?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
In the present study, a soil-derived mixed microbial consortium was developed for polyhydroxyalkanoates (PHAs) production via nutrient limitation. The enhanced consortium was then cultured continuously in sequential batch and the effect of different nitrogen to carbon ratios (N:C) on the yields and properties of produced PHAs and on the changes of the microbial population were investigated. In all cases, the produced polymers were identified as blends or co-polymers of 3-hydroxybutyrate (3HB) and 3-hydroxyvalerate (3HV) units, with 3HB being the dominant monomer. The degradation profile and the transition temperatures of the produced PHAs were further assessed and compared, as well as the molecular weights which ranged from 77.104 Da to 180.104 Da. In order to further investigate the effect of culturing conditions on the development and alteration of the microbial consortium, the Nile blue live staining was applied in the mixed cultures at the end of each operational period. Various fluorescent single colonies were selected based on differences in their morphology and were identified. Finally, the alterations in microbial diversity and community composition assessed via the RISA profiling method (rRNA Intergenic Spacer Analyses), resulted in the identification of a number of dominant organisms.
KEYWORDS:
Introduction
Plastics derived from petroleum may have provided very light, strong and economical materials for over 50 years but their extensive daily use as well as their challenging disposal have caused a cumulative environmental burden [Citation1]. Hence, there is an imperative need for the production of biodegradable and durable polymers from renewable resources. In this context, there are various approaches for the production of different types of bioplastics such as (a) the employment of chemical processes (i.e. the case of polycaprolactone/PCL), (b) combination of biological and thermo-chemical processes (i.e. the case of polylactide/PLA) or (c) entirely microbial processes (i.e. the case of poly-hydroxyalkanoates/PHAs) [Citation2]. The latter is of great interest nowadays due to the unique properties of the resulting polymers.
PHAs constitute a family of aliphatic polyesters that can be accumulated intracellularly as carbon reserves by more than 300 microorganisms under unbalanced growth conditions [Citation3]. For the sustainable production of such microbial bioplastics through the bioconversion of various substrates, using pure or mixed microbial cultures (PMC, MMC), the selection of appropriate strains and consortia is crucial in order to form selected types of PHAs with advanced properties [Citation4]. In its simplest form, PHA production via PMCs adopts a two-stage batch production process, with an inoculum of bacteria being introduced into a sterile solution of trace metal nutrients and a suitable carbon source and nutrients in the initial, i.e. growth phase. In the second stage, an essential element (such as N, P or O2) is deliberately limited and PHA accumulation takes place [Citation5]. The properties of the final polymer depend on the mix of carbon sources supplied during accumulation, the metabolic pathways bacteria currently use for the following conversion into precursors, and the substrate specificities of the enzymes involved [Citation6].
PHA production via MMCs has also been proposed as a low-cost production process, since no sterilization is necessary and the culture is able to adapt to various complex low-cost waste feedstocks. Most MMC production processes rely on ecological selection pressures that favor organisms with elevated PHA storage capacity, thus engineering the microbial consortium [Citation7]. One such method is the enrichment of the MMC in PHA-accumulating organisms, attained by subjecting it to a number of repeated aerobic feast/famine cycles, a process known as aerobic dynamic feeding (ADF) [Citation8]. Thus, a portion of the consortium is removed during each cycle, since the organisms that are able to store carbon during the feast conditions survive exploiting at a second stage, i.e. during the famine conditions, the stored carbon for new cells production (growth). A modified approach is to use alternating aerobic/anaerobic conditions, during which the polyphosphate and glycogen accumulating organisms (PAOs and GAOs), also capable for PHAs accumulation, are selectively enriched [Citation9] or to subject the MMC to alternating sufficiency/limitation of an essential for growth nutrient [Citation10–Citation12]. Although extensive research has been conducted on the valorization of different types of wastes via MMCs, further studies are required in order to enlighten the effect of key controlling factors. These include the feeding strategy (continuous or pulse, periodic supply of different precursors, etc.) and interactions among implicated microorganisms, as they may have quite different effects on the PHAs formation when compared to PHA production via PMCs, even under the same production conditions. Given that multiple organisms as well as multiple possible PHA production pathways could be present in an MMC environment, one of the key questions will be the compositional distribution of the produced polymer and the effect of these blend compositions on chemo-mechanical properties.
Based on the latter, the present study investigates the effect of nitrogen limitation during PHAs production from a mixed consortium initially derived from soil, on the microbial populations and on the properties of the final products. In this context, a synthetic medium with mixed volatile fatty acids was used as carbon source, and the fermentation process was carried out by operating a sequential batch reactor in cycles of alternating nitrogen (PHAs accumulation phase) and carbon limitation (growth phase). Ammonium sulfate was added during carbon limitation, in order to ensure nitrogen sufficiency for microbial growth. The sufficiency versus limitation of nitrogen was achieved by altering the ratio of N:C used.
Materials and methods
Seed culture and inoculum
For the startup of the bioreactor an enriched MMC derived from soil (clay soil, sample from Platani, Rio, Greece; 38° 18′ N/21° 49′ E) was used. The enrichment protocol that was followed was the same as described in the study of Ntaikou et al. [Citation4], in a draw-fill reactor.
Bioreactor, medium and growth conditions
The bioreactor was of 1.5 L working volume (Vw) and was operated in sequential batch mode (SB) with the carbon and nitrogen sources being supplied to the reactor separately. The same basal medium that was used for the enrichment of the initial seed culture was used [Citation4]. As carbon sources, sodium acetate, sodium buryrate and sodium propionate (all of analytical grade, Sigma Aldrich) were used in ratio 1:1:1 (on mass basis). As nitrogen source, (NH4)2SO4 (Sigma Aldrich) was used.
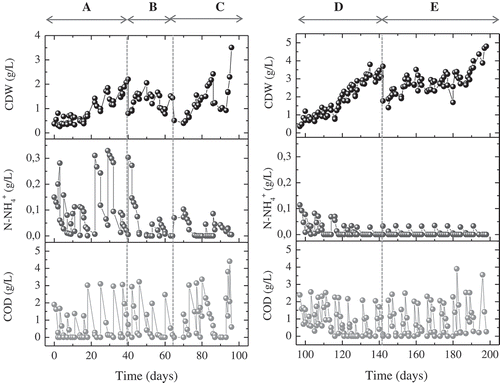
The overall operational period of the reactor was 207 days, and it was divided in 5 five distinct periods, designated period A, B, C, D, and E, during which the N:C ratios and the sequential C and N supply pattern were different. When the carbon source was supplied, the carbon sufficiency phase (accumulation of PHAs) started. During period A, the carbon phase lasted for 3 days (being supplied once per phase), during periods B and C, the carbon supply pattern was not periodic, whereas during periods D and E the carbon and nitrogen supply patterns were normalized with carbon being supplied twice per phase and lasting for 4 days, whereas the nitrogen sufficiency (growth) phase, which lasted 3 days, followed. In all cases, the supply of new feed was performed by removing 2/3 of the Vw of the reactor followed by adding equal volume of fresh medium. Between phases, a settling/decanting phase was applied for 1 h, in order for the microbial biomass to settle and remain in the reactor. The reactor was kept at 25 ± 1°C, with magnetic agitation of 200 rpm and constant aeration during accumulation and growth phases, with moisturized air via sterilized filter and volumetric rate of 0.83–1.2 L/min. Sampling was performed daily under constant agitation and OD, pH, cell dry mass (CDM), remaining carbon, measured in terms of dissolved Chemical Oxygen Demand (d-COD) and nitrogen, measured as N-NH4+ were followed versus time.
Isolation of microorganisms
At the end of each operational period, bacterial strains were isolated from the MMC by repeated streaking on agar plates. The solid medium was prepared by supplementing BSM with 20 g/L agar, sodium acetate, sodium butyrate and sodium propionate 0.5 g/L each, 0.2 g/L (NH4)2SO4 and 2 mg/L of the hydrophobic dye Nile Blue, previously dissolved in DMSO (2 mg per ml). Cultures were incubated at 23°C in order to apply the viable colony staining method [Citation13]. After 48 h, 72 h, and 96 h of incubation, agar plates were exposed to UV light (312 nm). Several fluorescent colonies were selected based on their morphological differences and were transferred to the same medium, without dye, to isolate single colonies.
DNA extraction and PCR amplification of isolates and mixed cultures
Total DNA extraction, using 106 cells on the average, was carried out using the Macherey-Nagel Tissue kit following the manufacturer’s protocol. The genetic diversity and similarity of the bacterial community was analyzed by amplification of the 16S rRNA marker (corresponding to positions 341–534 in E. coli), using eubacteria-specific primers, as described by Muyzer et al. [Citation14]. PCRs were carried out in 50 μL volumes (1 unit KAPA Taq DNA Polymerase, 1X KAPA PCR buffer A, 0.2 mM dNTPs, 1 mM MgCl, 0.5 μL DNA template, filled to 50 μL with sterile H2O). The thermocycling program for the touchdown PCR was as follows: initial denaturation was performed at 93°C for 5 min and then at 93°C for 1 min, followed by touchdown primer annealing from 65°C to 53°C (the annealing temperature was decreased 0.5°C every cycle for 25 cycles, to touchdown at 53°C), followed by extension at 68°C for 1 min (for each of the 25 cycles), with a final extension step at 68°C for 10 min. The PCR results were analyzed by horizontal electrophoresis in 1% agarose gel stained with ethidium bromide (1 μg/ml), after which they were inspected under UV light and photographed. Furthermore, in order to detect subtle differences in the amplified fragments, a second electrophoresis was performed in a 3% gel for 4 h.
In order to better delineate the content of the mixed cultures, the RISA technique [Citation15] was Applied. This method uses the Ribosomal 16S-23S Intergenic Spacer length heterogeneity between taxa to distinguish members of the community. The region is amplified via PCR and resolved on a polyacrylamide gel. The resulting complex banding pattern (a mixture of fragments) provides a profile of each microbial community studied. Each band corresponds to at least one organism. Specifically, the bacterial ISR (Intergenic Spacer Region) is located between the small and large subunits of rRNA. The primers C1 (5ʹ- TTGTACACACCGCCCGTCA-3ʹ) and C2 (5ʹ-GTACTTAGATGTTTCAGTTC-3ʹ) [Citation16] were used in order to amplify the respective region. After following the above-mentioned DNA extraction and PCR assay, the PCR products were separated in 8% polyacrylamide (29:1 acrylamide:bisacrylamide). Electrophoresis was ran for approximately 5 h at 60 V in 1X TBE buffer. PCR products’ sizes were estimated using TAKARA 50 bp ladder. After electrophoresis, the gel was stained using ethidium bromide for 10 min and was photographed with an ultraviolet transilluminator (BIORAD, Gel Doc XR+ system). In order to determine the dominant microorganisms, the prominent bands were excised from the polyacrylamide gel, re-amplified with PCR and purified using the Macherey-Nagel PCR cleanup, following the manufacturer’s protocol for acrylamide gel extraction. Finally, sequencing was conducted on an ABI 3700 capillary sequencer (Macrogen Europe, the Netherlands) using the primers (C1 and C2) of the amplification procedure.
Analytical techniques
Total and volatile suspended solids (TSS, VSS), d-COD (closed reflux method) and N-NH4+ (phenate method) were determined according to the Standard Methods [Citation17]. Short chain fatty acids ((SCFAs) acetate, propionate, butyrate) were quantified by gas chromatography (Varian CP-3800), with flame ionization detector and capillary column. Prior to analysis, samples were acidified with H2SO4 (0.6% v/v). The chemical composition of PHAs and the purity of the polymer samples was determined by measuring its methyl-ester derivatives using a gas chromatography analyzer (Varian CP-3800, FID) following the protocol described in the study of Kourmentza et al. [Citation18].
The recovery of PHAs from the microbial biomass at the end of each period was performed as described in the study of Ntaikou et al. [Citation4], by removing 1 L of mixed fermentation broth under agitation. It should be noted that the purification step with methanol was not performed in the case of samples A and B, which resulted to final products with lower purity than those of samples C, D and E. The recovery yield of produced polymer was estimated gravimetrically as weight of air-dried polymer samples/weight of lyophilized biomass × PHA content (wt %). The PHAs production yield per COD consumed (CODcons) was also determined according to the equation:
where CDM0 and CDMF are the cell dry weights of the recovered biomass in the beginning and the end of the accumulation phase of each experiment, respectively, estimated via the determination of volatile suspended solids (VSS), [%PHAs]0 and [%PHAs]F the % content of the bacterial biomass in PHAs, and CODcons the total COD consumed during the accumulation phase.
For further chemical and thermodynamic analysis, the PHAs pellets were casted with chloroform (20 ml/g polymer sample) at 70°C for 15 min. The solvent was slowly evaporated in proper vials at 70°C in order to receive PHAs films with thickness 0.15–0.25 mm.
Characterization of PHAs
Spectroscopic characterization of the recovered PHAs films was conducted via a Perkin-Elmer SPECTRUM BX FTIR Spectrometer in the mid-IR region, 4000–400 cm−1) at 4 cm−1 resolution using 32 scans, using 2 wt%.solution of purified PHAs in CHCl3. Thermogravimetric analysis (TGA) was performed via Labsys TM TG Setaram analyser in order to find out the thermal stability and degradation patterns of produced PHAs in the temperature range of 25-800°C. Specimens of ~10 mg were placed in an aluminum plate and were heated at a rate of 10°C/min in a nitrogen atmosphere. The molecular masses of the polymers, i.e. the mean molecular mass weight (Mw) mean molecular mass number (Mn) were determined via size exclusion chromatography (SEC) at 25°C with a Polymer Lab chromatograph comprising two Pl gel (5 μm) columns, a refractive index detector (RI) and a UV/VIS (254 nm) detector. Chloroform was used as an eluent at a flow rate of 1mL/min. Polystyrene standards with low polydispersity were used to generate calibration curves. Solutions of the products in CHCl3 at a concentration of 4 mg/mL were prepared. In the abovementioned solutions, a small quantity of toluene was added, so as to know when the experiment ended. The eluent (CHCl3) was filtered with a 5 μm filter, while all the prepared solutions were filtered using 0.45 μm filters. Differential scanning calorimetry (DSC) was performed using DSC 2920 CE, TA Instruments. Samples of 9–10 mg were weighed in an aluminum pan. Measurements were performed in a nitrogen atmosphere (50 mL min−1), in two heating steps from −30°C to 200°C and at a heating rate of 10°C min−1. The DSC endothermal peak values and areas of the second scan were used to evaluate the thermal properties of the PHAs, including the glass transition temperature (Tg, °C), the melting temperature (Tm, °C), the crystallization temperature (Tc, °C), the crystallization enthalpy (ΔHc) and the melting enthalpy (ΔHm). The DSC endothermal peak values and areas of the second scan were used to evaluate the thermal properties of the PHAs, including the glass transition temperature (Tg, °C), the melting temperature (Tm, °C), the crystallization temperature (Tc, °C), the crystallization enthalpy (ΔHc) and the melting enthalpy (ΔHmTm). Tc, °C temperatures were estimated from, respectively, the endothermic and exothermic peaks. The crystallinity (X) of the samples was also estimated assuming the enthalpy of fusion of 100% crystalline PHB to be 146 J/g [Citation19].
Results and discussion
Effect of nitrogen on PHAs yields
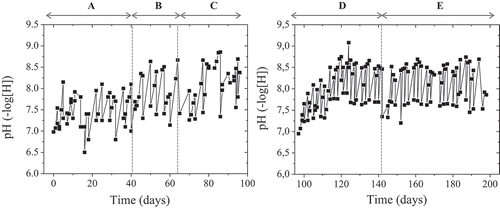
The sequential batch reactor was operated continually in five distinct operational periods (denoted as A, B, C, D, and E), as illustrated in and . During all operational periods, a mixture of equal quantities of sodium salts of acetate, butyrate, and propionate were used as carbon source. (NH4)2SO4 was used as the sole nitrogen source and the bioreactor was operated under the same conditions in terms of temperature, agitation, and aeration rate. Both carbon and nitrogen feeds were supplemented with a phosphate buffer in order to maintain the pH into the range of 7.5 ± 1 () which is considered favorable for PHAs production [Citation20]. The main differences among periods were the amount of nitrogen during the accumulating phases (carbon excess), whereas the carbon supply patterns and amounts of carbon being available for accumulation were also altered as described in section 2.2, thus resulting in different consumption of carbon (). As shown, the N:C ratios ranged from 17.7 to 5.2 during the accumulation phases of the operational periods A to C whereas during D and E periods the nitrogen limitation during accumulation phases was complete. The lowest PHA yield, measured either as g PHAs/g COD consumed (YPHAs/COD) or % PHAs/CDM (YPHAs/CDM) was estimated for the period A, during which there was the highest N:C ratio at the accumulation phase. It can be assumed thus, that when a sufficient amount of nitrogen is simultaneously available with carbon, a higher ratio of carbon source is forwarded to growth rather than PHAs production. This is in agreement with previous studies that revealed a significant enhancement of PHAs accumulation under nitrogen partial or complete limitation [Citation10,Citation21,Citation22]. Indeed, when mixed consortia are subjected to nutrient stress, they are able to metabolize acids towards acetyl-CoA, which is partially channeled to the tricarboxylic acid cycle (TCA) for growth and NAD(P)H production, and partially used for accumulation of PHAs [Citation5,Citation23]. The lower ratios of N:C and the complete limitation of N during the whole accumulation phase, resulted in considerably higher YPHAs/COD compared to that obtained in period A, in all cases. However, it can be noted that the obtained YPHAs/COD of periods B to D, exhibited a negligible increasing tendency. YPHAs/VSS seems to be mainly dependent on the initial amount of offered carbon, exhibiting 65% increase from period B to period C, during which the N:C ratio was similar but the available carbon was doubled. Similarly, Reddy and Mohan [Citation24], reported high levels of PHA accumulation by mixed consortia for higher substrate loads.
Table 1. N:C ratios and mean d-COD consumed during accumulation phases and final yields of recovered PHAs at the end of each period of the bioreactor operation.
Composition and properties of the recovered polymers
The monomeric composition, purity, mean molecular masses and polydispersity (P = Mw/Mn) of the produced polymers, are summarized in . In all cases, the produced polymers were identified as blends and/or co-polymers of 3-hydroxybutyrate (3HB) and 3-hydroxyvalerate (3HV) units, with 3HB being the dominant monomer. The generation of poly-3hydroxybutyrate-co-3hydroxyvalerate (P3HB-co-3HV) and its blend with P3HB from MMCs during their adaptation in nutrient limiting [Citation4,Citation18,Citation25] or stressful conditions [Citation26,Citation27] has previously been reported. The polymers recovered from periods C, D, E exhibited high purity, reaching 93%. The purity of the samples from periods A and B, was considerably lower (~80%), probably due to the simplified protocol used for their recovery from the lyophilized biomass, during which the purification step via methanol was omitted. The impurities of the polymer samples A and B are probably lipids and other soluble to organic solvents cell components that were not removed from biomass during washing with ethanol, and were actually detected in the SEC chromatographs via UV detector as low MW peaks (see supplementary data). The mean molecular masses of the samples were higher compared to those previously reported for MMC [Citation28] which range from 11.104 Da [Citation29] up to 327.104 Da [Citation30]. In the present study, the lowest Mw was noted for period A (77.104 Da) and the highest for period E (182.104 Da). It is reported that differences in PHAs composition are mainly affected by the types of carbon source used [Citation31]. However, since the type of carbon sources in the feed and their ratio were the same in all cases (acetate:propionate:butyrate = 1:1:1), the observed differences cannot be attributed to this parameter. One possible explanation is that different acids are consumed selectively for growth and others for accumulation due to dynamic changes of the microbial populations.
Table 2. Chemical composition, purity and molecular masses (Mw, mean molecular mass weigh, Mn, mean molecular mass number, P, polidispersity) of the recovered PHAs from the end of each period of the bioreactor operation.
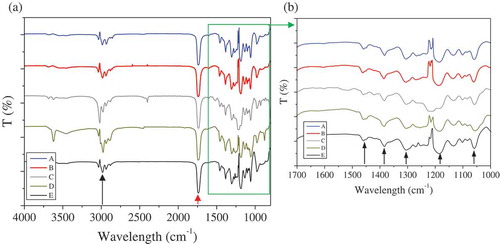
The FT-IR spectra of the polymers that were recovered at the end of all operational periods (A-E) via solvent extraction are illustrated in . The analysis revealed typical to PHA intense absorptions at 1724–1738 cm−1, corresponding C = O stretching groups, i.e. esters. Strong vibrations at 2925–2935 cm−1 which are characteristic of N-H or C-H and could be attributed to either proteins or PHAs are also seen. However, no peak is detected at 1650 cm−1, corresponding to C = O of amides, i.e. associated with proteins. Absorptions at 1450 cm−1 and at 1375 cm−1, corresponding to methylene groups (-CH2-) and methyl groups (-CH3-) respectively, are also apparent in all samples as well as other main peaks characterizing PHAs in the range of 1500–1000 cm−1 ()) indicating CH3 bending, CH2 wagging, C-O, C-C and C-O-C stretching [Citation32].
Figure 3. FT-IR spectra of the recovered PHAs from the end of each period of the bioreactor operation.
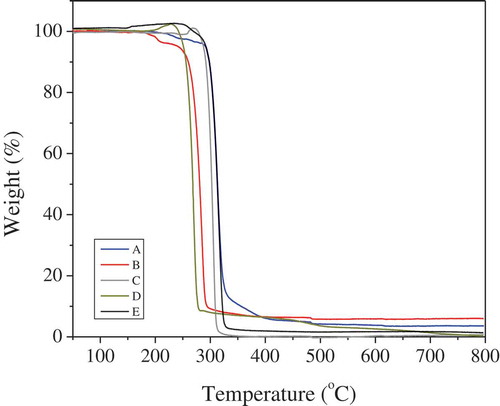
The degradation profiles of the recovered polymers were assessed via TGA, the curves of which are illustrated in . Specifically, the initial decomposition temperatures at which weight loss was 5 wt%, Td5 was 291.34oC, 241.79oC, 287.66° C, 251.16°C and 292.04°C for samples A, B, C, D, and E, respectively. The residual mass percentage at 800°C (R800), was calculated as 3.57%, 6.01%, 0.69%, 0.3% and 0.85% for samples A, B, C, D, and E, respectively, indicating that polymers that were recovered from periods A and B contain impurities that were not efficiently removed during the recovery process. This can also be due to the absence of methanol purification step for those two samples as mentioned above.
Figure 4. TGA results of the recovered PHAs from the end of each period of the bioreactor operation.
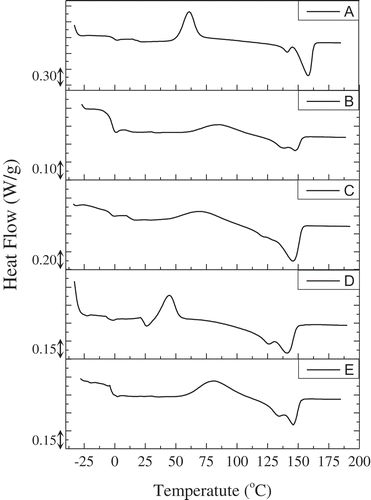
DSC curves of second heating scan of all samples are illustrated in , whereas summarizes the thermal properties obtained from the scans. Specifically, the polymer produced by cycle A exhibited two Tm, at 135°C and 158°C, with the second one being actually the highest observed among the tested samples. The presence of two Tm values could lead to the conclusion that the recovered bioplastic could be a blend of different PHAs along with co-polymers. It seems that during this initial cycle of bioreactor operation, where nitrogen was abundant, the microbial consortium was more heterogeneous since stress conditions were not strict, and large populations of different strains coexisted in the culture. In general, the Tm of all samples had values intermediate between those reported for P3HB Tm (168–180°C) and for P3HV Tm value (105–110°C) [Citation33], suggesting the co-polymeric nature of the produced polymer. The polymer with highest 3HB contents also exhibited the highest melting temperature (sample A with 91% 3HB), whereas the lowest Tm value was observed for samples C and D which had the lowest 3HB contents and highest HHV content. The higher ΔΗm values indicate the existence of polymer chains of high crystallinity in the sample. These chains are probably linear and symmetrically distributed. Moreover, it is seems that a higher content of 3HB leads to higher ΔΗm values, an observation that is in agreement with the literature [Citation9,Citation34].
Table 3. Transunion temperatures and enthalpies of the recovered PHAs from the end of each period of the bioreactor operation.
Genetic diversity and similarities of microbial populations
The genetic diversity and similarity of the bacterial community was analyzed by amplification of the 16S rRNA marker (corresponding to positions 341–534 in E. coli), using eubacteria-specific primers, as described by Muyzer et al. [Citation14]. Several single colonies from solid cultures of mixed consortia stained with Nile blue were selected and identified. Via this approach, various species of Pseudomonas sp, and strains of Stenotrophomonas maltophilia were recognized as dominant in almost every period of the bioreactor operation () and, as such, the information gained in terms of microbial diversity was not sufficient. For comparison purposes, isolates were further tested for PHAs production in batch cultures and yields were compared to those of the MMC of period E (preliminary results presented in supplementary data).
Table 4. Identification of bacteria strains that were isolated from the mixed consortia as formed at the end of each period of the bioreactor operation. The percentage of similarity and best match were determined using BLASTN against the NCBI non-redundant database.
In order to better delineate the content of the MMC the RISA technique [Citation15] was applied. The prominent bands after RISA, were excised from the polyacrylamide gel, purified and then a PCR was performed in order to multiply the PCR product concentration before sequencing analyses. Finally, sequencing was conducted. The electophoretic patterns () revealed a complex pattern in all bioreactor conditions. At a first glance, the pattern of the MMC of period A seems to be less complex that those of periods B, C, D, and E. It should be noted that though that the smaller number of bands exhibited at the gel electrophoresis may underestimate the true diversity, since unrelated microorganisms may have spacer regions of identical length and thus be represented in the RISA profile by a single band. Indeed, this assumption can be supported by the properties of the polymer recovered from period A, which were more diverse that those from other periods. Moreover, the attempt of identification of the bacteria corresponding to thick bands appearing at 1000-750 pb was unsuccessful, revealing that at least two different strains were present (bands marked with red, )). It can be assumed that in this area the dominant strains of Pseudomonas and Stenotrophomonas genus appear which were easily isolated and identified from the mixed culture. Indeed, in the solid cultures of the MMCs, several replicates of these microorganisms were isolated implying that these are the dominant stains in all cases. The RISA methodology, however, revealed the presence of microorganisms that were not successfully isolated via the Nile blue live staining as shown in . Those microorganisms appeared in the gel electrophoresis as distinct single bands (marked with yellow, )). It is of great interest that, among them, Klebsiella pneumonia was identified solely in period A and disappeared in the following operational periods of the bioreactor, as verified by the absence of the respective band in the electrophoresis gel. K. pneumonia is a facultative anaerobe, capable of simultaneous hydrogen and 1,3 propanediol production from glycerol under anaerobic conditions [Citation34] and is also reported as a PHAs producer from various substrates [Citation35].
Figure 6. RISA profile from the different periods of bioreactor operation (A, B, C, D, E) by the resolving of the ribosomal intergenic spacer region in 2% agarose gel (a) and 8% polyacrylamide (29:1 acrylamide:bisacrylamide) (b). The bands that were excised from the acrylamide gel are depicted. In yellow, the bands that were successfully sequences; in red, the bands that the sequencing revealed that the band excised included more than one microorganism. l = ladder, Takara wide range 50 bp.
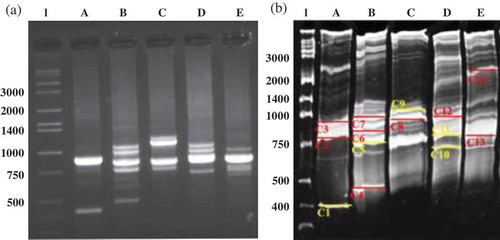
Table 5. Tentative identification of bacteria strains after applying the RISA method to the mixed cultures that were formed at the end of each period of the bioreactor operation. The percentage of similarity and best match were determined using BLASTN against the NCBI non-redundant database.
RISA is a method applied to the study of microbial communities and it is often referred to as community fingerprinting. The method uses PCR amplification of the high variable in length (and also nucleotide sequence) bacterial 16S-23S intergenic region between the small (16S) and large (23S) ribosomal subunits. Both types of variation have been extensively used to distinguish closely related bacterial species [Citation36]. The resulting PCR product will be a mixture of fragments contributed by several dominant community members which are visualized via acrylamide electrophoresis. The electophoretic patterns are complex and each band represents at least one organism. This type of analysis is very useful in visualizing the level of complexity in the microbial diversity when comparing differing environments, treatments, culture conditions, etc. [Citation15,Citation37–Citation39]. However, it should be noted that in some bacterial genomes the rRNA operon copy number can vary and the different copies may have different length; hence there could be more than one bands deriving from the same species depicted in the electrophoretic pattern. On the contrary, there are other bacterial species that, even if present in a microbial community, they will not yield a PCR fragment using RISA, usually because their ribosomal RNA genes are not organized in an operon [Citation36,Citation40].
Overall, it could be noted that RISA is a reliable method that can complement other used tools for the identification of the dominant microorganism to the species level. However, stronger molecular tools such as Next Generation Sequencing (NGS) are needed for the complete characterization of the microbial diversity and fingerprinting of complex microbial communities [Citation41,Citation42].
Conclusions
The present study reveals that alterations in N:C ratios during the accumulation phase in the SBR reactor, could significantly affect the composition and the properties of the produced PHAs. A high initial N:C ratio favors the diversity of microbial populations, thus leading to a more heterogenous final product, as revealed by DSC analysis. The findings implied that during initial running of the SBR with high N:C ratio, mixtures of homopolymers and co-polymers were formed. The strict limitation of nitrogen resulted in the formation of a more homogenous product, and was attributed to the elimination of some microbial strains and prevalence of certain ones, as revealed by RISA analysis. However, stronger molecular tools a such as NGS should be applied for further elucidating the role of the microbial strains of the obtained findings.
Supplemental Material
Download MS Word (2.8 MB)Acknowledgments
The authors would like to thank Dr. rer. nat. Constantin Flytzanis for providing the lab equipment for the RISA methodology.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
Notes on contributors
Ioanna Ntaikou
Ioanna Ntaikou is a biologist and received her PhD from the Department of Chemical Engineering, University of Patras. She is currently employed at FORTH/ICE-HT as a Senior Research Associate. Her research expertise is in applied microbiology processes for wastewater treatment and valorization via mixed and pure microbial cultures, focusing on the production of biofuels and bioplastics and modeling of biochemical processes.
Ioannis Koumelis
Ioannis Koumelis is a Chemical Engineer and was awarded a Diploma in the field of Materials Engineering from the department of Chemical Engineering, University of Patras. He received his MSc in the field of “Environment and Energy” of the same department. He currently works for ZOONOMI SA “Specialist Fish & Animal Feed Producers”, in the Quality Assurance Management Department.
Maria Kamilari
Maria Kamilari is a molecular ecologist working on functional differentiation and its evolutionary consequences using model and non model organisms, in order to address scientific questions related to adaptations to extreme conditions. She is currently a Marie Curie postdoctoral fellow at the Department of Biology, University of Denmark.
Zacharoula Iatridi
Zacharoula Iatridi is a Material Scientist and she received her MSc and PhD awards in Polymer Science and Technology, University of Patras. She currently works as a Post-doctoral researcher in the Laboratory of Polymers, Department of Chemical Engineering, University of Patras. Her research interests cover mainly the area of water-soluble polymers, focusing on the synthesis of comb-type copolymers, study of stimuli-responsive polymers and physicochemical and rheological properties of polymers.
Constantinos Tsitsilianis
Constantinos Tsitsilianis is full Professor and Director of the Polymer Lab of the Department of Chemical Engineering at the University of Patras. His research interest is in the area of macromolecular engineering, comprising synthesis and characterization of model macromolecules, nano-structured micelles and reversible physical hydrogels from stimuli responsive model block copolymers.
Gerasimos Lyberatos
Gerasimos Lyberatos is full Professor in the School of Chemical Engineering, National Technical University of Athens and collaborative faculty member of FORTH/ICE-HT. His research interests are in biochemical engineering, environmental technologies, bioprocess modeling, optimization and control.
References
- Rujnic-Sokele M, Pilipovic A. Challenges and opportunities of biodegradable plastics: A mini review. Waste Manag Res. 2017;35(2):132–140.
- Harting R, Johnston K, Petersen S. Correlating in vitro degradation and drug release kinetics of biopolymer-based drug delivery systems. Int J Biobased Plast. 2019;1(1):8–21.
- Anderson AJ, Dawes EA. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol Rev. 1990;54(4):450–472.
- Ntaikou I, Koumelis I, Tsitsilianis C, et al. Comparison of yields and properties of microbial polyhydroxyalkanoates generated from waste glycerol based substrates. Int J Biol Macromol. 2018;112:273–283.
- Kourmentza C, Ntaikou I, Kornaros M, et al. Production of PHAs from mixed and pure cultures of Pseudomonas sp. using short-chain fatty acids as carbon source under nitrogen limitation. Desalination. 2009;248(1–3):723–732.
- Byrom D. Plastics from Microbes. Mobley DP, ed. Munich: Hanser; 1994. p. 5–33.
- Daigger GT, Grady CPL Jr. The dynamics of microbial growth on soluble substrates. A unifying theory. Water Res. 1982;16(4):365–382.
- Beun JJ, Dircks K, Van Loosdrecht MCM, et al. Poly-β-hydroxybutyrate metabolism in dynamically fed mixed microbial cultures. Water Res. 2002;36(5):1167–1180.
- Bengtsson S, Pisco AR, Johansson P, et al. Molecular weight and thermal properties of polyhydroxyalkanoates produced from fermented sugar molasses by open mixed cultures. J Biotech. 2010;147:172–179.
- Silva F, Campanari S, Matteo S, et al. Impact of nitrogen feeding regulation on polyhydroxyalkanoates production by mixed microbial cultures. New Biotechnol. 2017;37:90–98.
- Valentino F, Karabegovic L, Majone M, et al. Polyhydroxyalkanoate (PHA) storage within a mixed-culture biomass with simultaneous growth as a function of accumulation substrate nitrogen and phosphorus levels. Water Res. 2015;77:49–63.
- Ntaikou I, Valencia Peroni C, Kourmentza C, et al. Microbial bio-based plastics from olive-mill wastewater: generation and properties of poly-hydroxyalkanoates from mixed cultures in a two-stage pilot scale system. J Biotechnol. 2014;18:138–147.
- Spiekermann P, Rehm BHA, Kalscheuer R, et al. A sensitive, viable-colony staining method using Nile red for direct screening of bacteria that accumulate polyhydroyalkanoic acids and other lipid storage compounds. Arch Microbiol. 1999;171:73–80.
- Muyzer G, de Waal EC, Uitterlinden AG. Profiling of complex microbial populations by analysis of polymerase chain denaturing gradient gel electrophoresis reaction-amplified genes coding for 16S rRNA. Appl Env Microbiol. 1993;59:695–700.
- Ciesielski S, Klimiuk E, Mozejko J, et al. Changes in microbial community structure during adaptation towards polyhydroxyalkanoates production. Polish J Microbiol. 2009;58:131–139.
- Dolzani L, Tonin C, Lagatolla L, et al. Identification of Acinetobacter isolates in the A. calcoaceticuss-A. baumannii complex by restriction analysis of the 16S-23S rRNA intergenic-spacer sewuences. J Clin Microbiol. 1996;33:1108–1113.
- APHA, AWWA & WEF. Standard methods for the examination of water and wastewater. 19th ed. Washington, DC: American Public Health Association; 1995.
- Kourmentza C, Ntaikou I, Lyberatos G, et al. PHA production by mixed and pure cultures of Pseudomonas sp. using synthetic and olive mill wastewater under nitrogen and dual nitrogen-oxygen limitation. Int J Biol Macromol. 2015;74:202–210.
- Barham PJ, Keller A, Otun EL, et al. Crystallization and morphology of a bacterial thermoplastic: poly-3-hydroxybutyrate. J Mater Sci. 1984;19:2781–2794.
- Kourmentza C, Kornaros M. Biotransformation of volatile fatty acids to polyhydroxyalkanoates by employing mixed microbial consortia: the effect of pH and carbon source. Biores Technol. 2016;222:388–398.
- Morgan-Sagastume F, Karlsson A, Johansson P, et al. Production of polyhydroxyalkanoates in open, mixed cultures from a waste sludge stream containing high levels of soluble organics, nitrogen and phosphorus. Water Res. 2010;44:5196–5211.
- Montiel-Jarillo G, Carrera J, Suarez-Ojeda ME. Enrichment of a mixed microbial culture for polyhydroxyalkanoates production: effect of pH and N and P concentrations. Sci Total Environ. 2017;583:300–307.
- Dias JML, Lemos PC, Serafim LS, et al. Recent advances in polyhydroxyalkanoate production by mixed aerobic cultures: from the substrate to the final product. Macromol Biosci. 2006;6:885–906.
- Reddy VM, Mohan VS. Effect of substrate load and nutrients concentration on the polyhydroxyalkanoates (PHA) production using mixed consortia through wastewater treatment. Biores Technol. 2012;114:573–582.
- Reddy VM, Kotamraju A, Mohan VS. Bacterial synthesis of polyhydroxyalkanoates using dark fermentation effluents: comparison between pure and enriched mixed cultures. Eng Life Sci. 2015;15(6):646–654.
- Martinez-Sanz M, Villano M, Oliveira C, et al. Characterization of polyhydroxyalkanoates synthesized from microbial mixed cultures and of their nanobiocomposites with bacterial cellulose nanowhiskers. New Biotechnol. 2014;31(4):64–376.
- Villano M, Beccari M, Dionisi D, et al. Effect of pH on the production of bacterial polyhydroxyalkanoates by mixed cultures enriched under periodic feeding. Process Biochem. 2010;45(5):714–723.
- Laycock B, Halley P, Pratt S, et al. The chemomechanical properties of microbial polyhydroxyalkanoates. Prog Polym Sci. 2014;39(2):397–442.
- Dai Y, Yuan ZG, Jack K, et al. Production of targetedpoly(3-hydroxyalkanoates) copolymers by glycogen accumulating organisms using acetate as sole carbon source. J Biotechnol. 2007;129:489–497.
- Patel M, Gapes DJ, Newman RH, et al. Physico-chemicalproperties of polyhydroxyalkanoate produced by mixed-culturenitrogen-fixing bacteria. Appl Microbiol Biotechnol. 2009;82:545–555.
- Gumel AM, Annuar MSM, Heidelberg T. Effects of carbon substrates on biodegradable polymer composition and stability produced by Delftia tsuruhatensis Bet002 isolated from palm oil mill effluent. Polym Degrad Stab. 2012;97:1227–1231.
- Arcos-Hernandez MV, Gurieff N, Pratt S, et al. Rapid quantification of intracellular PHA using infrared spectroscopy: an application in mixed cultures. J Biotechnol. 2010;150:372–379.
- Pereira SMF, Sanchez RJ, Rieumont J, et al. Synthesis of biodegradable polyhydroxyalcanoate copolymer from a renewable source by alternate feeding. Polymer Eng Sci. 2008;48:2051–2059.
- Dounavis AS, Ntaikou I, Kamilari M, et al. Production of bio-based hydrogen enriched methane from waste glycerol in a two stage continuous system. Waste Biomass Valor. 2016;7(4):677–689.
- Lopes MSG, Rocha RCS, Zanotto SP, et al. Screening of bacteria to produce polyhydroxyalkanoates from xylose. World J Microbiol Biotechnol. 2009;25(10):1751–1756.
- Fisher MM, Triplett EW. Automated approach for ribosomal intergenic spacer analysis of microbial diversity and its application to freshwater bacterial communities. Appl Environ Microbiol. 1999;65(10):4630–4636.
- Ciesielski S, Cydzik‐Kwiatkowska A, Pokoj T, et al. Molecular detection and diversity of medium‐chain‐length polyhydroxyalkanoates‐producing bacteria enriched from activated sludge. J Appl Microbiol. 2006;101(1):190–199.
- Toledo FL, Calvo C, Rodelas B, et al. Selection and identification of bacteria isolated from waste crude oil with polycyclic aromatic hydrocarbons removal capacities. Syst Appl Microbiol. 2006;29(3):244–252.
- Clarridge JE. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin Microbiol Rev. 2004;17(4):840–862.
- Kovacs A, Yacoby K, Gophna U. A systematic assessment of automated ribosomal intergenic spacer analysis (ARISA) as a tool for estimating bacterial richness. Res Microbiol. 2010;161(3):192–197.
- Gao W, Zheng C, Lei Y, et al. Analysis of bacterial communities in white clover seeds via high-throughput sequencing of 16S rRNA gene. Curr Microbiol. 2019;76(2):187–193.
- Tufail S, Munir S, Jamil N. Variation analysis of bacterial polyhydroxyalkanoates production using saturated and unsaturated hydrocarbons. Braz J Microbiol. 2017;48(4):629–636.