Abstract
Objectives: MicroRNA-155 (miR-155) regulates activation of T cells. However, its relationship with T-cell immune activation level in human immunodeficiency virus (HIV) patients remains unclear.
Methods: We recruited 103 HIV-1 infected patients with combination antiretroviral therapy (cART) and 79 cART naïve patients. The miR-155 levels in circulatory T cells were detected by quantitative reverse transcription–PCR. T cell immune activation was detected by the expression of CD38 via flow cytometry.
Results: The levels of miR-155 in the total peripheral blood mononuclear cells, CD4+, and CD8+ T cells from HIV-1 patients were increased (p < 0.01). cART naïve patients exhibited much higher miR-155 levels in CD4 + and CD8+ T cells than patients with cART(p < 0.01). The percentage of CD4 + CD38+ T and CD8 + CD38+ T cells was also increased in cART naïve patients (p < 0.01). MiR-155 level was positively related to immune activation of T cells.
Conclusion: Our findings suggest that miR-155 levels in circulating T cells of HIV-1 patients are increased and associated with T cell activation.
Introduction
Human immunodeficiency virus type 1 (HIV-1) mainly destroys CD4+ T lymphocytes and leads to depletion of CD4+ T cells and disturbed T cell homeostasis, resulting in acquired immune deficiency syndrome (AIDS)Citation1. Nonspecific systemic immune activation is a marked consequence of HIV-1 infectionCitation2,Citation3. Many HIV-1 infected patients exhibit persistent inflammation and chronic immune activation even after years of effective therapyCitation2–5. In addition, many non-AIDS-related diseases of HIV-1 patients are associated with immune activation, such as metabolic syndrome, atherosclerosis, osteoporosis, kidney failure, liver steatosis, and neurocognitive disordersCitation6–8.Thus, the activated phenotype of CD8+ and CD4+ T cells defined by the expression of CD38 is one of the best biomarkers for disease progression in HIV-1 patientsCitation3. To some extent, CD38 surpasses the prognostic value of the conventional clinical markers of CD4+ T cell counts and HIV viral loadCitation2,Citation3. However, in resource-limited areas, the flow cytometry based detection of CD38 expression is unattainable because of the high cost, expensive devices and lack of experienced laboratory personnel.
MicroRNAs are small non-coding RNAs that functions in RNA silencing and post-transcriptional regulation of gene expressionCitation9–11. Many microRNAs can be used as disease biomarkersCitation10. Compared with other laboratory tests, such as flow cytometry, detection of microRNAs is much easier and cheaper, and samples can be stored at −80 C for long time. Thus it is necessary to find microRNAs that can be used as markers of T cell immune activation in HIV-1 infection. MicroRNA-155 (miR-155) is a newly discovered microRNA that regulates the immune response at multiple levelsCitation12. It is found to be greatly increased in activated CD8+ T cells, and may regulate CD8+ T cell responsesCitation13–15. MiR-155 also plays a vital role in T cell responses against infectionsCitation16. By now, there are few studies on the relationship between miR-155 expression and T cell activation in HIV-1 infection. Here, we investigated levels of miR-155 in peripheral T cells from HIV-1 patients with or without combination antiretroviral therapy (cART), and analyzed the correlation between miR-155 levels and immune activation of T cells. Our results showed that the expression levels of miR-155 in both CD4+ and CD8 + T cells of HIV-1 patients were increased, which was positively related with the enhanced immune activation of T cells. Therefore, our findings indicate that miR-155 may be a potential biomarker for immune activation of both CD4+ and CD8 + T cells in HIV-1 infection.
Methods
Study population
Patients with HIV-1 were recruited between the period of June 2018 to May 2019. HIV-1 infection was diagnosed on the basis of positive results from serological and HIV-1RNA detection assays. The subjects were excluded if they had received systemic antibiotics, vaccination or any immunomodulatory drug or haven any immune disease in the previous 3 months. A total of 51 age-, gender- and race-matched HIV negative controls were also recruited.
Ethical approval
This study received approval from the Ethics Review Boards. All subjects were volunteers and provided written informed consent prior to involvement.
Flow cytometry
The CD38 expressions on CD4+ and CD8+ T cells were analyzed using flow cytometry with a FACS can flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA) and commercial flow cytometry assay kits (BD Biosciences). Cell staining was performed at room temperature using a cocktail of the following fluorochrome-conjugated antibodies with the recommend dose by the manuals:Anti-CD3-PerCP-CY5.5 (Cat.No.: 561478), anti-CD4-FITC (Cat.No.: 555346), anti-CD8-APC-Cy7(Cat.No.: 561967) and anti-CD38-PE (Cat.No.: 555460). Briefly, 50 µL whole blood was added to the antibody cocktail, mixed and then incubated for 20 min in the dark at room temperature. Then red blood cells were lysed with lysis buffer, and the rest cells were washed three times with 1 mL of phosphate-buffered saline. The percentages of CD3 + CD4 + CD38+ and CD3 + CD8 + CD38 + T cells and mean fluorescence intensity (MFI) of CD38 were detected.
Detection of miR-155
About 5 mL whole blood from the participants was collected into tubes containing EDTA. Peripheral blood mononuclear cells (PBMCs) were isolated by a density gradient centrifugation method using Ficoll-Paque PLUS (GE Healthcare Life Science, Marlborough, USA). CD4+ and CD8+ T lymphocytes were purified from PBMCs using MACS human CD4 and CD8 microbeads for positive selection (Miltenyi Biotec, Bergisch Gladbach, Germany), respectively. The purity of CD4+ and CD8+ T lymphocytes was >90%. The total RNA was extracted using TRIzol (Thermo Fisher Scientific, Inc., Waltham, MA, USA). The quantitative analysis of miR-155 was performed as described beforeCitation17, using reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis with a Bulge-Loop™ miRNA qRT-PCR Starter kit(Guangzhou RiboBio, Co., Ltd. Guangzhou, China) and the hsa-miR-155 qRT-PCR primer set (Guangzhou RiboBio, Co., Ltd.). A U6 small nuclear RNA primer set (Guangzhou RiboBio, Co., Ltd.) was used as the internal control. The experiments were performed according to the protocol provided in the manufacturer of the kit using a 10 μL reaction system. Briefly, the miRNA RT reaction mix included 1 μL RNA template,1μL miRNA RT primer, 2 μL 5X reverse transcription buffer, 2 μL RTase Mix, 4 μL RNase-free water. The mix was incubated at 42 °C 60 min, and 70°C10 min. The miRNA qPCR reaction system was a mixture of 10 μl SYBR Green Master Mix, 0.8 μl of miRNA forward primer, 0.8 μl of miRNA reverse primer,2μl RT product, and the final volume was taken to 20 μl with water. Real-time PCR was performed for 40 cycles of denaturation (95 °C, 45 s), annealing (62 °C, 30 s), and extension (72 °C, 30 s), and double-stranded DNA was measured at 86 °C after each cycle. Each sample was repeated three times. The relative expression levels of miRNA were calculated by 2ΔΔCT method.
Statistical analysis
Statistical analyses were performed using SPSS for Windows version 20.0 (IBM SPSS, Armonk, NY, USA). Student’s t-test was used to compare between two groups and one-way analysis of variance was used when comparing more than three groups. χ2 was used for categorical variables. The correlation was tested using Spearman’s correlation test. All tests were two-tailed. p < 0.05 was considered to indicate a statistically significant difference.
Results
Clinical information of subjects
A total of 79 cART-naïve HIV-1-infected patients and 103 patients receiving cART were recruited in this study. The mean HIV-1 infection time for the patients with cARTwas11.56 ± 4.19 years, which was significantly longer than that for the cART naïve patients (5.82 ± 3.15 years; p < 0.05). The patients with an average 7.22 ± 1.68 years of cART had a significantly lower viral load, and a higher CD4+ T cell count and CD4/CD8 ratio, compared with cART naïve patients(p < 0.01).The regimens for patients receiving cART were primarily d4T + 3TC + NVP and AZT + 3TC + NVP (82 cases; 79.61%). The detailed participant data is presented in .
Table 1 Demographic and clinical data of the participants
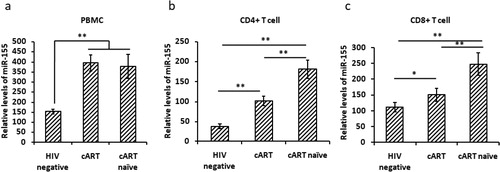
Increased miR-155 levels in peripheral CD4+ and CD8+ T cells of HIV-1 patients
We first isolated CD4+ and CD8+ T cells from PBMCs and detected miR-155 expression levels in PBMC, CD4+ and CD8+ T cells. We found significantly higher miR-155 levels in PBMC, CD4+ and CD8+ T cells of HIV-1 patients compared with HIV negative controls(p < 0.01) (). Furthermore, the miR-155 levels in CD4+ and CD8+ T cells of cART naïve patients were higher than those of patients receiving cART (p < 0.01) (). These results indicate that miR-155 levels are increased in PBMC, CD4+ and CD8+ T cells of HIV-1 patients, and cART can restore the miR-155 expression in both CD4+ and CD8+ T cells.
High levels of immune activation were found in HIV-1 patients
Immune activation of CD4+ and CD8+ T cells was detected by testing both the percentage and MFI of T cells expressing CD38. The results showed that percentage of CD4+ CD38+ and CD8 + CD38+ T cells in cART naïve patients was much higher than that in patients with cART and HIV negative controls (p < 0.01) (). Percentage of CD8+ T cells in patients with cART was higher than that in HIV negative controls (p < 0.01) (), but no difference in the percentage of CD4 + CD38+ T cells was found between patients with cART and HIV negative controls.
Figure 2. Immune activation of peripheral T cells in HIV-1-infected patients. *p < 0.05; **p < 0.01. cART, combination antiretroviral therapy.
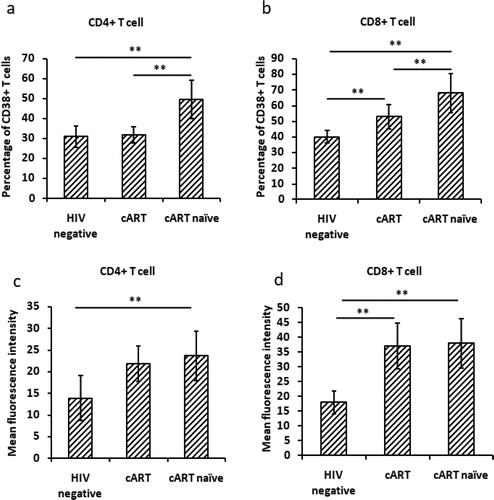
For the MFI of CD38, HIV patients had much higher MFI on both CD4+ and CD8+ T cells than HIV negative controls, although the p value for CD38 MFI on CD4 + T cells between patients with cART and negative controls was more than 0.05 ().
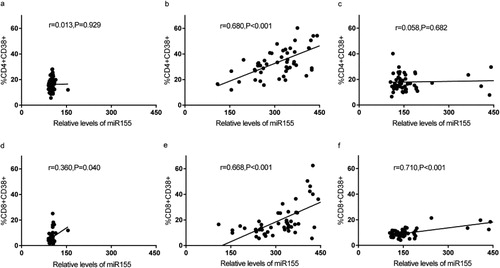
MiR-155 expression positively correlates with immune activation in T cells ofHIV-1 patients
Finally, we analyzed the correlation between immune activation and miR-155 expression in CD4+ and CD8+ T cells. We found that immune activation, based on CD38 expression, of CD4+ and CD8+ T cell subsets was positively correlated withmiR-155 expression in HIV-1 patients without cART (, 3e). In HIV negative controls, 3d)and patients with cART (, 3f), correlations were found between CD8 + CD38+ T cells and miR-155 expression. No statistically significant relationship between MFI of CD38 and miR-155 existed among the three groups of participants.
Discussion
Biomarkers are widely used in clinical practice and they are useful for predicting, diagnosing or monitoring disease. Reliable biomarkers based on PCR methods are suitable for application in resource limited areas because of the low cost and ease to use. CD38 is a widely used biomarker for T cell immune activation in infectious disease, such as HIV-1/AIDS and HBV infectionCitation3,Citation8, but the detection is based on flow cytometry which needs expensive devices and experienced personnel. In this study, we investigated the potential of miR-155 as a biomarker for immune activation of T cells in HIV-1 patients.
The activation of T cells, especially CD8+ T cells, plays an important role in virus clearance, yet excessive activation of CD8+ T cells can also lead to immune-mediated damage and aggravation of disease progressionCitation18,Citation19. Studies found that nonspecific systemic immune activation is the main cause that aggravates HIV-1 infection to AIDSCitation3–8. Even cART can effectively suppress HIV-1 replication and reduce the morbidity and mortality associated with HIV-1 infection, but fails to restore immune functionCitation20. In our study, we found high levels of immune activation (CD38 expression) of CD4+ and CD8+ T cells in cART naïve patients. Immune activation of CD4+ T cells in patients with cART was restored to normal levels, but CD8+ T cell activation was still higher than HIV negative controls, which was consistent with other studiesCitation3,Citation13.
MicroRNAs are promising biomarkers in molecular diagnostics because they are important for many diseases and are stable in common clinical samples, such as blood and urineCitation21,Citation22.MiRNAs play crucial roles in the process of maturation, proliferation, migration, differentiation, and function in the immune systemCitation23. Among the most abundant miRNAs present in immune cells, miR-155 was shown to be critical for T cell fitness and effector functionality, as the selective deletion of miR-155 leads to severe defects in immune responsesCitation24. MiR-155 has been reported to impact disease progression of HIV-1 infection by regulating the transformation of naïve Tregs and naïve CD4 subsets into activated subsetsCitation25. In addition, miR-155 exerts an anti-HIV-1 effect by targeting several HIV-1 dependency factors involved in post-entry and pre-integration events, leading to severely diminished HIV-1 infectionCitation26. Jin et alCitation27 reported that miR-155 levels in the peripheral blood of HIV-1 patients are associated with T cell immune response, including immune activation and exhaustion. They found a positive relation between miR-155 and CD8+ T cell activation, but did not find any relationship between miR-155 and CD4+ T cell activation. In our study, we found miR-155 levels in peripheral blood (including PBMC, CD4+ and CD8+ T cells) of HIV-1 patients both with and without cART were increased significantly. cART can restore the miR-155 levels partially in CD4+ and CD8+ T cells of HIV-1 patients. The miR-155 levels in CD4+ and CD8+ T cells of HIV-1 patients were consistent with their increased T cell activation, which was confirmed by the positive correlation between miR-155 expression and CD4+ and CD8+ T cell activation levels. Our results indicate that miR-155 is a potential biomarker of T cell immune activation.
MiR-155 can affect T cell immune response by regulating multiple genes associated with type I IFN signaling and responseCitation14,Citation26. In addition, the frequency of CD8+ T cells producing IFN-γ was reduced in the absence of miR-155Citation28. In herpes virus infection, miR-155 was reported to be an important regulator in the generation and maintenance of both effector and effector memory CD8+ T cellsCitation29. Existing studies indicate that the impact of miR-155 on CD4+ T cell function is mainly illustrated by the differentiation of T helper cellsCitation30. The defects observed in the CD4+ T cell population include a Th2-skewing bias, as well as the inability to produce functional Th17 cells, resulting in a resistance to the induction of experimental autoimmune encephalomyelitis and experimental rheumatoid arthritisCitation31. Our study indicated that the immune activation can also be impacted by miR-155. The underline mechanisms of miR-155 regulating T cell response need to be further studied.
In summary, our study found increased expression levels of miR-155 in peripheral T cells of HIV-1 patients, which was positively correlated with enhanced immune activation of T cells. cART can partially restore both immune activation and miR-155 expression of T cells. Our findings indicate that miR-155 expression is a potential biomarker for immune activation of T cells in HIV-1 infected patients. However, there are two limitations in our study. Firstly, we didn’t detect HLA-DR expression on T cells, which is also reported as a biomarker for T cell activationCitation3. The HLA-DR + CD38+ double positive T cells were thought to be more specific for T cell activationCitation3,Citation7. Secondly, we only examined miR-155 levels and CD38 expression on total CD4+ and CD8+ T cells. CD38 was reported to be differentially expressed on different subtypes of T cells (e.g. low expression on central memory subtype vs high expression on effective memory subtypes)Citation3,Citation7,Citation13. Further studies are needed to explore the relationship between miR-155 and immune activation of T cell subtypes.
Contributions of authors
Zhenghao Zhang (first author)was the principal investigators who designed the research, supervised data and sample collection, conducted the analyses and drafted the manuscript. Yong Wu and Jiangnan Chen recruited volunteers, collected samples. Fangqing Hu and Xuefang Chen supervised the performance of flow cytometry and q-PCR. Wenfang Xu (corresponding author) coordinated the study and revised the manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Nou E, Lo J, Grinspoon SK. Inflammation, immune activation, and cardiovascular disease in HIV. Aids. 2016;30(10):1495–1509.
- Hazenberg MD, Stuart JWTC, Otto SA, et al. T-cell division in human immunodeficiency virus (HIV)-1 infection is mainly due to immune activation: a longitudinal analysis in patients before and during highly active antiretroviral therapy. Blood. 2000;95(1):249–255.
- Jin C, Zhang F, Wu L, et al. Immune activation and CD127 expression on T lymphocyte subsets of a Chinese cohort of pediatric AIDS patients with different viral responses. Curr HIV Res. 2012;10(7):584–591.
- Guihot A, Dentone C, Assoumou L, et al. Residual immune activation in combined antiretroviral therapy-treated patients with maximally suppressed viremia. Aids. 2016;30(2):327–330.
- Benito JM, Zabay JM, Gil J, et al. Quantitative alterations of the functionally distinct subsets of CD4 and CD8 T lymphocytes in asymptomatic HIV infection: changes in the expression of CD45RO, CD45RA, CD11b, CD38, HLA-DR, and CD25 antigens. J Acquir Immune Defic Syndr Hum Retrovirol. 1997;14(2):128–135.
- Ji S, Jin C, Höxtermann S, et al. Prevalence and Influencing Factors of Thyroid Dysfunction in HIV-Infected Patients. Biomed Res Int. 2016; 2016:1–11.
- Jin C, Ji S, Xie T, et al. Severe dyslipidemia and immune activation in HIV patients with dysglycemia. HIV Clin Trials. 2016;17(5):189–196.
- Dentone C, Fenoglio D, Schenone E, et al. Increased CD38 expression on T lymphocytes as a marker of HIV dissemination into the central nervous system. HIV Clin Trials. 2015;16(5):190–196.
- Jin C, Peng X, Liu F, et al. MicroRNA-181 expression regulates specific post-transcriptional level of SAMHD1 expression in vitro . Biochem Biophys Res Commun. 2014;452(3):760–767.
- Rice AP. Roles of microRNAs and long-noncoding RNAs in human immunodeficiency virus replication. Wiley Interdiscip Rev RNA. 2015;6(6):661–670.
- Jin C, Peng X, Xie T, et al. Detection of the long noncoding RNAs nuclear-enriched autosomal transcript 1 (NEAT1) and metastasis associated lung adenocarcinoma transcript 1 in the peripheral blood of HIV-1-infected patients. HIV Med. 2016;17(1):68–72.
- Seddiki N, Brezar V, Ruffin N, et al. Role of miR-155 in the regulation of lymphocyte immune function and disease. Immunology 2014;142(1):32–38.
- Jin C, Cheng L, Lu X, et al. Elevated expression of miRNA-155 is associated with the differentiation of CD8 + T cells in patients with HIV-1. Mol Med Rep. 2017;16(2):1584–1589.
- Gracias DT, Stelekati E, Hope JL, et al. The microRNA miR-155 controls CD8(+) T cell responses by regulating interferon signaling. Nat Immunol. 2013;14(6):593–602.
- Salaun B, Yamamoto T, Badran B, et al. Differentiation associated regulation of microRNA expression in vivo in human CD8+ T cell subsets. J Transl Med. 2011;9:44.
- Lind EF, Elford AR, Ohashi PS. Micro-RNA 155 is required for optimal CD8+ T cell responses to acute viral and intracellular bacterial challenges. J Immunol. 2013;190(3):1210–1216.
- Jin C, Peng X, Liu F, et al. Interferon-induced sterile alpha motif and histidine/aspartic acid domain-containing protein 1 expression in astrocytes and microglia is mediated by microRNA-181a. Aids. 2016;30(13):2053–2064.
- Peng H, Li Q-L, Hou S-H, et al. Association of genetic polymorphisms in CD8+ T cell inhibitory genes and susceptibility to and progression of chronic HBV infection. Infect Genet Evol. 2015; 36:467–474.
- Ye B, Liu X, Li X, Kong H, Tian L, Chen Y. T-cell exhaustion in chronic hepatitis B infection: current knowledge and clinical significance. Cell Death Dis. 2015;6:e1694.
- Jin CZ, et al. Different plasma levels of interleukins and chemokines: comparison between children and adults with AIDS in China. Chin Med J (Engl). 2009;122(5):530–535.
- Martinez B, Peplow PV. MicroRNAs as biomarkers of diabetic retinopathy and disease progression. Neural Regen Res. 2019;14(11):1858–1869.
- Melak T, Baynes HW. Circulating microRNAs as possible biomarkers for coronary artery disease: a narrative review. EJIFCC 2019;30(2):179–194.
- Giri BR, Mahato RI, Cheng G. Roles of microRNAs in T cell immunity: Implications for strategy development against infectious diseases. Med Res Rev. 2019;39(2):706–732.
- Tao Y, Ai R, Hao Y, et al. Role of miR-155 in immune regulation and its relevance in oral lichen planus. Exp Ther Med. 2019;17(1):575–586.
- Seddiki N, Swaminathan S, Phetsouphanh C, Kelleher AD. miR-155 is differentially expressed in Treg subsets, which may explain expression level differences of miR-155 in HIV-1 infected patients. Blood 2012;119(26):6396–6397.
- Cheng YQ, Ren JP, Zhao J, et al. MicroRNA-155 regulates interferon-γ production in natural killer cells via Tim-3 signalling in chronic hepatitis C virus infection. Immunology 2015;145(4):485–497.
- Jin C, Cheng L, Höxtermann S, et al. MicroRNA-155 is a biomarker of T-cell activation and immune dysfunction in HIV-1-infected patients. HIV Med. 2017;18(5):354–362.
- Imaizumi T, Tanaka H, Tajima A, et al. IFN-γ and TNF-α synergistically induce microRNA-155 which regulates TAB2/IP-10 expression in human mesangial cells. Am J Nephrol. 2010;32(5):462–468.
- Tsai CY, Allie SR, Zhang W, Usherwood EJ. MicroRNA miR-155 affects antiviral effector and effector Memory CD8 T cell differentiation. J Virol. 2013;87(4):2348–2351.
- Hu J-Y, Zhang J, Ma J-Z, et al. MicroRNA-155-IFN-γ feedback loop in CD4(+)T cells of erosive type oral lichen planus. Sci Rep. 2015;5:16935.
- Singh UP, Murphy AE, Enos RT, et al. miR-155 deficiency protects mice from experimental colitis by reducing T helper type 1/type 17 responses. Immunology 2014;143(3):478–489.