Abstract
To understand the pathogenesis of low level viraemia (LLV) in HIV-infected patients on boosted protease inhibitors (PI/b), we enrolled 34 subjects with a median HIV-RNA 79 copies/mL and followed them for 15 months. Samples for next generation sequencing were collected at three time-points. Two showed resistance-associated mutations (RAMs) in the protease gene, while 95–100% had RAMs in the gag gene, which evolved in approximately a quarter of subjects, suggesting a potential clinical role of these kind of mutations.
Introduction
Low level persistent HIV viraemia (LLV) in HIV–infected individuals on combination antiretroviral therapy (cART) is not uncommonly seen in clinical practice. This has been associated with virological failure in some, but not in all cases.Citation1,Citation2 It usually occurs with low adherence to cART but it could also be consequent to suboptimal drug concentrations or residual shedding from viral reservoirs, after clonal activation and expansion of long-lived HIV-infected cells.Citation3–6 Clinical management is uncertain.
In clinical practice, the detection of resistance-associated mutations (RAM) in a subject with LLV is limited by the cut-off for successful genome amplification (which varies between 200 and 400 copies/mL) of standard validated genotypic assays and by their ability to characterize only viral sequences represented in more than 15–20% of the viral population.Citation7
Boosted protease inhibitors (PI/b) are often preferred in patients with higher risks of poor adherence and resistance development, due to their high genetic barrier. Several studies have shown that virological failure on a PI/b-containing regimen is rarely associated with the development of RAM in the protease gene.Citation8–12 However, recent studies showed that mutations in the substrate of the viral protease (the gag polyprotein) may confer resistance to the PI/b even in absence of mutations in the protease gene.Citation13,Citation14
The aim of this study was to characterize HIV viral populations through next generation sequencing (NGS) and to detect mutations in the substrate of the viral protease gag polyprotein in PLWH presenting with LLV while on PI/b containing antiretroviral regimens.
Methods
This study was conducted in three large HIV clinical centres in London, UK, between 2015 and 2018. It was approved by a National Ethics committee and registered at www.clinicaltrials.gov (NCT02354209).
Individuals receiving PI/b-containing cART either as a first or a switch regimen, with HIV-RNA between 41 and 1000 copies/mL on two consecutive occasions after a previous undetectable viral load (HIV-RNA <40 copies/mL) or after at least six months’ treatment with PI/b, if never achieved complete viral suppression, were included. Patients with less than 95% adherence to cART, assessed with Morisky questionnaire (score less than 2) or according to clinician judgment where the questionnaire was not available, were excluded.
Plasma samples were collected at study baseline, at three to six months and 12 months post- enrolment and analysed by NGS assay at Imperial College London, London, UK.Citation15 The cut-off for detection of minority variants was validated at 2%. Where available, stored plasma collected before the initiation of PI/b containing cART was also analysed.
Descriptive statistics were used to present data. Non parametric Kruskall-Wallis analysis for paired samples was used to determine the significance of the variation of HIV-RNA at different time points.
Results
We enrolled 34 patients, 90% were males and median age (inter quartile range, IQR) was 46 (39–57) years. One subject with a viral load of 131714 c/mL at baseline was excluded. At study baseline, median (IQR) HIV-RNA was 79 (39-128) copies/mL. PCR amplification was successful in 18 of the 24 available samples (75%). NGS showed the presence of RAMs in the protease gene of one subject (M48I, 95% of viral population and I50V, 97% of viral population) and in the gag polyprotein gene of 17/18 (94%) subjects (in all cases, the mutated sequence was found in more than 20% of the viral population). At 3–6 months, median (IQR) HIV-RNA was 82 (59-166) copies/mL. Samples were available from 25 study subjects and PCR was successful in 14 (56%). In the patient with major protease RAMs at baseline, these were maintained at 3–6 months. However, no other patients developed RAMs in the PR gene. Nevertheless, at 3–6 months, all 14 individuals harboured gag RAMs.
At 12 months, median (IQR) HIV-RNA was 65 (52-87), with no significant variation compared to the two previous time points (p = 0.44). Twelve/22 (55%) samples were successfully amplified. One subject, whose plasma RNA failed to amplify at both baseline and 3–6 month time-point, had two protease RAMs in the 12 month sample (M48I and I50V). Again, all samples amplified contained RAMs in the gag polyprotein gene.
The most commonly encountered mutations in the gag polyprotein gene were in the region of the matrix (MA) codon (R76K, Y79F, T81A) and in the p1/p6 cleavage site (L449P and P453T) (, ).
Figure 1 Proportion of mutated viral populations detected with the next generation assay at different time points. Lower limit of detection (2%) is indicated in the graph, points below the 2% lines indicate non mutated virus sequences.
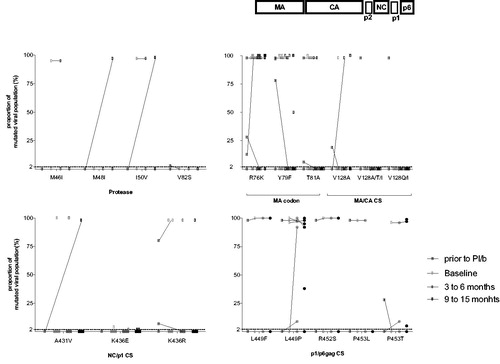
Table 1 Summary of data. VL: viral load (copies/mL), RAMs: resistance associated mutations. protease RAMs are highlighted in bold.
Patients with available paired samples showed variations (acquisition or loss of mutations in the gag polyprotein gene) over time: two out of 12 (16%) showed changes between baseline and 3–6 months and two out of eight (25%) between 3–6 months and 12 months ().
Sixteen stored plasma samples preceding the initiation of PI/b containing cART were available for analysis. Median (IQR) HIV-RNA was 248067 (76201-725850) copies/mL. While only one sample showed a protease RAM (V82S) represented in 3% of the viral population and disappearing in subsequent samples, just over 80% of the samples (13/16, 81%) had at least one RAM in the gag polyprotein gene. During the study, five/10 (50%) paired follow-up samples varied in viral sequence (one consisting of the acquisition of two RAMs in the protease gene).
Discussion
Our study shows that, in a small cohort of individuals with LLV (between 40 and 1000 copies/mL), despite excellent adherence to a PI/b-containing cART, two individuals presented RAMs in the protease gene, whereas almost all had mutated GAG polyprotein sequences. During the 15 months follow-up, HIV-RNA results remained stable, despite of the fact that most individuals did not modify their cART. Only a small proportion of subjects, between 16 and 25% out of those with amplifiable samples (n = 12 and n = 8, respectively), acquired or lost mutations in the gag polyprotein during the study period. While the significance of this variation is uncertain and the number of subjects with paired samples that we could analyse was limited, it is striking that half the samples collected showed different RAMs compared to their paired archived samples taken prior to the introduction of PI/b. This may suggest a that mutations in the gag gene are associated with low level viraemia in certain individuals. In previous studies, it has been suggested that they may confer resistance to PI/b in clinical practice.Citation13,Citation14,Citation16 However, further research is needed to confirm the clinical meaning of such mutations, especially considering that in our small study, the gag gene mutations seemed to lead to LLV rather than full clinical virological failure over 12 months.
Acknowledgments
The authors would like to thank all the subjects who took part in the study and the research teams at Chelsea and Westminster hospital and St Stephen AIDS trust, St Mary’s Hospital and St Thomas Hospital.
Additional information
Funding
References
- Venter WDF, Moorhouse M, Grobbee DE, et al. Effect of HIV-1 low-level viraemia during antiretroviral therapy on treatment outcomes in WHO-guided South African treatment programmes: a multicentre cohort study. Lancet Infect Dis. 2017;18(2):188–197.
- Joya C, Won SH, Okulicz J, et al. Low level viremia is associated with virologic failure in a large military cohort. Clin Infect Dis. 2015;61(3):5072.
- Tobin NH, Learn GH, Holte SE, et al. Evidence that low-level viremias during effective highly active antiretroviral therapy result from two processes : Expression of archival virus and replication of virus evidence that low-level viremias during effective highly active antiretroviral therapy. J Virol. 2005;79(15):9625–9634.
- Nettles RE, Kieffer TL, Kwon P, et al. Intermittent HIV-1 viremia (blips) and drug resistance in patients receiving HAART. JAMA. 2005;293(7):817–829.
- Vancoillie L, Hebberecht L, Dauwe K, et al. Longitudinal sequencing of HIV-1 infected patients with low-level viremia for years while on ART shows no indications for genetic evolution of the virus. Virology. 2017;510:185–193.
- Auyeung K, Watson B, Nohpal A, et al. A single untimed plasma drug concentration measurement during low-level HIV viremia predicts virologic failure. Clin Microbiol Infect. 2016;22(12):1004.e9–1004.e16.
- Parikh UM, McCormick K, Van Zyl G, Mellors JW. Future technologies for monitoring HIV drug resistance and cure. Curr Opin HIV AIDS. 2017;12(2):182–189.
- De Meyer S, Lathouwers E, Dierynck I, et al. Characterization of virologic failure patients on darunavir/ritonavir in treatment-experienced patients. AIDS 2009;23(14):1829–1840.
- Dolling DI, UK Collaborative HIV Cohort Study (UK CHIC), Dunn DT, et al. Low frequency of genotypic resistance in HIV-1-infected patients failing an atazanavir-containing regimen: A clinical cohort study. J Antimicrob Chemother. 2013;68(10):2339–2343.
- Lataillade M, Chiarella J, Yang R, et al. Prevalence and clinical significance of HIV drug resistance mutations by ultra-deep sequencing in antiretroviral-naïve subjects in the CASTLE study. PLoS One. 2010;5(6):e10952–e10957.
- Simen B, St. John E, Kozal M, et al. Virologic failures on initial boosted-PI regimen infrequently possess low-level variants with major PI resistance mutations by ultra-deep sequencing. PLoS One. 2012;7(2):e30118.
- Perrier M, Visseaux B, Landman R, et al. No impact of HIV-1 protease minority resistant variants on the virological response to a first-line PI-based regimen containing darunavir or atazanavir. J Antimicrob Chemother. 2018;73(1):173–176.
- Fun A, Wensing AMJ, Verheyen J, Nijhuis M. Human immunodeficiency virus gag and protease: partners in resistance (Figure 1). 2012;1–14. http://www.retrovirology.com/content/pdf/1742-4690-9-63.pdf
- Perrier M, Castain L, Regad L, et al. HIV-1 protease, Gag and gp41 baseline substitutions associated with virological response to a PI-based regimen. J Antimicrob Chemother. 2019;74(6):1–14.
- Rockstroh J, Boesecke C, McClure MO, et al. Use of next-generation sequencing in the CHAT study (acute HCV in HIV): effect of baseline resistance-associated NS3 variants on treatment failure. HIV Clin Trials. 2018;4336:1–6.
- Delfraissy J, Delaugerre C, Gupta RK. Phenotypic characterization of virological failure following lopinavir/ritonavir monotherapy using full-length gag– protease genes. J Antimicrob Chemother. 2014;69(12):3340–3348.
