Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is RNA virus, responsible for coronavirus disease 2019 (COVID-19), which has now taken the form of a pandemic. Even though the world has undertaking all possible measures to impede the spread of this droplet infection, still the prevalence of COVID-19 is on the rise and has infected more than 208 countries all over the globe. Reviewing the website and global articles from PubMed, Scopus and other cites on COVID-19 a global pandemic, this review article provides an update on the all major aspects of coronavirus such as origin, epidemiology, mode of transmission, structure and molecular characterization, characterization of spike protein, clinical features including cutaneous manifestation, radiological features, molecular testing, histopathology and the current preventive approaches. Importantly, review also provides an update on current projections of Saudi Arabia on COVID-19 pandemic.
Keywords:
Background
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is an RNA virus responsible for causing an infection termed as coronavirus disease 2019 (COVID-19), which has taken the form of a pandemic. The first reported case of SARS-CoV-2 was from the People’s Republic of China in December 2019 in a city named Wuhan, which is the capital of Hubei province, which is considered as an industrial hub of China. World Health Organization (WHO) declared SARS-CoV-2 as a public health emergency of international concern on the 30th of January 2020, and later on the 11th of March 2020, it was given the status of a pandemic (World Health Organization (WHO); Zhu et al., Citation2020). The origin of SARS-CoV-2 is still a mystery; however, it is established that it is a zoonotic virus (Zhu et al., Citation2020). Previously six viruses belonging to the coronavirus family were responsible for causing infection in humans, SARS-CoV-2 is mentioned as the 7th member of the coronavirus family, which has infected humans after the Middle East Respiratory Coronavirus (MERS-CoV) (Zhu et al., 2020). UpToDate, the therapeutic measures to deal with this viral infection are only supportive, and implementing prevention strategies aimed at plummeting the spread of this virus among the community is the only available option (Alabdulmonem, Shariq, & Rasheed, Citation2020). This review provides an update on the all major aspects of coronaviruses such as origin, epidemiology, mode of transmission, structure and molecular characterization, characterization of spike protein and its significance, clinical features such as cutaneous manifestation, radiological features, molecular testing, histopathology and the current preventive approaches. Not only have these, review also provides an update on current programs of Saudi Arabia on COVID-19 pandemic.
Origin of coronaviruses, genetic view and sites of infections
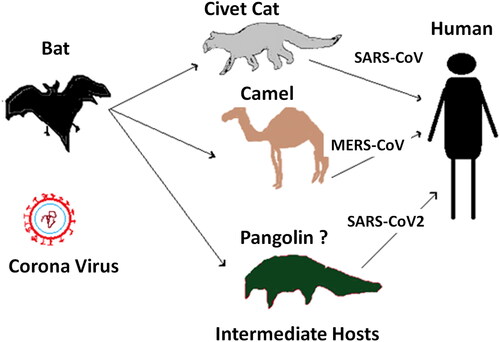
CoVs principally belong to the group of zoonotic viruses that cause infections primarily among animals and, under usual situations, usually do not cause disease in the human population. Under certain circumstances, genetic mutations occur in these viruses, which can make it adaptive to survival within the human body leading to infections in humans. Various studies revealed that previously occurring members of Coronavirus family, SARS CoV and MERS CoV are both originated from bats (Lau et al., Citation2010). It was also found in previously conducted studies that CoV also have an intermediate host which is animal before it infects humans, in case of SARS CoV bat was the primary host where is palm civets served as an intermediate host from where it was transmitted to humans and in case of MERS CoV bats, and camels served as primary and intermediate host respectively (Müller et al., Citation2014). The zoonotic source of SARSCoV2 is not confirmed yet; however, certain molecular studies revealed that the genetic makeup of this virus has genetic sequence similarities to CoVs, which occur in various species of bats, labelling them as the primary host for this virus (Zhou et al., Citation2020). Furthermore, molecular studies conducted on the genomic configuration of SARS CoV2 also concluded the similarities of this virus with CoVs that are present in pangolins giving a thought that pangolins might be the intermediate host for this virus (Li, Zai et al., Citation2020). Recently, Shereen et al. described the key suspected reservoirs and mode of transmission of SARS-CoV-2 to humans (Shereen, Khan, Kazmi, Bashir, & Siddique, Citation2020). Based on the previously proposed assumptions of mode of transmission of novel SARS-CoV-2 to humans (Lau et al., Citation2010; Li, Zai et al., Citation2020; Müller et al., Citation2014; Shereen et al., Citation2020; Zhou et al., Citation2020), we described the zoonotic transmission of SARS-CoV-2 to humans in simplified ways in . However, the pangolin serving as an intermediate host is still doubtable, and further studies at the molecular level as well as research investigations are required for conformity of the intermediate source for this virus as it is a mandatory step in order to bring to an end for this pandemic.
Figure 1. Proposed zoonotic transmission of SARS-CoV-2 to humans. The proposed assumptions of mode of transmission of novel SARS-CoV-2 to humans were drawn from the previous studies (Lau et al., Citation2010; Li, Zai et al., Citation2020; Müller et al., Citation2014; Shereen et al., Citation2020; Zhou et al., Citation2020).
The outlines features of novel SARS-CoV-2 was found to be very much similar to the previously reported coronavirus MERS-CoV (Barry, Al Amri, & Memish, Citation2020), which originated from Saudi Arabia in 2012 and led to fatal outcomes due to severe pneumonia and acute renal failure in Saudi city Jeddah and other regions of the world including Republic of Korea in 2015 (Mizumoto, Saitoh, Chowell, Miyamatsu, & Nishiura, Citation2015). Several studies demonstrated a fatal association between the infection found in camels and COVID infection in humans (Alagaili et al., Citation2014; Hemida et al., Citation2014; Sabir et al., Citation2016). In 2016, Sabir et al. reported that camels serve as a reservoir for the diversification of the MERS-CoVs and are the one of the most important sources of human infections with this virus (Sabir et al., Citation2016). These findings were also supported by another study performed by Alagaili et al. showed significant evidences from a geographic and temporal survey of camels in Saudi Arabia that the MERS-CoV strains were circulating in camels and were distributed in all over the regions of Saudi Arabia which were phylogenetically classified into clades that correlate with the outbreaks of the disease among humans (Alagaili et al., Citation2014). Furthermore, another study at molecular levels showed that nasal and rectal swab from camels were found to be positive for the presence of MERS-CoV sequences (Hemida et al., Citation2014). In support of these, studies also reported that the genomic characterization of the MERS-CoV strains from camels revealed a similarity with the virus found in infected humans (Alagaili et al., Citation2014; Sabir et al., Citation2016). International Committee on Taxonomy of Viruses (ICTV) classified the members of Coronaviridae family into four genera which are alpha (α), beta (β), gamma (γ) and delta (δ) CoV (Wong, Li, Lau, & Woo, Citation2019). Out of these groups, α and β CoV can infect mammals, whereas γ and δ CoV cause infection only in birds, and to a lesser extent in swine (Wong et al., Citation2019; Yin & Wunderink, Citation2018). It is also worth knowing that severity of infection also depends on the type of CoV which has infected human, two species belonging to α − CoVs (HCoV-229E and HCoV-NL63) whereas two species of β-CoVs (HCoV-HKU1 and HCoV-OC43) are comparatively less pathogenic and on infecting the human they present with mild upper respiratory symptoms that are analogous to the common cold (Yin & Wunderink, Citation2018). However, SARS-CoV-1 and MERS-CoV that belongs to β-CoVs are highly pathogenic and can lead to severe life-threatening fatal pneumonia (Yin & Wunderink, Citation2018). Previously these two β-CoVs viruses lead to fatal outbreaks resulting in a high mortality rate. SARS-CoV1 originated from Guangdong Province of China in November 2002 and spread rapidly to Beijing, Hong Kong, Vietnam, Singapore and Canada till March 2003 (Leung et al., Citation2004). Novel SARS-CoV2 is the recent most member of CoV family and belongs to β-CoVs that infect humans and has the potential to cause severe life-threatening respiratory tract infections (Li, Liu, Yu, Tang, & Tang, Citation2020).
Epidemiology
A meta-analysis based on the data from eight laboratory centres revealed that males are more infected by COVID-19 as compared to the female population with a ratio of 23,871:22,377, previously occurring outbreaks due to MERS-CoV which belonged to coronavirus family also infected male population more than female (Channappanavar et al., Citation2017). Another study conducted on 41 cases reported from China demonstrated that 30 patients out of them were male (73%) (Channappanavar et al., Citation2017). These findings may be attributed to the fact that the female population has potent innate and adaptive immune responses (Jaillon, Berthenet, & Garlanda, Citation2019). Most of the severe and critical cases are elderly (≥65-year-old) and people with certain underlying medical conditions, especially if not well controlled, including chronic lung diseases, cardiovascular diseases, diabetes, and immunocompromised patients (Centers for Disease Control and Prevention (CDC), Citation2020). The overall case fatality rate is 2.3% and ranged from 0.9% to 7.2% (Wu & McGoogan, Citation2020). The mortality rate of COVID-19, which has remained at 2% since the beginning of the pandemic till now whereas, in contrast, the mortality rate from MERS-CoV was comparatively higher and varied from region to region. In Saudi Arabia, it was 42%, whereas in South Korea, it was estimated to be 19%, ranging from 7% among younger age groups to 40% among older adults, particularly those who were above 60 years of age and the SARS -CoV-1 had an overall mortality rate of 11% (Leung et al., Citation2004; Mizumoto et al., Citation2015).
Mode of transmission and basic reproduction number
In the early stage of the pandemic, several health agencies, including CDC and WHO relayed on the MERS-COV and other coronavirus transmissions for COVID-19 (CDC; WHO). The virus can be transmitted either directly or indirectly. Directly via respiratory droplets produced during coughing, sneezing, or even talking. Droplets only travel for a short distance but not for more than 6 feet. Indirectly, when droplets fall on objects, then touch by the other then touching their mouth, nose, or eyes. As the pandemic continued to grow rapidly, several studies conducted to determine if the virus is airborne. Their finding indicates that the virus remains suspended in the air for more distance than it was believed. The speed of this virus spreading suggests that air play a role in transmission (Lewis, Citation2020). The basic reproduction number (R0) is known as the average number of secondary cases produced by a single infection in a susceptible population. It’s an indication of how the infectious agent capable of been transmitted to others (Delamater, Street, Leslie, Yang, & Jacobsen, Citation2019). If the R0 more than one, the outbreak will continue happening, studies reported that SARS-CoV-2 has a higher R0 in comparison with SARS and MERS-CoV (Delamater et al., Citation2019). Liu et al. reviewed 12 studies for R0 and were reported to be 3.28 with a median of 2.79, which was significantly higher as reported previously by WHO (Liu, Gayle, Wilder-Smith, & Rocklöv, Citation2020), indicating that basic reproduction number of the secondary cases produced by a single infection in a susceptible population was significantly enhanced.
Structure and molecular characterisation
CoV is composed of a single-stranded positive-sense RNA genome ranging approximately from 26 to 32 kilobases in length. It is surrounded by an envelope composed of lipid bilayer from which the anchored surface proteins are spiking out, mimicking it to the shape of a crown; hence the name Corona is given to it. Phylogenetic analysis of this virus verified that 80% of its nucleotide identity is similar to SARS-CoV (Lu et al., Citation2020). Quasispecies has prior been reported for SARS-CoV and MERS-CoV, signifying that these beta coronaviruses may be comprised of a multifaceted and vibrant distribution of intimately related variants in vivo that is similar to various RNA viruses (Xu, Zhang, & Wang, Citation2004). On analyzing sequence variability in the clinical sample of patients infected by SARS-CoV-2 viral quasispecies was found to be present (Capobianchi et al., Citation2020). Lu’s phylogenetic analysis conducted on SARS CoV-2 revealed that it is related to the subgenus Sarbecovirus. The similarity of about 89% in the genomic structure of SARS CoV-2 to bat derived coronavirus species, bat-SL-CoVZXC21, and bat-SL-CoVZC45 that were responsible for the SARS outbreak in 2003 have been found (Lu et al., Citation2020). Whereas the resemblance of the genomic structure of CoV occurring in the same two bat species with SARS-CoV and MERS-CoV was found to be 79% and 50%, respectively (Capobianchi et al., Citation2020; Lu et al., Citation2020; Xu et al., Citation2004; Zaki, van Boheemen, Bestebroer, Osterhaus, & Fouchier, Citation2012; Zhu et al., 2020). A study in which phylogenetic analysis was conducted on SARS-CoV-2 and naturally occurring CoV strains in a species of bat called as Rhinolophus or horseshoe bats revealed 98.7% nucleotide resemblance in the gene encoding for the enzyme called as partial RNA-dependent RNA polymerase (RdRp) of the coronavirus of bat strain BtCoV/4991 (GenBank KP876546, 370 bp sequence of RdRp) and 87.9% of similarity in the nucleotide sequence in bat-SL-CoVZC45 and bat-SL-CoVZXC21 (Zhou et al., Citation2020). Studies conducted to explore the evolutionary analysis confirms that on the basis of N, S, and ORF1a/1b genes SARS-CoV-2 is undeniable, a novel virus which has autonomously been transferred from animals to human (Wu et al., Citation2020). Recently several investigators described the overall structure of novel SARSCoV-2 which is very much similar to other members of the CoVs family (Astuti & Ysrafil, Citation2020; Shereen et al., Citation2020). Based on these studies, the outline structure of novel SARSCoV-2 is drawn and summarized in . It is comprised of an envelope containing nucleocapsid core enclosing the genomic RNA. The nucleocapsid core is composed of two diverse types of proteins that are protruding out of the envelope in the form of spikes, and these are termed as haemagglutinin esterase (HE) and glycoprotein trimmer (S) (Astuti & Ysrafil, Citation2020; Shereen et al., Citation2020). The genome of this single-stranded RNA virus is comprised of ten open reading frames (ORFs) with two hundred and sixty-five nucleotides at the 5′ end region and two hundred nice nucleotides present at 3′ end (Astuti & Ysrafil, Citation2020; Shereen et al., Citation2020). Recently, Grifoni et al., described the sequence homology of novel SARS-CoV-2 and MERS-CoV through bioinformatics approach (Astuti & Ysrafil, Citation2020). Based on these studies (Astuti & Ysrafil, Citation2020; Grifoni et al., Citation2020; Shereen et al., Citation2020), we summarized the similarities between the RNA genome of SARS-CoV-2 with MERS-CoV in simplified ways in .
Figure 2. The structure of novel SARS-CoV-2. The virion has nucleocapsid composed of genomic RNA and phosphorylated nucleocapsid (N) protein, which is buried inside phospholipid bilayers and covered by the spike glycoprotein trimmer (S). The membrane (M) protein hemagglutinin-esterase (HE) and the envelope (E) protein are located among the S proteins in the virus envelope. The structural detail of SARS-CoV-2 was derived from the recently published studies (Astuti & Ysrafil, Citation2020; Shereen et al., Citation2020).
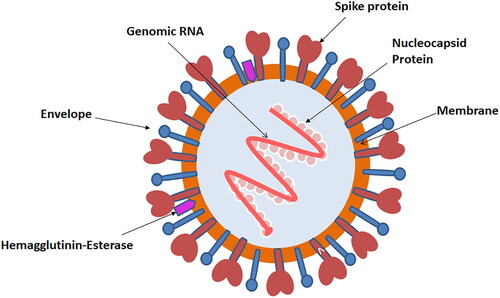
Table 1. Similarities between novel SARS-CoV-2 genome structure with MERS-CoV.
Characterization of spike protein and its significance
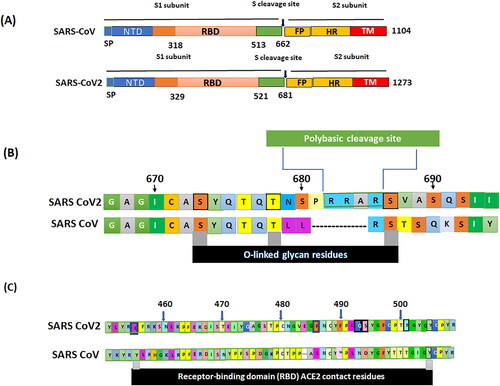
Currently, there is no specific antiviral treatment or vaccine available, which can contribute to the management of SARS-CoV2. Understanding the sequence of spike protein (S) and its receptor may be the key to discovering appropriate treatment or a vaccine. Spike proteins (S) are structural glycoproteins that are situated on the envelope and functions in adherence by binding the virus to angiotensin-converting enzyme 2 receptor (ACE2) that are present on host cells (He, Tao, Yan, Huang, & Xiao, Citation2020). The spikes on the novel SARS‐CoV‐2 was recently described by Andersen, Rambaut, Lipkin, Holmes, & Garry (Citation2020). Based on these investigations, the data on spike on the novel SARS‐CoV‐2 are summarized in . The S glycoprotein of SARS-CoV2 is composed of 1273 amino acids, whereas that of SARS-CoV is made up of 1104 amino acids. S glycoprotein of both of these viruses contains an amino (N) terminal and a carboxyl (C)‐terminal termed as S1 and S2 subunits, respectively (). S1 subunit is composed of a receptor‐binding domain (RBD) that is made up of two hundred amino acid molecules, furthermore, within the RBD two subdomains are located which are the core and external subdomains, the core subdomain functions in the formation of S trimer particles whereas the external subdomain is responsible for adhering with the ACE 2 receptor (Lu et al., Citation2013). ACE 2 receptors are present widely in various organs of the human body, especially on the heart, lung, liver, kidney, testis, and intestine. These receptors contribute to normal physiological functions as well as in regulating blood pressure (Anguiano, Riera, Pascual, & Soler, Citation2017). Binding of SARS-CoV-2 to the ACE 2 receptors is mediated by a polybasic cleavage site that is located in between S1 and S2 subunits of the S glycoprotein (). This cleavage site is named as furin. Acquisition of the furin cleavage site may be considered to be a gain of function that had lead SARS-CoV2 to commence its current pandemic.
Figure 3. Spike on the novel SARS‐CoV‐2. (A) Structural diagrams of spike glycoproteins of SARS‐CoV, and SARS‐CoV‐2 focus on the putative maturation sites. All spike domains of coronaviruses contain S1 subunit, including signal peptide (SP), N-terminal domain (NTD), receptor-binding domain (RBD), and S2, including fusion peptide (FP), and the transmembrane domain (TM). (B) Acquisition of polybasic cleavage site (furin) and O-linked glycans. Both the polybasic cleavage site and the three adjacent predicted O-linked glycans are unique to SARS-CoV-2. (C) The spike protein SARS-CoV-2, was aligned against the SARS-CoV. Key residues in the spike protein that make contact to the ACE2 receptor are marked with RBD in SARS-CoV-2 and SARS-CoV. The spikes on the novel SARS-CoV-2 was recently described by Andersen et al. (Citation2020).
Role of immune therapy in the treatment of COVID-19 patients
Immune-based therapies for the treatment of patients with COVID-19 are currently under study or are being in use. There are two main types of immune therapies currently under consideration, the first one is using convalescent plasma which contain antibodies against the novel SARS-CoV-2 that directly target the virus. Whereas the other is based on to enhance the body immune responses against the infection using the various pharmacological inhibitors against proinflammatory cytokines including inhibitors for interleukin-1 (IL-1) or interleukin-6 (IL-6) (Huang et al., Citation2020; Mascaretti, De Angelis, & Berti, Citation2020; Mehta et al., Citation2020; Schulert & Grom, Citation2014; Shakoory et al., Citation2016). In Saudi Arabia, plasma therapy is currently being in use for treating COVID-19 patients in various health facilities. As part of the therapy, plasma is occupied from the patients who were recovered from COVID-19 infection and transfuse into newly infected patients (Huang et al., Citation2020). Studies conducted on serum of infected patients suffering from COVID-19 moderate to severe showed excessive production of cytokine and arised a clinical condition termed as macrophage activation syndrome (MAS) (Huang et al., Citation2020; Mascaretti et al., Citation2020; Schulert & Grom, Citation2014; Shakoory et al., Citation2016). It is characterized by the massive inflammation that presents as high grade fever, liver dysfunction, coagulopathy and increase in serum ferritin levels along with involvement of pulmonary system (Huang et al., Citation2020; Mascaretti et al., Citation2020; Schulert & Grom, Citation2014; Shakoory et al., Citation2016). Studies also revealed that viruses are capable of triggering of MAS as was found in patients with COVID-19 (Shakoory et al., Citation2016). The severity of clinical symptoms of COVID-19 patients was found to be associated with excessive production of proinflammatory cytokines such as IL-1 and IL-6 (Mehta et al., Citation2020; Schulert & Grom, Citation2014; Shakoory et al., Citation2016). Moreover, it is also reported that the regulation of signal transduction in immune cells reducing the symptoms associated with COVID-19 (Schulert & Grom, Citation2014). Furthermore, IL-6 and IL-1β inhibitors along with Janus kinase (JAK) inhibitors, are considered to be a part of immune therapy that play a vital role in inhibiting the release of cytokines (Schulert & Grom, Citation2014). Therefore, immune therapy may be one of the best therapeutic options for COVID-19 patients.
Clinical presentation interms of cutaneous manifestation
Skin manifestations can be a clue in diagnosing COIVD-19. The mostly described skin manifestation in a case series of 22 Italian patients is varicella-like exanthem that appears three days after the onset of the systemic symptoms and disappears on day 8, without subsequent scars. It manifests as a scattered exanthem that constantly involves the trunk with mild/absent pruritus, which differentiated it from true Varicella (Marzano et al., Citation2020). Others report drug hypersensitivity, non-specific erythematous rash, urticaria, dusky acrocyanosis, dry gangrene, transient unilateral livedo reticularis, and a dengue-like petechial rash (Manalo, Smith, Cheeley, & Jacobs, Citation2020).
Pregnancy and children
Currently, there is no known difference in clinical presentation and incidence between pregnant and any other adult of reproductive age. While in children, the presentation is similar to other viral respiratory infections, and usually less severe than adults with over 90% asymptomatic, mild, or moderate cases with no significant gender preference as recently reported by WHO and CDC (CDC; WHO). These manifestations mainly include fever, fatigue, cough and runny nose with no auscultatory abnormalities or hypoxaemia, while critical cases in children present with ARDS or respiratory failure are found in infants (CDC; WHO).
Imaging
It is now well documented that the imaging such as chest x-ray (CXR), computed tomography (CT), etc. are typically considered as the first-line imaging modalities of choice with the advantage of portability to reduce the risk of transmission (Xie et al., Citation2020; Ye, Zhang, Wang, Huang, & Song, Citation2020). In additions, the Chinese CDC has also set four epidemiological clinical criteria that link to the confirmed cases COVID-19 or a part of symptomatic cluster of these patients, among them CXR was considered as one of the top clinical criteria for COVID-19 patients (Barry et al., Citation2020). Based on these, several countries including Saudi Arabia followed the same criteria through which existence of pneumonia and/or novel SARS-CoV-2 has been check out through CXR (Barry et al., Citation2020; Ye et al., Citation2020). At the same time, it is also important to point out that many radiological features including CXR, CT for the novel COIVD-19 infected pneumonia are non-specific and might overlap with the findings of a typical pneumonia, which eventually resemble MERS and SARS (Xie et al., Citation2020). Despite of these possibilities, the role of imaging via CXR or CT is crucial for the management of COVID-19 patients, which is not just limited to establish the diagnosis but also important to determine the severity, disease progression, and also for therapeutic efficacy (Rubin et al., Citation2020; Wong et al., Citation2020). However, the value of imaging is diminished by other factors such as the risk of unjustified radiation, COVID-19 transmission, and consumption of protective equipment (Wong et al., Citation2020; Ye et al., Citation2020). Hence, the role of imaging is variable and can be affected by the available resources and public health directives. In April 2020, a multinational consensus statement from the Fleischer Society on the role of chest imaging during the COVID-19 pandemic (Xie et al., Citation2020). In general, the CXR is less sensitive than CT in particular for early changes, although the sensitivity and specificity can be altered by the onset of imaging and severity of the symptoms. The most common CXR findings are consolidation, followed by ground-glass opacification with peripheral, lower zone, and bilateral predominance (Rubin et al., Citation2020; Wong et al., Citation2020; Ye et al., Citation2020). The chest CT findings are also variable in different patients and stages of the disease, the typical CT findings in patients with COVID-19 are bilateral peripheral areas of ground-glass opacities with or without consolidations based on the severity (Rubin et al., Citation2020; Wong et al., Citation2020; Ye et al., Citation2020). Hence, multifocal lesions with predominant distribution in the posterior middle and lower zones of the lungs should prompt the diagnosis of COVID-19 in a proper clinical setting.
Molecular testing
As the initial signs and symptoms of COVID-19 resemble the ordinary flu, which is fever, flu, and cough, that is the reason it must be diagnosed in a suspected case without delay. Diagnosis of this virus also will play a role in designing a critical strategy for tracking the virus, understanding the epidemiology, facilitating case management, and to suppress its spread in the community. Various diagnostic techniques are available which can aid in the detection of this virus, these include both serological and molecular methods, and yet Real-time reverse transcription-polymerase chain reaction (rRT-PCR) has proved itself to be a sensitive and cost-effective diagnostic tool for detection of this single-stranded RNA virus (Jeon, Seo, Oh, Kingsley, & Choi, Citation2017). WHO has recommended the use of rRT-PCR for suspected cases of COVID-19 since January 17, 2020 (WHO). For those regions or health care facilities who have no testing capacity or have inadequate skill to test suspected cases are encouraged to transport the suspected samples to WHO reference laboratories that are providing confirmatory testing (Jeon et al., Citation2017; Xu et al., Citation2020; WHO). Although it’s an admirable technique, but there are certain limitations associated with it as it requires advanced laboratory instruments as well as a high standard biosafety laboratory with well-trained staff. Furthermore, this testing facility is not available in every clinical diagnostic laboratory, so time is worn out during the transport of sample, performance, and delivering the results, which can be up to 2 or 3 days. As COVID-19 is a public health emergency, this time-consuming critical process of sample detecting is extremely inconvenient (Jeon et al., Citation2017). Moreover, RT-PCR tests can confer negative results, particularly during an early stage of infection, as well as co detection of other coronaviruses (CoVs) are commonly encountered, and the role of positive PCR results is not always explicitly exhibited (Xie et al., Citation2020). Due to these reasons, a substitute molecular technique is essential in this critical period for the detection of COVID-19. Recently a novel molecular technique has been introduced termed as the Loop-mediated isothermal amplification (LAMP) reaction. It is an amplification technique with a high level of competence, sensitivity, specificity, and speediness under isothermal circumstances as compared to PCR (Jeon et al., Citation2017; Xie et al., Citation2020). It can produce up to 109 copies of target DNA in minimum time duration of approximately lesser than an hour at a constant temperature of 65 °C. That is the reason almost immediately after it was introduced. It took its place as a popular isothermal amplification method in the field of molecular diagnosis (Jeon et al., Citation2017; Xie et al., Citation2020). summarizes the detailed comparison between polymerase chain reaction (PCR) and loop-mediated isothermal amplification (LAMP) techniques for the diagnosis of COVID-19 patients. The details described in this table for the diagnosis of COVID-19 patients were derived from the recently published report by Nguyen, Duong Bang, and Wolff (Citation2020).
Table 2. Comparison between Polymerase Chain Reaction (PCR) and Loop-Mediated Isothermal Amplification (LAMP) for the diagnosis of novel COVID-19 infection.
Histopathology
Although most of COVID-19 lung biopsies/resections shows the same histopathologic morphological changes, these changes are not specific or characteristic for COVID-19 since they are also present in other diseases like SARS and MERS. The lung biopsy of the cases shows diffuse alveolar damage (DAD) morphology, especially in the advanced stages of the disease (Xu et al., Citation2020). These changes are time-dependent, which vary from early stage to late advance one. In the early stage, most of the biopsies show bilateral congestion and pulmonary edoema associated with spherical/globular intra-alveolar proteinaceous material with only focal and patchy mononuclear inflammatory infiltrates (mainly lymphocytes) as well as mild pneumocytes type-II hyperplasia (Xu et al., Citation2020). As the case becomes progressive, the histopathologic morphology changes. Here there will be marked bilateral but patchy diffuse alveolar damage (DAD) with hyaline membrane thickening and deposition, areas of haemorrhage, interstitial edoema, fibroblastic proliferation, intra-alveolar fibrin exudate, fibrinoid vascular necrosis, severe pneumocytes type-II hyperplasia with desquamation, and presence of multinucleated syncytial giant cells with large bizarre nuclei, amphophilic granular cytoplasm, and prominent nucleoli and the later cells have not been reported to show neither intra-cytoplasmic inclusions nor intra-nuclear ones (Xu et al., Citation2020).
Management of COVID-19 in Saudi Arabia
Prediction of COVID-19 outbreak in Saudi Arabia
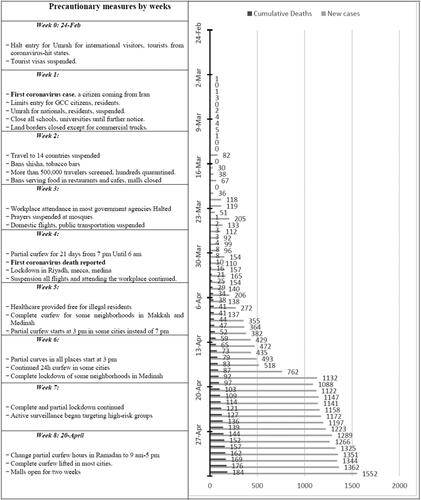
Few sources available to predict the future of the pandemic and when it will end in Saudi Arabia (Abou Elkhir, Citation2020; Kulveit & Lagerros, Citation2020; Luo, Citation2020). We identified three sources, two based on the susceptible-infected-recovered (SIR) model and the third based on several models. summarize their results for Saudi Arabia. Moreover, Al-Tawfiq and Memish also have recently described some of the important preventive and therapeutic measures for COVID-19 not only for Saudi population but also for the population from the whole eastern mediterranean region, which have also been discussed below (Al-Tawfiq & Memish, Citation2020).
Table 3. Saudi government projection models for COVID-19 pandemic.
Containment and mitigation measures by Saudi Arabia
Unlike Italy and the United States, Saudi Arabia started implementing containment measures before the diagnosis of the first case on 2 March 2020. Such strong measures reduced the burden on the health system and helped to flatten the curve (Yezli & Khan, Citation2020). Temporary Umrah pilgrimage ban was announced and made effective on 27 February 2020, until writing this article. Further announcements were made in the following weeks include suspension of all educational activities, ban group prayers, withhold attending workplace for government employees, and partial curfew on all the country with a complete curfew on some cities (Yezli & Khan, Citation2020). The following table summarizes most of the important policies applied by the Saudi government ().
Preventive measures by Saudi Arabia
Saudi government has also followed the similar preventive measures as instructed by WHO and CDC in their recent reports (CDC; WHO). Briefly, the prevalence of COVID-19 can be achieved by essential preventive measures which include washing the hands regularly with soap and water for at least 20 seconds, maintain social distancing among people which is the most effective ways to reduce the spread of illness during an outbreak, covering coughs and sneezes, use of surgical face mask (CDC). In healthcare facilities the preventive measures that should take place which includes establishing a surveillance system for COVID-19 in healthcare and community settings, applying steps regarding standard and droplet precautions for all patients by complying to a set of practices like hand hygiene, use of personal protective equipment, respiratory hygiene/cough etiquette, isolation of suspected cases (CDC).
Potential treatment option followed by Saudi Arabia
The therapeutic management for treating COVID-19 has not been available yet. Saudi Arabia has managed the COVID-19 patients with the following measures such as x-ray, oxygen saturation with the appropriate measures, including isolation and symptomatic reliefs, plasma therapy and by the use of several anti-viral drugs as the whole world are now using for COVID-19 patients (CDC; Rubin et al., Citation2020; Wang et al., Citation2020; WHO; Wong et al., Citation2020). Currently, supportive treatment is being delivered depending upon the severity of disease, along with the implication of isolation protocols and other preventive strategies (Jaillon et al., Citation2019; WHO). Recovery time in mild cases is about 2 weeks while 3–6 weeks in severe and critical cases. Remdesivir is a nucleotide analog of adenosine that was actually used to treat the Ebola virus but it was found to be effective for the COVID-19 management (Wang et al., Citation2020). Other than remdesivir, several other drugs are also in currently in use for the treatment of COVID-19 patients (“https://www.moh.gov.sa/Ministry/Media Center/Publications/Documents/MOH-therapeutic-protocol-for-COVID-19.pdf”; “https://saudigazette.com.sa/article/592690/SAUDI-ARABIA/Health-Ministry-Plasma-from-recovered-COVID-19-patients-used-in-three-regions”; Wang et al., Citation2020). Drugs targeting the receptors to which SARS-CoV2 adhere, such as 3CLpro inhibitor and vinyl sulphone protease inhibitor, have shown activity against this virus; however, clinical trials are still under process (Wang et al., Citation2020). Oral oseltamivir and lopinavir/ritonavir as well ganciclovir intravenously has also been used as a therapeutic measure in few patients and several studies also evaluated the role of IV immunoglobulin and the role of steroids; however, their role in the management is uncertain (Wang et al., Citation2020). Chloroquine used as an antimalarial drug also has shown efficacy in treating viral infections, the antiviral effects of chloroquine are because it inhibits the viral uncoating as well as inhibiting post-translational modifications of newly formed proteins mainly by inhibiting the glycosylation hence the viral replication is inhibited (Wang et al., Citation2020). A study conducted by Wang et al. showed promising results in the treatment of COVID-19 when the combination of chloroquine and remdesivir was used (Wang et al., Citation2020). Favipiravir is another antiviral drug which acts as an inhibitor of viral RNA-dependent RNA polymerase and also currently using in Saudi Arabia for treating COVID-19 patients (“https://www.moh.gov.sa/Ministry/MediaCenter/Publications/Documents/MOH-therapeutic-protocol-for-COVID-19.pdf”; “https://saudigazette.com.sa/article/592690/SAUDI-ARABIA/Health-Ministry-Plasma-from-recovered-COVID-19-patients-used-in-three-regions”). Moreover, another drug dexamethasone has also been included as a therapeutic option for infected patients which require non-invasive or invasive ventilation (Yezli & Khan, Citation2020). Interestingly, the triple combination therapy, comprising Lopinavir, Ritonavir, and IL-1β for 14-days has also tested and also included in the therapeutic options for the treatment of severely infected COVID-19 patients in Saudi Arabia and was also found to be effective for reducing clinical symptoms (CDC; Wang et al., Citation2020; WHO). On the other hand, the convalescent plasma has now been well considered as a passive immunotherapy for the treatment of COVID-19 patients, this type of therapy has usually chosen when there are no specific vaccines or drugs available for emerging infection‐related diseases (Tobaiqy et al., Citation2020). This therapy was found to be a promising therapeutic options for the treatment of COVID-19 patients and is also currently using in Saudi Arabia (“https://www.moh.gov.sa/Ministry/MediaCenter/Publications/Documents/MOH-therapeutic-protocol-for-COVID-19.pdf”; “https://saudigazette.com.sa/article/592690/SAUDI-ARABIA/Health-Ministry-Plasma-from-recovered-COVID-19-patients-used-in-three-regions”). A very recent case series reported from China showed perfect recovery of severely infected COVID-19 patients after adminstration of convalescent plasma (Tobaiqy et al., Citation2020). Convalescent plasma has now been proposed for the treatment of COVID‐19 infected critically sick patients (CDC; WHO). However, the data still limited, and further clinical trials are required to prove their efficacies.
On-going strategies for vaccine development against COVID-19 infection
Over ten million individuals from all over the globe have now been confirmed positive for COVID-19 infection and the infectivity rate is continuously on the rise but still a stable drug or a vaccine that potentially fight against this fatal infection is not available (“An updated guide to the coronavirus drugs and vaccines in development,” 2020; Schaefer, Tam, Savulescu, & Voo, Citation2020; Sinha, Citation2020). The development of vaccine against this novel SARS-COV-2 will be the ultimate solution to handle this global pandemic. The main purpose behind the viral vaccine development is to enhance the immune system of an individual against a future viral attack. This can be done by pre-exposing the individual to either weakened/killed virus or by preexposing of virus structural parts, which leads the individual body to develop a defence mechanism (“An updated guide to the coronavirus drugs and vaccines in development,” 2020; Schaefer et al., Citation2020). Scientists all over the globe are currently following the same strategies, about 19 top laboratories including Imperial College London, Sanofi Pasteur, France, etc. are currently working on more than 120 vaccine programs (“An updated guide to the coronavirus drugs and vaccines in development,” 2020; Schaefer et al., Citation2020; Sinha, Citation2020). In addition, several laboratories such as the Moderna Inc., MA, USA are trying to develop antibodies against the spike proteins of the novel SARS-COV-2 and several other laboratories such as CureVac, German Enterprise, etc. are aimed to develop an RNA-based vaccine (“An updated guide to the coronavirus drugs and vaccines in development,” 2020; Schaefer et al., Citation2020). All of these vaccines developing programs are currently under clinical trials at different phases (“An updated guide to the coronavirus drugs and vaccines in development,” 2020). We are hoping that the vaccines against the novel SARS-COV-2 become available within 10 months or so, but it will completely depends on the success of newly develop vaccine(s) in all three phases of clinical trials.
Conclusions
A complete genomic characterization of this novel virus is vital from a diagnostic point as well as to assess the viral evolution. The variability and genetic structure of this novel corona virus resulting in the progression of quasispecies in patients infected can be extremely supportive in knowing the background for this virus as well as its adaptation in a new host. Therefore, further studies are required at the molecular level in order to disentangle the significance of intrapatient inconsistency in the SARS-CoV-2 evolutionary trajectory. Although it is an obligate parasite and transmits by droplets, it cannot replicate exterior to living cells; however, it can linger in the viable form on various environmental surfaces such as doorknobs for a certain time and the contaminated surfaces are assumed to be a vital source in the spread of this virus. The incubation period of this virus ranges from 2 to 14 days after which the infected person presents with cough, fever and shortness of breath, which can proceed by severe pneumonia, particularly in old age patients or those who are suffering from a chronic debilitating disease such as diabetes. Diagnosis depends on serological and molecular-based techniques, rapid point of care diagnostic tests, and serological assays are hastily emerging. The early and efficient health policies made by the Saudi government led to an efficient containment of this pandemic in comparison with developed countries' policies. Patients usually present with fever, cough, fatigue, and dyspnoea. The associated laboratory findings are leukopoenia, raised CRP and LDH. Chest X-ray mostly shows infiltrates, while CT show ground‐glass opacity and patchy bilateral shadowing. Molecular testing with nucleic acid amplification such as PCR test is required to confirm the diagnosis in appropriate patients with suspected infection and ideally through nasopharyngeal specimen swab testing. Patients with normal x-ray and oxygen saturation can be managed as an outpatient with the appropriate measures, including isolation and symptomatic reliefs. Furthermore, convalescent plasma transfusion has also been suggested and successfully implemented on some patients. Moreover, remdesivir so far was found to be most promising treatment options but the data are still limited and further clinical trials are required to prove its efficacy.
Authors’ contributions
All authors involved in conceptualization, review design, literature searching and drafting of the review article. All authors have read and approved the final article.
Ethics approval
Not applicable.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Abou Elkhir, O. (2020). Predicting the peak of the epidemic curve of COVID-19 in selected Arab countries. Medium Tachy Health. Retrieved May 5, 2020, from https://medium.com/@osama.abouelkhir/predicting-the-peak-of-the-epidemic-curve-of-covid-19-in-selected-arab-countries-202eb9c3585e
- Alabdulmonem, W., Shariq, A., & Rasheed, Z. (2020). COVID-19: A global public health disaster. International Journal of Health Sciences, 14(3), 7–8.
- Alagaili, A. N., Briese, T., Mishra, N., Kapoor, V., Sameroff, S. C., Burbelo, P. D., … Lipkin, W. I. (2014). Middle East respiratory syndrome coronavirus infection in dromedary camels in Saudi Arabia. mBio, 5(2), e00884-14. doi:10.1128/mBio.00884-14
- Al-Tawfiq, J. A., & Memish, Z. A. (2020). COVID-19 in the Eastern Mediterranean Region and Saudi Arabia: Prevention and therapeutic strategies. International Journal of Antimicrobial Agents, 55(5), 105968. doi:10.1016/j.ijantimicag.2020.105968
- (2020). An updated guide to the coronavirus drugs and vaccines in development, by Damian Garde, STAT News, March.
- Andersen, K. G., Rambaut, A., Lipkin, W. I., Holmes, E. C., & Garry, R. F. (2020). The proximal origin of SARS-CoV-2. Nature Medicine, 26(4), 450–452. doi:10.1038/s41591-020-0820-9
- Anguiano, L., Riera, M., Pascual, J., & Soler, M. J. (2017). Circulating ACE2 in cardiovascular and kidney diseases. Current Medicinal Chemistry, 24(30), 3231–3241. doi:10.2174/0929867324666170414162841
- Astuti, I. & Ysrafil. (2020). Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): An overview of viral structure and host response. Diabetology & Metabolic Syndrome, 14(4), 407–412. doi:10.1016/j.dsx.2020.04.020
- Barry, M., Al Amri, M., & Memish, Z. A. (2020). COVID-19 in the shadows of MERS-CoV in the Kingdom of Saudi Arabia. Journal of Epidemiology and Global Health, 10(1), 1–3. doi:10.2991/jegh.k.200218.003
- Capobianchi, M. R., Rueca, M., Messina, F., Giombini, E., Carletti, F., Colavita, F., … Bartolini, B. (2020). Molecular characterization of SARS-CoV-2 from the first case of COVID-19 in Italy. Clinical Microbiology and Infection. doi:10.1016/j.cmi.2020.03.025.
- Centers for Disease Control and Prevention (CDC). (2020). People who are at higher risk for severe illness. Retrieved May 5, 2020, from https://www.cdc.gov/coronavirus/2019-ncov/need-extra-precautions/people-at-higher-risk.html?CDC_AA_refVal=https%3A%2F%2Fwww.cdc.gov%2Fcoronavirus%2F2019-ncov%2Fspecific-groups%2Fpeople-at-higher-risk.html
- Channappanavar, R., Fett, C., Mack, M., Ten Eyck, P. P., Meyerholz, D. K., & Perlman, S. (2017). Sex-based differences in susceptibility to severe acute respiratory syndrome coronavirus infection. Journal of Immunology, 198(10), 4046–4053. doi:10.4049/jimmunol.1601896
- Delamater, P. L., Street, E. J., Leslie, T. F., Yang, Y. T., & Jacobsen, K. H. (2019). Complexity of the basic reproduction number (R(0)). Emerging Infectious Diseases, 25(1), 1–4. doi:10.3201/eid2501.171901
- Grifoni, A., Sidney, J., Zhang, Y., Scheuermann, R. H., Peters, B., & Sette, A. (2020). A Sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe, 27(4), 671–680.e2. doi:10.1016/j.chom.2020.03.002
- He, J., Tao, H., Yan, Y., Huang, S. Y., & Xiao, Y. (2020). Molecular mechanism of evolution and human infection with SARS-CoV-2. Viruses, 12(4), 428. doi:10.3390/v12040428
- Hemida, M. G., Chu, D. K., Poon, L. L., Perera, R. A., Alhammadi, M. A., Ng, H. Y., … Peiris, M. (2014). MERS coronavirus in dromedary camel herd, Saudi Arabia. Emerging Infectious Diseases, 20(7), 1231–1234. doi:10.3201/eid2007.140571
- https://saudigazette.com.sa/article/592690/SAUDI-ARABIA/Health-Ministry-Plasma-from-recovered-COVID-19-patients-used-in-three-regions
- https://www.moh.gov.sa/Ministry/MediaCenter/Publications/Documents/MOH-therapeutic-protocol-for-COVID-19.pdf
- Huang, C., Wang, Y., Li, X., Ren, L., Zhao, J., Hu, Y., … Cao, B. (2020). Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet, 395(10223), 497–506. doi:10.1016/S0140-6736(20)30183-5
- Jaillon, S., Berthenet, K., & Garlanda, C. (2019). Sexual dimorphism in innate immunity. Clinical Reviews in Allergy & Immunology, 56(3), 308–321. doi:10.1007/s12016-017-8648-x
- Jeon, S.B., Seo, D.J., Oh, H., Kingsley, D., & Choi, C. (2017). Development of one-step reverse transcription loop-mediated isothermal amplification for canorovirus detection in oysters. Food Control, 73, 1002–1009. doi:10.1016/j.foodcont.2016.10.005
- Kulveit, J., & Lagerros, J. (2020). COVID-19 forecasting. EpidemicForecasting.org. [cited 2020 Apr 29]. Retrieved May 5, 2020, from http://epidemicforecasting.org/
- Lau, S. K., Li, K. S., Huang, Y., Shek, C. T., Tse, H., Wang, M., … Yuen, K. Y. (2010). Ecoepidemiology and complete genome comparison of different strains of severe acute respiratory syndrome-related Rhinolophus bat coronavirus in China reveal bats as a reservoir for acute, self-limiting infection that allows recombination events. Journal of Virology, 84(6), 2808–2819. doi:10.1128/JVI.02219-09
- Leung, G. M., Hedley, A. J., Ho, L. M., Chau, P., Wong, I. O., Thach, T. Q., … Lam, T. H. (2004). The epidemiology of severe acute respiratory syndrome in the 2003 Hong Kong epidemic: An analysis of all 1755 patients. Annals of Internal Medicine, 141(9), 662–673. doi:10.7326/0003-4819-141-9-200411020-00006
- Lewis, D. (2020). Is the coronavirus airborne? Experts can't agree. Nature, 580(7802), 175. doi:10.1038/d41586-020-00974-w
- Li, H., Liu, S. M., Yu, X. H., Tang, S. L., & Tang, C. K. (2020). Coronavirus disease 2019 (COVID-19): Current status and future perspectives. International Journal of Antimicrobial Agents, 55(5), 105951. doi:10.1016/j.ijantimicag.2020.105951
- Li, X., Zai, J., Zhao, Q., Nie, Q., Li, Y., Foley, B. T., & Chaillon, A. (2020). Evolutionary history, potential intermediate animal host, and cross-species analyses of SARS-CoV-2. Journal of Medical Virology, 92(6), 602–611. doi:10.1002/jmv.25731
- Liu, Y., Gayle, A. A., Wilder-Smith, A., & Rocklöv, J. (2020). The reproductive number of COVID-19 is higher compared to SARS coronavirus. Journal of Travel Medicine, 27(2), Taaa021. 10.1093/jtm/taaa021.
- Lu, G., Hu, Y., Wang, Q., Qi, J., Gao, F., Li, Y., … Gao, G. F. (2013). Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature, 500(7461), 227–231. doi:10.1038/nature12328
- Lu, R., Zhao, X., Li, J., Niu, P., Yang, B., Wu, H., … Tan, W. (2020). Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. The Lancet, 395(10224), 565–574. doi:10.1016/S0140-6736(20)30251-8
- Luo, J. (2020). When will COVID-19 end? Data-driven prediction. SUTD Data-Driven Innovation Lab. Retrieved May 5, 2020, from https://ddi.sutd.edu.sg/%0D
- Manalo, I. F., Smith, M. K., Cheeley, J., & Jacobs, R. (2020). A dermatologic manifestation of COVID-19: Transient livedo reticularis. Journal of the American Academy of Dermatology, 83(2), 700. doi:10.1016/j.jaad.2020.04.018.
- Marzano, A. V., Genovese, G., Fabbrocini, G., Pigatto, P., Monfrecola, G., Piraccini, B. M., … Calzavara-Pinton, P. (2020). Varicella-like exanthem as a specific COVID-19-associated skin manifestation: Multicenter case series of 22 patients. Journal of the American Academy of Dermatology, 83(1), 280–285. doi:10.1016/j.jaad.2020.04.044.
- Mascaretti, L., De Angelis, V., & Berti, P. (2020). The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic and Transfusion Medicine: Reflections from Italy. Blood Transfusion = Trasfusione del Sangue, 18(2), 77–78. doi:10.2450/2020.0071-20
- Mehta, P., McAuley, D. F., Brown, M., Sanchez, E., Tattersall, R. S., Manson, J. J., & HLH Across Speciality Collaboration, UK. (2020). COVID-19: Consider cytokine storm syndromes and immunosuppression. The Lancet, 395(10229), 1033–1034. doi:10.1016/S0140-6736(20)30628-0
- Mizumoto, K., Saitoh, M., Chowell, G., Miyamatsu, Y., & Nishiura, H. (2015). Estimating the risk of Middle East respiratory syndrome (MERS) death during the course of the outbreak in the Republic of Korea, 2015. International Journal of Infectious Diseases, 39, 7–9. doi:10.1016/j.ijid.2015.08.005
- Müller, M. A., Corman, V. M., Jores, J., Meyer, B., Younan, M., Liljander, A., … Drosten, C. (2014). MERS coronavirus neutralizing antibodies in camels, Eastern Africa, 1983-1997. Emerging Infectious Diseases, 20(12), 2093–2095. doi:10.3201/eid2012.141026
- Nguyen, T., Duong Bang, D., & Wolff, A. (2020). 2019 Novel coronavirus disease (COVID-19): Paving the road for rapid detection and point-of-care diagnostics. Micromachines, 11(3), 306. doi:10.3390/mi11030306
- Rubin, G. D., Ryerson, C. J., Haramati, L. B., Sverzellati, N., Kanne, J. P., Raoof, S., … Leung, A. N. (2020). The role of chest imaging in patient management during the COVID-19 pandemic: A multinational consensus statement from the Fleischner Society. Chest, 158(1), 106–116. doi:10.1016/j.chest.2020.04.003
- Sabir, J. S., Lam, T. T., Ahmed, M. M., Li, L., Shen, Y., Abo-Aba, S. E., … Guan, Y. (2016). Co-circulation of three camel coronavirus species and recombination of MERS-CoVs in Saudi Arabia. Science, 351(6268), 81–84. doi:10.1126/science.aac8608
- Schaefer, G. O., Tam, C. C., Savulescu, J., & Voo, T. C. (2020). COVID-19 vaccine development: Time to consider SARS-CoV-2 challenge studies? Vaccine, 38(33), 5085–5088. doi:10.1016/j.vaccine.2020.06.007
- Schulert, G. S., & Grom, A. A. (2014). Macrophage activation syndrome and cytokine-directed therapies. Best Practice & Research. Clinical Rheumatology, 28(2), 277–292. doi:10.1016/j.berh.2014.03.002
- Shakoory, B., Carcillo, J. A., Chatham, W. W., Amdur, R. L., Zhao, H., Dinarello, C. A., … Opal, S. M. (2016). Interleukin-1 receptor blockade is associated with reduced mortality in sepsis patients with features of macrophage activation syndrome: Reanalysis of a prior phase III trial. Critical Care Medicine, 44(2), 275–281. doi:10.1097/CCM.0000000000001402
- Shereen, M. A., Khan, S., Kazmi, A., Bashir, N., & Siddique, R. (2020). COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses. Journal of Advanced Research, 24, 91–98. doi:10.1016/j.jare.2020.03.005
- Sinha, D. K. (2020). COVID-19: Vaccine development and therapeutic strategies. Health, Medicine and Research. https://indiabioscience.org/columns/general-science/covid-19-vaccine-development-and-therapeutic-strategies
- Tobaiqy, M., Qashqary, M., Al-Dahery, S., Mujallad, A., Hershan, A. A., Kamal, M. A., & Helmi, N. (2020). Therapeutic management of COVID-19 patients: A systematic review. Infection Prevention in Practice, 2(3), 100061. doi:10.1016/j.infpip.2020.100061
- Wang, M., Cao, R., Zhang, L., Yang, X., Liu, J., Xu, M., … Xiao, G. (2020). Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Research, 30(3), 269–271. doi:10.1038/s41422-020-0282-0
- Wong, A. C. P., Li, X., Lau, S. K. P., & Woo, P. C. Y. (2019). Global epidemiology of bat coronaviruses. Viruses, 11(2), 174. doi:10.3390/v11020174
- Wong, H. Y. F., Lam, H. Y. S., Fong, A. H., Leung, S. T., Chin, T. W., Lo, C. S. Y., … Ng, M. Y. (2020). Frequency and distribution of chest radiographic findings in COVID-19 positive patients. Radiology, 296(2), E72–E78. doi:10.1148/radiol.2020201160
- World Health Organization (WHO). Coronavirus disease (COVID-19) pandemic. Retrieved May 05, 2020, from https://www.who.int/emergencies/diseases/novel-coronavirus-2019
- Wu, F., Zhao, S., Yu, B., Chen, Y. M., Wang, W., Song, Z. G., … Zhang, Y. Z. (2020). Author Correction: A new coronavirus associated with human respiratory disease in China. Nature, 580(7803), E7. doi:10.1038/s41586-020-2202-3
- Wu, Z., & McGoogan, J. M. (2020). Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: Summary of a report of 72‐314 cases from the Chinese Center for Disease Control and Prevention. JAMA, 323(13), 1239–1242. doi:10.1001/jama.2020.2648
- Xie, X., Zhong, Z., Zhao, W., Zheng, C., Wang, F., & Liu, J. (2020). Chest CT for typical 2019-nCoV pneumonia: Relationship to negative RT-PCR testing. Radiology, 296(2), E41–E45. doi:10.1148/radiol.2020200343
- Xu, D., Zhang, Z., & Wang, F.-S. (2004). SARS-associated coronavirus quasispecies in individual patients. The New England Journal of Medicine, 350(13), 1366–1367. doi:10.1056/NEJMc032421
- Xu, Z., Shi, L., Wang, Y., Zhang, J., Huang, L., Zhang, C., … Wang, F. S. (2020). Pathological findings of COVID-19 associated with acute respiratory distress syndrome. The Lancet. Respiratory Medicine, 8(4), 420–422. doi:10.1016/S2213-2600(20)30076-X
- Ye, Z., Zhang, Y., Wang, Y., Huang, Z., & Song, B. (2020). Chest CT manifestations of new coronavirus disease 2019 (COVID-19): A pictorial review. European Radiology, 30(8), 4381–4389. doi:10.1007/s00330-020-06801-0
- Yezli, S., & Khan, A. (2020). COVID-19 social distancing in the Kingdom of Saudi Arabia: Bold measures in the face of political, economic, social and religious challenges. Travel Medicine and Infectious Disease, 37, 101692. doi:10.1016/j.tmaid.2020.101692
- Yin, Y., & Wunderink, R. G. (2018). MERS, SARS and other coronaviruses as causes of pneumonia. Respirology, 23(2), 130–137. doi:10.1111/resp.13196
- Zaki, A. M., van Boheemen, S., Bestebroer, T. M., Osterhaus, A. D., & Fouchier, R. A. (2012). Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. The New England Journal of Medicine, 367(19), 1814–1820. doi:10.1056/NEJMoa1211721
- Zhou, P., Yang, X. L., Wang, X. G., Hu, B., Zhang, L., Zhang, W., … Shi, Z. L. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 579(7798), 270–273. doi:10.1038/s41586-020-2012-7
- Zhu, N., Zhang, D., Wang, W., Li, X., Yang, B., Song, J., … China Novel Coronavirus Investigating and Research Team. (2020). A novel coronavirus from patients with pneumonia in China, 2019. The New England Journal of Medicine, 382(8), 727–733. doi:10.1056/NEJMoa2001017.