Anetekhai MA, Cummings S, Abanikanda OTF, Turner GF, van Oosterhout C, Hänfling B. 2013. Mismatch between molecular (mtDNA) and morphological classification of Macrobrachium prawns from Southern Nigeria: Cryptic freshwater species and brackish water morphotypes. Aquaculture 410–411:25–31.
- Katzmann W, Laubier L. 1975. Paraonidae (Polychètes Sédentaires) de l’Adriatique. Annalen des Naturhistorischen Museums in Wien. Serie B, Für Botanik und Zoologie 79:567–588.
- Kimura M. 1980. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution 16:111–120.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33:1870–1874.
- Kvist S. 2016. Does a global barcoding gap exist in Annelida? Mitochondrial DNA 27:2241–2252.
- Langeneck J, Barbieri M, Maltagliati F, Castelli A. 2019. Molecular phylogeny of Paraonidae. Molecular Phylogenetics and Evolution 136:1–13.
- Langeneck J, Scarpa F, Maltagliati F, Sanna D, Barbieri M, Cossu P, Mikac B, Curini-Galletti M, Castelli A, Casu M. 2020. A complex species complex: The controversial role of ecology and biogeography in the evolutionary history of Syllis gracilis Grube, 1840 (Annelida, Syllidae). Journal of Zoological Systematics and Evolutionary Research 58:66–78.
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948.
- Laubier L, Ramos J. 1974. Paraonidae (Polychètes Sédentaires) de Méditerranée. Bulletin du Muséum National d’Histoire Naturelle 113:1097–1148.
- Lobo J, Teixeira MAL, Borges LMS, Ferreira MSG, Hollatz C, Gomes PT, Sousa R, Ravara A, Costa MH, Costa FO. 2016. Starting a DNA barcode reference library for shallow water polychaetes from the Southern European Atlantic coast. Molecular Ecology Resources 16:298–313.
- Lovell LL. 2002. Paraonidae (Annelida: Polychaeta) of the Andaman Sea, Thailand. Phuket Marine Biological Center Special Publication 24:33–56.
- Luttikhuizen PC, Dekker R. 2010. Pseudo-cryptic species Arenicola defodiens and Arenicola marina (Polychaeta: Arenicolidae) in Wadden Sea, North Sea and Skagerrak: Morphological and molecular variation. Journal of Sea Research 63:17–23.
- Mallet J. 2007. Species, concept of. In: Levin S et al, editors. Encyclopedia of biodiversity. Cambridge, Massachussets, the U.S.A: Academic Press. Vol. 5, pp. 427–440.
- Maltagliati F, Camilli L, Lardicci C, Castelli A. 2001. Evidence for morphological and genetic divergence in Perinereis cultrifera (Polychaeta: Nereididae) from two habitat types at Elba Island. Journal of the Marine Biological Association UK 81:411–414.
- Maltagliati F, Peru AP, Casu M, Rossi F, Lardicci C, Curini-Galletti M, Castelli A. 2000. Is Syllis gracilis (Polychaeta: Syllidae) a species complex? An allozyme perspective. Marine Biology 136:871–879.
- Martinelli M, Cadalanu R, Floris A, Santoni M, Rossi F, Lardicci C, Castelli A. 1997. Distribuzione dei policheti in alcuni stagni della Sardegna. Biologia Marina Mediterranea 6:399–402.
- Meyer A, Bleidorn C, Rouse GW, Hausen H. 2008. Morphological and molecular data suggest a cosmopolitan distribution of the polychaete Proscoloplos cygnochaetus Day, 1954 (Annelida, Orbiniidae). Marine Biology 153:879–889.
- Modica MV, Russini V, Fassio G, Oliverio M. 2017. Do larval types affect genetic connectivity at sea? Testing hypothesis in two sibling marine gastropods with contrasting larval development. Marine Environmental Research 127:92–101.
- Napolitano E, Iacono R, Ciuffardi T, Reseghetti F, Poulain P-M, Notarstefano G. 2019. The Tyrrhenian Intermediate Water (TIW): Characterization and formation mechanisms. Progress in Oceanography 170:53–68.
- Neal L, Wiklund H, Muir AI, Linse K, Glover AG. 2014. The identity of juvenile Polynoidae (Annelida) in the Southern Ocean revealed by DNA taxonomy, with notes on the status of Herdmanella gracilis Ehlers sensu Augener. Memoirs of Museum Victoria 71:203–216.
- Nygren A. 2014. Cryptic polychaete species: A review. Zoologica Scripta 43:172–183.
- Nygren A, Pleijel F. 2011. From one to ten in a single stroke – Resolving the European Eumida sanguinea (Phyllodocidae, Annelida) species complex. Molecular Phylogenetics and Evolution 58:132–141.
- Orlić M, Gačić M, La Violette PE. 1992. The currents and circulation of the Adriatic Sea. Oceanologica Acta 15:109–124.
- Paradis E. 2010. pegas: An R package for population genetics with an integrated–modular approach. Bioinformatics 26:419–420.
- Paradis E, Claude J, Strimmer K. 2004. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics 20:289–290.
- Pérès JM, Picard J. 1964. Nouveau manuel de bionomie benthique. Recueil des Travaux de la Station marine d’Endoume 31:5–137.
- Pleijel F, Rouse G, Nygren A. 2009. Five colour morphs and three new species of Gyptis (Hesionidae, Annelida) under a jetty in Edithburg, South Australia. Zoologica Scripta 38:89–99.
- Posada D, Crandall KA. 2001. Intraspecific gene genealogies: Trees grafting into networks. Trends in Ecology & Evolution 16:37–45.
- Puillandre N, Lambert A, Brouillet S, Achaz G. 2012. ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Molecular Ecology 21:1864–1877.
- Quiroz-Martinez B, Schmitt FG, Dauvin JC. 2012. Statistical analysis of polychaete population density: Dynamics of dominant species and scaling properties in relative abundance fluctuations. Nonlinear Processes in Geophysics 19:45–52.
- Righi S, Maletti I, Maltagliati F, Castelli A, Barbieri M, Fai S, Prevedelli D, Simonini R. 2019. Morphometric and molecular characterization of an expanding Ionian population of the fireworm Hermodice carunculata (Annelida). Journal of the Marine Biological Association UK 99:1569–1577.
- Ronquist F, Teslenko M, van der Mark P, Ayres D, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2011. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61:539–542.
- Sanna D, Biagi F, Alaya HB, Maltagliati F, Addis A, Romero A, De Juan J, Quignard JP, Castelli A, Franzoi P, Torricelli P, Casu M, Carcupino M, Francalacci P. 2013. Mitochondrial variability of the pipefish Syngnathus abaster. Journal of Fish Biology 82:856–876.
- Sjölin E, Erséus C, Källersjö M. 2005. Phylogeny of Tubificidae (Annelida, Clitellata) based on mitochondrial and nuclear sequence data. Molecular Phylogenetics and Evolution 35:431–441.
- Strelzov VE. 1973. Polychaete worms of the family Paraonidae Cerruti, 1909 (Polychaeta, Sedentaria). Akademia Nauk. Moscow 170.
- Struck TH, Feder JL, Bendiksby M, Birkeland S, Cerca J, Gusarov VI, Kistenich S, Larsson K-H, Liow LH, Nowak MD, Stedje B, Bachmann L, Dimitrov D. 2018. Finding evolutionary processes hidden in cryptic species. Trends in Ecology & Evolution 33:153–163.
- Sur HI, Özsoy E, Ünlüata Ü. 1993. Simultaneous deep and intermediate depth convection in the northern Levantine Sea, winter 1992. Oceanologica Acta 16:33–43.
- Syomin V, Sikorski A, Bastrop A, Köhler N, Stradomsky B, Fomina E, Matishov D. 2017. The invasion of the genus Marenzelleria (Polychaeta: Spionidae) into the Don River mouth and the Taganrog Bay: Morphological and genetic study. Journal of the Marine Biological Association UK 97:975–984.
- Tavaré S. 1986. Some probabilistic and statistical problems in the analysis of DNA sequences. Lectures on Mathematics in the Life Sciences 17:57–86.
- Tebble N. 1959. On a collection of polychaetes from the Mediterranean coast of Israel. Bulletin of the Research Council of Israel B8:9–30.
- Templeton AR, Crandall KA, Sing CF. 1992. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram Estimation. Genetics 132:619–633.
- Trabelsi M, Gilles A, Fleury C, Mâamouri F, Quignard JP, Faure E. 2002. Atherina punctata and Atherina lagunae (Pisces, Atherinidae), new species found in the Mediterranean Sea. 2. Molecular Investigations of Three Atherinid Species. Comptes Rendus Biologies 325:1119–1128.
- Vaidya G, Lohman DJ, Meier R. 2011. SequenceMatrix: Concatenation software for the fast assembly of multi–gene datasets with character set and codon information. Cladistics 27:171–180.
- Vignes F, Barbone E, Breber P, D’Adamo R, Roselli L, Ungaro N, Focardi S, Renzi M, Basset A. 2009. Spatial and temporal description of the dystrophic crisis in Lesina lagoon during summer 2008. Transitional Waters Bulletin 2:47–62.
- Villamor A, Costantini F, Abbiati M. 2014. Genetic structuring across marine biogeographic boundaries in rocky shore invertebrates. Plos One 97:e101135.
- Virgilio M, Abbiati M. 2004. Habitat discontinuity and genetic structure in populations of the estuarine species Hediste diversicolor (Polychaeta: Nereididae). Estuarine, Coastal and Shelf Science 61:361–367.
- Weber AAT, Merigot B, Valiere S, Chenuil A. 2015. Influence of the larval phase on connectivity: Strong differences in the genetic structure of brooders and broadcasters in the Ophioderma longicauda species complex. Molecular Ecology 24:6080–6094.
- Zhang J, Kapli P, Pavlidis P, Stamatakis A. 2013. A general species delimitation method with applications to phylogenetic placements. Bioinformatics 29:2869–2876.