Abstract
The development of BCR-ABL1 tyrosine kinase inhibitors (TKIs) markedly improved the prognosis of patients with chronic myeloid leukemia (CML). Approximately 50% of patients who achieve deep molecular response (DMR) remain in treatment-free remission (TFR) even after discontinuation of TKIs. Although TKIs may achieve clinical “cure” after TKI treatment for specific periods, there are no reliable biomarkers for predicting the response to TKIs and the probability of TFR in CML. An increase in natural killer (NK) cells in the peripheral blood of TKI-treated CML patients is correlated with better outcomes, suggesting that TKIs induce antitumor NK cell immunity against CML cells. Killer immunoglobulin-like receptors (KIRs) are highly polymorphic NK cell receptors that play important roles in the regulation of immune responses. The identification of allelic polymorphisms of KIRs by next-generation sequencing uncovered novel aspects of KIRs. Here we summarize the current knowledge of the genetic and immunological aspects of KIRs and discuss the association between allelic polymorphisms of KIRs and TKI-treated CML.
1. Killer immunoglobulin-like receptors
Natural killer (NK) cells are a unique lymphocyte subset characterized by the presence of large granules in the cytoplasm. NK cells function in innate immunity and eliminate virus-infected and tumor cells by releasing perforin or granzyme. NK cells induce antibody-dependent cellular cytotoxicity, which consists of the secretion of cytotoxic cytokines such as IFN-γ following antibody conjugation. Although the factors determining the cytotoxic activity of NK cells are not fully understood, it may be regulated by the balance of receptor-mediated activating and inhibitory signals, namely the prevalence of activating over inhibitory signals leads to NK cell activation [Citation1,Citation2].
The cytotoxicity of NK cells is regulated by killer immunoglobulin-like receptors (KIRs), which are NK cell surface receptors belonging to the immunoglobulin superfamily. KIRs have many subtypes that show abundant allelic polymorphisms. Histocompatibility leukocyte antigen (HLA) class I molecules function as KIR ligands and act as an inhibitory signal for NK cells: target cells expressing HLA ligands are not attacked by NK cells, whereas the absence of self HLAs (missing self, ) promotes NK cell-mediated lysis. KIRs are classified into inhibitory KIRs, which contain an adaptor molecule called immunoreceptor tyrosine-based inhibitory motif, and activating KIRs, which contain an immunoreceptor tyrosine-based activating motif [Citation3]. Most NK cells express several inhibitory KIRs, and their role has been investigated extensively, whereas the function of activating KIRs remains unclear.
Figure 1. NK cells do not damage cells expressing self MHC class I (A), whereas they damage cells deficient in or with reduced expression of MHC class I (B:missing self, modified from Reference [Citation36]).
![Figure 1. NK cells do not damage cells expressing self MHC class I (A), whereas they damage cells deficient in or with reduced expression of MHC class I (B:missing self, modified from Reference [Citation36]).](/cms/asset/d8adc33a-8ad8-4fbf-8762-bf0a4137ab8c/timm_a_1796062_f0001_c.jpg)
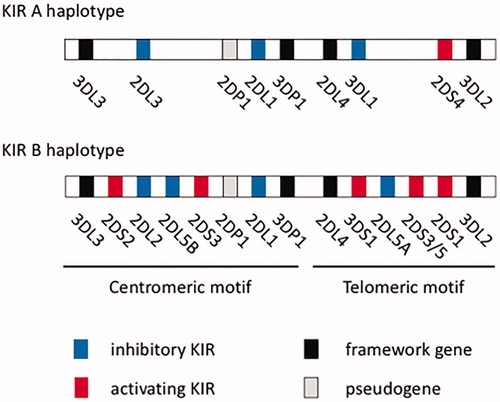
KIR genes are located in the long arm of human chromosome 19 and include 9–15 activating and inhibitory genes. They are classified into centromere and telomere genes, and KIR haplotypes are defined according to the content of specific KIR genes. Haplotype A is defined by the presence of nine framework genes, namely KIR2DL4, KIR3DL2, KIR3DL3, KIR3DP1, pseudogenes KIR2DP1 and KIR2DL1, KIR2DL3, KIR2DS4 and KIR3DL1, whereas the existence of other genes defines haplotype B [Citation4] (). Haplotype A contains one activating receptor gene, KIR2DS4, and haplotype B contains several activating KIR genes. Each individual has two haplotypes, and haplotype AA is the most frequent in Japan, whereas the frequency of haplotype BB is low [Citation5,Citation6].
2. Functional and clinical significance of polymorphisms of KIRs and HLAs
HLA genes are located in human chromosome 6 and are inherited separately from KIR genes. HLA gene polymorphisms determine the intensity of signaling through activating and inhibitory KIRs. The clinical significance of HLA gene polymorphisms for viral infections such as human immunodeficiency virus (HIV) and human papilloma virus infection, and their association with malignancies and autoimmune diseases such as rheumatic arthritis and diabetes mellitus have been investigated extensively [Citation7–11]. Initial findings suggested that a strong NK cell-activating signal is associated with autoimmune diseases, whereas a substantial inhibitory signal is associated with infectious diseases; however, studies testing this hypothesis show inconsistent results. Research on allogeneic hematopoietic stem cell transplantation for hematologic malignancies suggests that the outcomes of transplantation depend on donor and recipient combinations of HLAs and KIRs [Citation12,Citation13], although definitive conclusions have not been reached.
Licensing is an important mechanism underlying the regulation of NK cell immunity. During licensing or education, inhibitory KIRs interact with self HLAs and mediate the attack of HLA-deficient cells by NK cells, leading to potent cytotoxic effects against target cells () [Citation14,Citation15]. Although NK cells lacking inhibitory KIRs against self HLAs should be theoretically activated, this does not occur due to the absence of licensing. The combination of HLAs and KIRs determines the licensing status, although its clinical significance, especially for determining NK cell immunity, remains to be elucidated [Citation16–18].
Figure 3. NK cells recognize self MHC class I, leading to MHC class I-deficient cell killing (A), which does not occur in the absence of recognition (B) (modified from Reference [Citation36]).
![Figure 3. NK cells recognize self MHC class I, leading to MHC class I-deficient cell killing (A), which does not occur in the absence of recognition (B) (modified from Reference [Citation36]).](/cms/asset/9e028692-0e1d-42a0-877b-06090729e223/timm_a_1796062_f0003_c.jpg)
Different KIRs have specific immunological functions; however, studies report inconsistent results depending on the disease and pathology. KIR proteins are expressed at different levels, and the licensed/unlicensed status is not consistent among NK cells. Furthermore, the involvement of the CD94/NKG2 complex and other NK cell receptors in the regulation of immunity in addition to KIRs makes it difficult to determine the clinical significance of KIRs. To comprehensively elucidate the immunological functions of KIRs, a novel paradigm may be necessary.
3. Allelic polymorphisms of KIRs
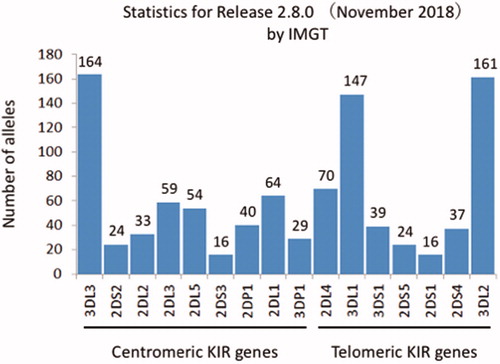
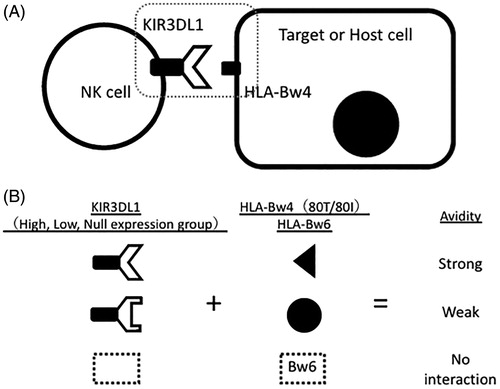
Advances in molecular biology including the development of next-generation sequencing revealed allelic polymorphisms of KIRs (), which has contributed to our understanding of the function of each KIR allele. For example, KIR3DL1 has more than 100 alleles associated with different protein expression levels. Binding avidity and its associated cytotoxicity are defined by the combination of allelic polymorphisms of KIR3DL1 and HLA-Bw4 [Citation6,Citation19–22]. HLA-Bw4 epitopes have an amino acid dimorphism at position 80 (isoleucine 80I and threonine 80 T) [Citation23]. This suggests that allelic combinations of KIR3DL1 and HLA-Bw4 define their binding avidity and the intensity of inhibitory signals mediated by KIR3DL1 () [Citation19,Citation21].
Figure 5. (A) Allelic combination of KIR3DL1 and HLA-Bw4 determines their interaction avidity and subsequently the magnitude of NK cell cytotoxicity against target cells. (B) Depending on the matching status of KIR3DL1 alleles and HLA-Bw4/Bw6, their avidity is defined strong, weak, or no.
In vitro findings obtained using cell lines were confirmed in clinical studies. For example, high expression levels of KIR3DL1 and Bw4-80I are associated with slow HIV expansion and a lower incidence of progression to acquired immunodeficiency syndrome (AIDS) [Citation24]. These results may be explained as follows: the combination of alleles leads to NK cell licensing. In addition, HIV infection downregulates HLA-B protein expression, which induces NK cell attack (missing self). On the other hand, the KIR3DL1*004 allele, which is not expressed on the cell surface, and KIR3DS1, an activating KIR, are associated with a low incidence of progression to AIDS [Citation24,Citation25]. These results suggest that the lack of inhibitory signals through KIR3DL1 and the presence of activating signals promote NK cell activation and eradication of HIV.
The roles of KIRs in malignancies have been investigated. KIRs modulate the efficacy of anti-GD2 antibodies against neuroblastoma [Citation26] and affect the outcomes of auto- and allogeneic hematopoietic stem cell transplantation for acute myelogenous leukemia [Citation27,Citation28]. These results are consistent with the effect of decreased or no binding avidity between KIR3DL1 and HLA-B on better outcome, and suggest that the lack of a KIR3DL1 inhibitory signal is associated with higher NK cell activity, although this needs to be verified.
The results of clinical studies focusing on KIR gene content are inconsistent. Next-generation sequencing enabled allelic genotyping of KIRs, which may uncover their immunological functions and change the existing paradigm.
4. Chronic myeloid leukemia (CML) and NK cells
CML is a hematologic malignancy arising from the Philadelphia chromosome or translocation t(9;22) (q34;q11). The prognosis of CML was improved by the advent of imatinib, an ATP-competitive ABL tyrosine kinase inhibitor (TKI), in 2001 [Citation29], and further improved by the development of second- and third-generation TKIs. Although CML is not a life-threatening disease, several issues need to be addressed. First, whether lifelong administration of TKIs is necessary remains unclear. Second, non-specific inhibition of tyrosine kinases by TKIs can lead to edema and serious cardiovascular adverse events. Third, long-term administration of TKIs limits quality of life (QOL) [Citation30], and its cost cannot be ignored.
Because TKIs do not damage leukemia stem cells [Citation31], TKI discontinuation was initially not recommended. However, several clinical trials assessed the safety and feasibility of TKI discontinuation in patients showing a good molecular response. Forty-one percent of patients treated with the first-generation TKI imatinib who show a two-year deep molecular response (DMR) do not experience relapse (treatment-free remission, TFR) after TKI discontinuation [Citation32]. A trial assessing the discontinuation of the second-generation TKI dasatinib showed that 48% of the patients showing a one-year DMR achieved one-year TFR [Citation33]. The proportions of patients achieving TFR were similar in other trials, suggesting that a specific period of TKI treatment may lead to “clinical cure” of CML [Citation34].
Interferon-treated patients and those showing an increase in NK cells during TKI treatment have a high probability of TFR. Maintenance of DMR after discontinuation of imatinib is associated with increased IFN-γ-producing activated NK cells, and many patients who show increased NK cells during dasatinib therapy achieve TFR [Citation33,Citation35]. This suggests that antitumor immunity mediated by the activation of NK cells leads to TFR. However, the detailed underlying mechanism and the factors associated with NK cell activation remain to be specified.
5. KIRs in CML
Licensed/unlicensed status according to KIR and HLA polymorphisms may be correlated with the outcome and feasibility of TFR in patients with TKI-treated CML [Citation36]. Genotyping of HLAs and KIRs in 59 cases of CML in the chronic phase (CML-CP) showed that KIR haplotypes were associated with DMR in imatinib- or nilotinib-treated patients [Citation37]. Among 17 cases with haplotype AA, 87.5% achieved MR4.5, whereas it was achieved in only 44.4% of haplotype Bx cases except those with haplotype AA (n = 42). The same group analyzed the probability of TFR with no relapse even after discontinuation of TKIs [Citation38]. These authors analyzed the frequency of TFR at 36 months after discontinuation of TKIs in 36 cases that achieved MR4.5, and the results showed that TFR was maintained in 85.7% of haplotype AA cases and 45.5% of haplotype Bx cases. In addition, KIR3DS1/3DL1 and the ligand HLA-Bw4 are associated with relapse after TKI discontinuation. Marin et al. analyzed complete cytogenetic response (CCyR), progression-free survival (PFS), and overall survival (OS) rates in 174 CML-CP cases treated with imatinib for 2 years. The KIR2DS1 gene was negatively associated with CCyR (76.9 vs. 87.9%, p = 0.003), PFS (85.3 vs. 98.1%, p = 0.007), and OS (94.4 vs. 100%, p = 0.015) [Citation39]. Similar analyses in 130 dasatinib-treated CML-CP cases showed no clinical impact of the KIR2DS1 gene on CCyR and major molecular response [Citation40]. A recent study reported that the KIR2DL5B gene is associated with reduced efficacy of TKIs, although this needs further verification [Citation41].
The association between KIR gene polymorphisms and the efficacy of TKIs has been demonstrated, and studies consistently show an association between haplotype A and better prognosis. However, verification is under way, and the KIR gene with the greatest impact needs to be identified.
6. Allelic polymorphisms of KIRs in TKI-treated CML
Polymorphisms of KIRs may determine the activation or inhibition of NK cells, which may define the efficacy of TKIs against CML. However, this association is difficult to determine, and the significance of allelic polymorphisms of KIRs remains unclear. We used next-generation sequencing to perform allelic genotyping of KIRs and HLAs in 76 CML-CP cases and analyzed their association with the efficacy of TKIs. We showed that several specific alleles of KIR2DL4, KIR2DS4, KIR3DL1, and KIR3DL2 were positively associated with DMR, indicating increased efficacy of TKIs [Citation42] (). It is paradoxical that KIR haplotype A was associated with better prognosis in several previous reports, because this haplotype consists mainly of inhibitory KIRs. It may be attributed to this haplotype containing KIR2DL4, KIR2DS4, KIR3DL1, and KIR3DL2 (), specific alleles of which were associated with better prognosis in our analysis. Haplotype analysis of KIRs revealed that these alleles are strongly linked, indicating that they are common haplotypes. Binding avidity between KIR3DL1 and HLA-B may determine NK cell activity. Our analysis showed that patients with no interacting pairs had a better outcome than those with strong interacting pairs, indicating that the lack of interacting pairs may be associated with stronger antitumor NK cell immunity, which is consistent with previous reports in other diseases. In cases with weak-interacting pairs, the KIR3DL1*005 allele was strongly associated with better outcomes, suggesting the importance of this allele (). Though its immunological significance has not been clarified yet, KIR3DL1*005 allele may represent competent NK cell immunity due to diminished suppression of them. We also found that achieving DMR was associated with higher NK cell activity in vitro (), suggesting that the magnitude of NK cell immunity may actually contribute to eradicating CML cells in vivo. However, estimation of allelic KIR haplotypes using Haplo.stats software pointed that KIR3DL1*005 allele is linked with KIR2DL4*005/011 and KIR2DS4*007/*010 alleles (), which suggests that further investigations are needed to explore which allele is the most important. TKIs suppress regulatory T cells, which may derive from Src kinase inhibition [Citation43]. As a result, TKIs promote NK cell activity [Citation44]. We found that the combinations of KIRs and HLAs may determine the magnitude of antitumor NK cell immunity (). Allelic genotyping may predict the efficacy of TKIs and the outcome of TKI discontinuation.
Figure 6. Achievement of deep molecular response (DMR) in TKI-treated CML is associated with allelic polymorphisms of KIRs. KIR2DL4*005/001 or *008, KIR2DS4*003 or *007/010, KIR3DL1*005, and KIR3DL2*009 or *010 were associated with better prognosis (modified from Reference [Citation42]).
![Figure 6. Achievement of deep molecular response (DMR) in TKI-treated CML is associated with allelic polymorphisms of KIRs. KIR2DL4*005/001 or *008, KIR2DS4*003 or *007/010, KIR3DL1*005, and KIR3DL2*009 or *010 were associated with better prognosis (modified from Reference [Citation42]).](/cms/asset/a89e95dc-90ce-4927-ab35-2d839cca74c6/timm_a_1796062_f0006_b.jpg)
Figure 7. Cases of achievement of DMR according to the binding avidity between KIR3DL1 alleles and HLA-B subtypes are shown. A: Patients lacking binding avidity (n = 12) showed better outcomes than individuals with strong interaction avidity (n = 35). Among cases with weak binding avidity, individuals with KIR3DL1*005 showed excellent prognosis (modified from Reference [Citation42]).
![Figure 7. Cases of achievement of DMR according to the binding avidity between KIR3DL1 alleles and HLA-B subtypes are shown. A: Patients lacking binding avidity (n = 12) showed better outcomes than individuals with strong interaction avidity (n = 35). Among cases with weak binding avidity, individuals with KIR3DL1*005 showed excellent prognosis (modified from Reference [Citation42]).](/cms/asset/3a7d3945-b99d-4a84-9188-489083e872e1/timm_a_1796062_f0007_b.jpg)
Figure 8. NK cell activity of TKIs-treated CML patients against HLA class I-deficient K562 cells was analyzed as CD107a degranulation and intracellular IFN-γ production by flow cytometry. The patients who achieved DMR showed higher NK cell activity. Representative data (A) and aggregated data (B, C) are shown (modified from Reference [Citation42]).
![Figure 8. NK cell activity of TKIs-treated CML patients against HLA class I-deficient K562 cells was analyzed as CD107a degranulation and intracellular IFN-γ production by flow cytometry. The patients who achieved DMR showed higher NK cell activity. Representative data (A) and aggregated data (B, C) are shown (modified from Reference [Citation42]).](/cms/asset/a7958386-f06a-4f71-a741-337c898c86d8/timm_a_1796062_f0008_b.jpg)
Figure 9. TKIs damage not only CML cells harboring the bcr-abl gene, but also Foxp3+ regulatory T cells, which indirectly activate NK cells. The cytotoxicity against CML cells may be determined by the combinations of allelic polymorphisms of HLAs in CML cells and KIRs in NK cells.
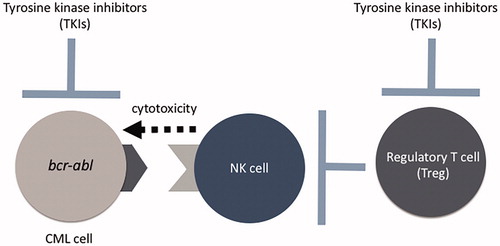
Table 1. Estimated allelic haplotypes harboring KIR3DL1*005.
7. Conclusions
Predicting the probability of TFR after discontinuation of TKIs could reduce medical costs and contribute to the maintenance of QOL. Although the immunological functions of KIRs are specific to each KIR, binding avidity between KIR3DL1 and HLA-B has a clinical impact on NK cell immunity in several diseases. In the near future, allelic genotyping of KIRs and HLAs may uncover novel aspects of NK cell immunity, which would contribute to the development of novel immunotherapy methods based on antitumor NK cell immunity.
Disclosure statement
S.K. received research funding and lecture fees from Bristol-Myers Squibb, Pfizer, Otsuka Pharmaceutical, Ohara Pharmaceutical, Chugai Pharmaceutical, Kyowa Kirin, Astellas Pharma Inc., Taiho Pharmaceutical, Nissan Chemical, and Novartis Pharmaceuticals. H.U. received research funding from Ohara Pharmaceutical.
References
- Caligiuri MA. Human natural killer cells. Blood. 2008;112(3):461–469.
- Vivier E, Raulet DH, Moretta A, et al. Innate or adaptive immunity? The example of natural killer cells. Science. 2011;331(6013):44–49.
- Held W, Mariuzza RA. Cis interactions of immunoreceptors with MHC and non-MHC ligands. Nat Rev Immunol. 2008;8(4):269–278.
- Middleton D, Gonzelez F. The extensive polymorphism of KIR genes. Immunology. 2010;129(1):8–19.
- Saunders PM, Pymm P, Pietra G, et al. Killer cell immunoglobulin-like receptor 3DL1 polymorphism defines distinct hierarchies of HLA class I recognition. J Exp Med. 2016;213(5):791–807.
- Yawata M, Yawata N, Draghi M, et al. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J Exp Med. 2006;203(3):633–645.
- Carrington M, Wang S, Martin MP, et al. Hierarchy of resistance to cervical neoplasia mediated by combinations of killer immunoglobulin-like receptor and human leukocyte antigen loci. J Exp Med. 2005;201(7):1069–1075.
- Habegger de Sorrentino A, Sinchi JL, Marinic K, et al. KIR-HLA-A and B alleles of the Bw4 epitope against HIV infection in discordant heterosexual couples in Chaco Argentina. Immunology. 2013;140(2):273–279.
- Hawes SE, Kiviat NB. Are genital infections and inflammation cofactors in the pathogenesis of invasive cervical cancer? J Natl Cancer Inst. 2002;94(21):1592–1593.
- McGeough CM, Berrar D, Wright G, et al. Killer immunoglobulin-like receptor and human leukocyte antigen-C genotypes in rheumatoid arthritis primary responders and non-responders to anti-TNF-α therapy. Rheumatol Int. 2012;32(6):1647–1653.
- van der Slik AR, Koeleman BP, Verduijn W, et al. KIR in type 1 diabetes: Disparate distribution of activating and inhibitory natural killer cell receptors in patients versus HLA-matched control subjects. Diabetes. 2003;52(10):2639–2642.
- Sekine T, Marin D, Cao K, et al. Specific combinations of donor and recipient KIR-HLA genotypes predict for large differences in outcome after cord blood transplantation. Blood. 2016;128(2):297–312.
- Venstrom JM, Pittari G, Gooley TA, et al. HLA-C-dependent prevention of leukemia relapse by donor activating KIR2DS1. N Engl J Med. 2012;367(9):805–816.
- Anfossi N, Andre P, Guia S, et al. Human NK cell education by inhibitory receptors for MHC class I. Immunity. 2006;25(2):331–342.
- Kim S, Poursine-Laurent J, Truscott SM, et al. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature. 2005;436(7051):709–713.
- Tanimine N, Tanaka Y, Kobayashi T, et al. Quantitative effect of natural killer-cell licensing on hepatocellular carcinoma recurrence after curative hepatectomy. Cancer Immunol Res. 2014;2(12):1142–1147.
- Orr MT, Murphy WJ, Lanier LL. 'Unlicensed' natural killer cells dominate the response to cytomegalovirus infection. Nat Immunol. 2010;11(4):321–327.
- Du J, Lopez-Verges S, Pitcher BN, et al. CALGB 150905 (Alliance): Rituximab broadens the antilymphoma response by activating unlicensed NK cells. Cancer Immunol Res. 2014;2(9):878–889.
- Carr WH, Pando MJ, Parham P. KIR3DL1 polymorphisms that affect NK cell inhibition by HLA-Bw4 ligand. J Immunol. 2005;175(8):5222–5229.
- Gardiner CM, Guethlein LA, Shilling HG, et al. Different NK cell surface phenotypes defined by the DX9 antibody are due to KIR3DL1 gene polymorphism. J Immunol. 2001;166(5):2992–3001.
- O'Connor GM, Guinan KJ, Cunningham RT, et al. Functional polymorphism of the KIR3DL1/S1 receptor on human NK cells. J Immunol. 2007;178(1):235–241.
- Pando MJ, Gardiner CM, Gleimer M, et al. The protein made from a common allele of KIR3DL1 (3DL1*004) is poorly expressed at cell surfaces due to substitution at positions 86 in Ig domain 0 and 182 in Ig domain 1. J Immunol. 2003;171(12):6640–6649.
- Gumperz JE, Barber LD, Valiante NM, et al. Conserved and variable residues within the Bw4 motif of HLA-B make separable contributions to recognition by the NKB1 killer cell-inhibitory receptor. J Immunol. 1997;158(11):5237–5241.
- Martin MP, Qi Y, Gao X, et al. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet. 2007;39(6):733–740.
- Martin MP, Gao X, Lee JH, et al. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet. 2002;31(4):429–434.
- Forlenza CJ, Boudreau JE, Zheng J, et al. KIR3DL1 allelic polymorphism and HLA-B epitopes modulate response to anti-GD2 monoclonal antibody in patients with neuroblastoma. J Clin Oncol. 2016;34(21):2443–2451.
- Boudreau JE, Giglio F, Gooley TA, et al. KIR3DL1/HL A-B subtypes govern acute myelogenous leukemia relapse after hematopoietic cell transplantation. JCO. 2017;35(20):2268–2278.
- Marra J, Greene J, Hwang J, et al. KIR and HLA genotypes predictive of low-affinity interactions are associated with lower relapse in autologous hematopoietic cell transplantation for acute myeloid leukemia. J Immunol. 2015;194(9):4222–4230.
- Jabbour E, Kantarjian H. Chronic myeloid leukemia: 2016 update on diagnosis, therapy, and monitoring. Am J Hematol. 2016;91(2):252–265.
- Rea D. Management of adverse events associated with tyrosine kinase inhibitors in chronic myeloid leukemia. Ann Hematol. 2015;94(Suppl 2):S149–S158.
- Michor F, Hughes TP, Iwasa Y, et al. Dynamics of chronic myeloid leukaemia. Nature. 2005;435(7046):1267–1270.
- Mahon FX, Rea D, Guilhot J, et al.; Intergroupe Français des Leucémies Myéloïdes Chroniques. Discontinuation of imatinib in patients with chronic myeloid leukaemia who have maintained complete molecular remission for at least 2 years: the prospective, multicentre Stop Imatinib (STIM) trial. Lancet Oncol. 2010;11(11):1029–1035.
- Imagawa J, Tanaka H, Okada M, et al.; DADI Trial Group. Discontinuation of dasatinib in patients with chronic myeloid leukaemia who have maintained deep molecular response for longer than 1 year (DADI trial): A multicentre phase 2 trial. Lancet Haematol. 2015;2(12):e528–e535.
- Mahon FX. Discontinuation of tyrosine kinase therapy in CML. Ann Hematol. 2015;94(Suppl 2):S187–S193.
- Mizoguchi I, Yoshimoto T, Katagiri S, et al. Sustained upregulation of effector natural killer cells in chronic myeloid leukemia after discontinuation of imatinib. Cancer Sci. 2013;104(9):1146–1153.
- Ureshino H, Shindo T, Tanaka H, et al. Chronic myeloid leukemia and NK cell immunity. Rinsho Ketsueki. 2017;58(4):381–388.
- La Nasa G, Caocci G, Littera R, et al. Homozygosity for killer immunoglobin-like receptor haplotype A predicts complete molecular response to treatment with tyrosine kinase inhibitors in chronic myeloid leukemia patients. Exp Hematol. 2013;41(5):424–431.
- Caocci G, Martino B, Greco M, et al. Killer immunoglobulin-like receptors can predict TKI treatment-free remission in chronic myeloid leukemia patients. Exp Hematol. 2015;43(12):1015–1018.e1.
- Marin D, Gabriel IH, Ahmad S, et al. KIR2DS1 genotype predicts for complete cytogenetic response and survival in newly diagnosed chronic myeloid leukemia patients treated with imatinib. Leukemia. 2012;26(2):296–302.
- Ali S, Sergeant R, O'Brien SG, et al. Dasatinib may overcome the negative prognostic impact of KIR2DS1 in newly diagnosed patients with chronic myeloid leukemia. Blood. 2012;120(3):697–698.
- Yeung DT, Tang C, Vidovic L, et al. KIR2DL5B genotype predicts outcomes in CML patients treated with response-directed sequential imatinib/nilotinib strategy. Blood. 2015;126(25):2720–2723.
- Ureshino H, Shindo T, Kojima H, et al. Allelic polymorphisms of KIRs and HLAs predict favorable responses to tyrosine kinase inhibitors in CML. Cancer Immunol Res. 2018;6(6):745–754.
- Fei F, Yu Y, Schmitt A, et al. Dasatinib inhibits the proliferation and function of CD4 + CD25+ regulatory T cells. Br J Haematol. 2009;144(2):195–205.
- Hekim C, Ilander M, Yan J, et al. Dasatinib changes immune cell profiles concomitant with reduced tumor growth in several murine solid tumor models. Cancer Immunol Res. 2017;5(2):157–169.