ABSTRACT
The Kobe University Macroalgal Culture Collection (KU-MACC), as a section of the Kobe University Research Center for Inland Seas, distributes cultures of macroalgae to the public under the umbrella of the National BioResource Project of Ministry of Education, Culture, Sports, Science and Technology and Japan Agency for Medical Research and Development, in collaboration with the Microbial Culture Collection of the National Institute of Environmental Studies. KU-MACC principally holds macroalgae (Phaeophyceae, Rhodophyta and Ulvophyceae), but also holds some important related taxa of Schizocladiophyceae and Charophyceae. They are unialgal strains maintained as vegetative thalli and by cryo-preservation. Stock cultures are grown in culture chambers under various temperature conditions in transparent polypropylene vials containing Provasoli’s Enriched Seawater (PES) or PESI medium. The collection includes the “genome strains” of Ectocarpus siliculosus, Chondrus crispus, Chara braunii and their related stains. Depending on the phylogenetic group, DNA sequences of the nuclear rDNA ITS region, chloroplast rbcL, and mitochondrial cox3 genes are determined and used for taxonomy and as genetic tags. Based on the genetic data, molecular phylogenies of Ulvophyceae, Phaeophyceae and Rhodophyta are presented. In the rbcL molecular phylogeny of Ulvophyceae, due to rather limited coverage of culture strains, only members of Ulothricales, Ulvales and Bryopsidales are represented by KU-MACC strains, whereas 10 orders are recognized in the class. The molecular phylogeny of Phaeophyceae based on the cox3 gene showed a tree topology similar to the published multigene molecular phylogeny except for the position of Ishige okamurae, although the bootstrap supports of most nodes were low: Ishigeales branched near the base of Phaeophyceae next to Discosporangiales in the multigene molecular phylogeny, but in the cox3 tree, it showed a long branch and was sister to Fucales. In the molecular phylogeny of Rhodophyta, 14 orders are represented by KU-MACC strains covering about half of currently recognized Bangiophycean and Florideophycean orders.
Introduction
The Kobe University Macroalgal Culture Collection (KU-MACC; )) was established in 2003 as a section of the Kobe University Research Center for Inland Seas (KURCIS) and distributes cultures to the public under the umbrella of the National BioResource Project of the Ministry of Education, Culture, Sports, Science and Technology and the Japan Agency for Medical Research and Development, in collaboration with the Microbial Culture Collection of the National Institute of Environmental Studies (NIES). KU-MACC principally holds macroalgae (Phaeophyceae, Rhodophyta and Ulvophyceae), but also holds some important related taxa such as Schizocladiophyceae and Charophyceae. They are unialgal strains maintained as vegetative thalli (Kawai, Motomura, & Okuda, Citation2005) and by cryo-preservation (Heesch et al., Citation2012). Stock cultures are grown in culture chambers of different temperature conditions (5°C, 10°C, 15°C and 20°C) under LED/cold-cathode lamp/fluorescent tube illumination (long day; 16:8 h light:dark) (). By reducing the light intensity, vegetative growth and maturation are suppressed in order to reduce the required frequency of medium changes and reproduction by formation of reproductive cells. Unialgal cultures are maintained in transparent polypropylene vials of 7 ml volume (Assist Tube, Sarstedt, Nümbreche, Germany; ) containing Provasoli’s Enriched Seawater (PES) medium for Rhodophyta and Ulvophyceae (Starr & Zeikus, Citation1993) and a modified PES medium for Phaeophyceae and Schizocladiophyceae (PESI; Tatewaki, Citation1966). Cryopreservation of cultures is conducted using a controlled-rate cooler (Kryo 320-1.7, Planer, Sunbury-On-Thames, UK; )). Cryopreserved cultures are stored in CyroSytem 6000 Cryogenic Sample Storage (Taylor Wharton, Bolivar, Ohio, USA) units. The cryopreserved stains of KU-MACC and NIES are mutually backed up ().
Figure 1. Kobe University Macroalgal Culture Collection (KU-MACC). (a) Top page of web site, (b) illuminated incubator for stock culture, (c) transparent box for tubes of stock culture, (d) stock culture, (e) programable-rate freezer for cryopreservation, (f) liquid nitrogen sample storage tanks for cryopreserved cultures of KU-MACC and (g) backup of NIES.

The collection is based on the culture strains collected and isolated by the researchers of Kobe University Research Center for Inland Sea (Hiroshi Kawai, Takeaki Hanyuda, Shinya Uwai et al.) and strains established and donated by Drs Dieter G. Müller, Akira F. Peters (Germany), Eric C. Henry, Robert Waaland (USA), Willem Prud’homme van Reine (The Netherlands), John West (Australia) and the researchers of the Graduate School of Science, Hokkaido University (Satoshi Shimada, Kazuhiro Kogame et al.). More than 1100 strains of about 300 species in 35 orders covering a large portion of the macroalgal lineages are preserved and are available for distribution.
The collection includes the strains used in full-genome sequencing projects of Ectocarpus siliculosus (Cock et al., Citation2010), Chondrus crispus (Collén et al., Citation2013), Chara braunii (Nishiyama et al., Citation2018) and their related stains. Culture strains are available for order through the collection website (http://ku-macc.nbrp.jp/locale/change?lang=en), where information about the strains (taxonomy, collection information, presence/absence of genetic tag and cryopreserved stocks, culture conditions, publications using the strain) and their photographic images () are available.
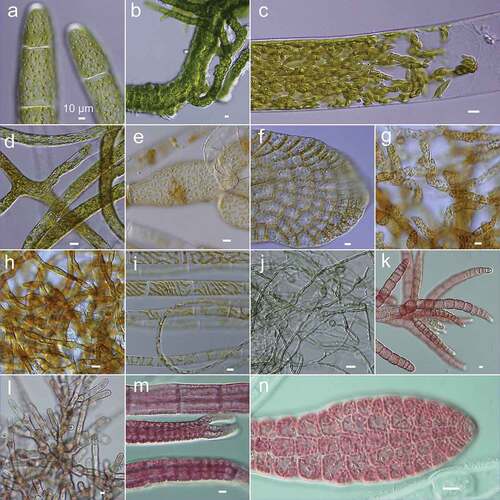
Figure 2. Micrographs of selected strains in KU-MACC. (a) KU-3047, Acrosiphonia spinescens (b) KU-1613, Ulva linza (c) KU-2994, Bryopsis corticulans (d) KU-1518, Codium hubbsii (e) KU-1152, Choristocarpus tenellus (f) KU-1072, Dictyopteris latiuscula (g) KU-1451, Macrocystis pyrifera (h) KU-1372, Ectocarpus siliculosus (i) KU-1335, Ectocarpus sp. (j) KU-1859, Pyropia tenera (k) KU-3141, Dasya sinicola (l) KU-2598, Galaxaura sp. (m) KU-948, Polysiphonia senticulosa (n) KU-971, Delesseria serrulata. Scale = 10 µm.
Depending on the phylogenetic groups, DNA sequences of the nuclear rDNA ITS region (Rhodophyta and Ulvophyceae), chloroplast rbcL (Rhodophyta and Ulvophyceae) and mitochondrial cox3 (Phaeophyceae) genes are determined and used for taxonomy and as genetic tags. Here, we present molecular phylogeny based on the culture strains housed in KU-MACC and published sequences analysed by maximum likelihood (ML) methods of Ulvophyceae, Phaeophyceae and Rhodophyta.
Materials and methods
Culture strains housed in the KU-MACC (indicated as KU-####) were used for genetic analyses (Tables S1–S3). Genomic DNA was extracted from fresh specimens or silica gel-dried specimens using the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) and the QuickExtract Plant DNA Extraction Solution (Epicentre, Madison, Wisconsin, USA) following the manufacturers’ instructions. Polymerase chain reaction (PCR) amplifications of the mitochondrial cox3 gene for strains of Phaeophyceae, and the chloroplast rbcL gene for strains of Rhodophyta and Ulvophyceae, were carried out using the KOD FX (ToYoBo, Osaka, Japan) PCR enzyme and the TaKaRa PCR Thermal Cycler Dice (Takara Bio, Kusatsu, Japan). Primers used for PCR and sequencing are listed in Table S4. After PEG purification (Lis, Citation1980), PCR products were sequenced using a CE DTCS Quick Start Kit (Beckman Coulter, Fullerton, California, USA) and a CEQ8000 DNA analysis system (Beckman Coulter) according to the manufacturer’s instructions, or sequenced by a DNA sequencing service (FASMAC, Atsugi, Japan). For the molecular phylogenetic analyses, both published and newly determined sequence data were used (Tables S1–S3). Molecular phylogenetic trees for three datasets (dataset 1 (Phaeophyceae and Schizocladiophyceae): 229 OTUs, cox3 gene, 687 bp; dataset 2 (Rhodophyceae): 245 OTUs, rbcL gene, 1,285 bp; dataset 3 (Ulvophyceae): 82 OTUs, rbcL gene, 1,320 bp) were constructed by ML analysis. Alignments were prepared using the program MAFFT v.6 (Katoh & Toh, Citation2008) and then manually adjusted prior to phylogenetic analyses. For ML analyses, we used RAxML GUI v.1.31 (Silvestro & Michalak, Citation2012) run to conduct 1000 Rapid Bootstrap searches followed by an ML search, with the GTR + G model for each codon.
Results and discussion
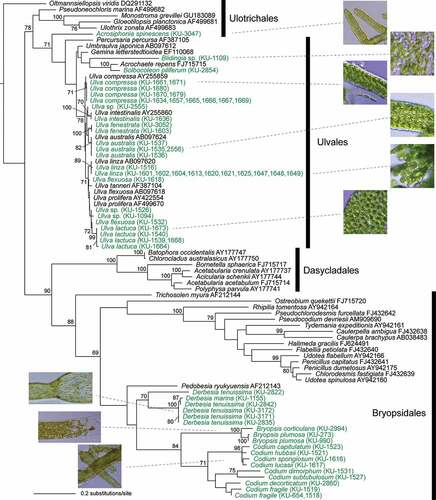
In the rbcL molecular phylogeny of Ulvophyceae, due to rather limited coverage of culture strains, only members of Ulothricales, Ulvales and Bryopsidales were represented by KU-MACC strains (), whereas 10 orders are recognized in the class (Guiry & Guiry, Citation2020). In the tree, in Bryopsidales, Bryopsis spp. formed a clade with Codium of medium support (84%) which was sister to Derbesia. Ulothricales was represented by Acrosiphonia spinescens as a KU-MACC strain, but the monophyly of the order was not supported in the rbcL tree. Ulvales included Blidingia sp., Bulbocoleon piliferum and Ulva spp. As to the low coverage of the culture strains of Ulvophyceae, coenocytic species, which are more common in tropical and sub-tropical habitats, generally require higher culture temperatures (< 20°C), frequent changes of culture media and culture transfer, so their stable maintenance tends to be more difficult compared to those of Phaeophyceae and Rhodophyta.
Figure 3. Maximum likelihood (ML) molecular phylogenetic tree of Ulvophyceae based on chloroplast rbcL gene sequences of KU-MACC culture strains (indicated by green letters) and published sequences (black letters).
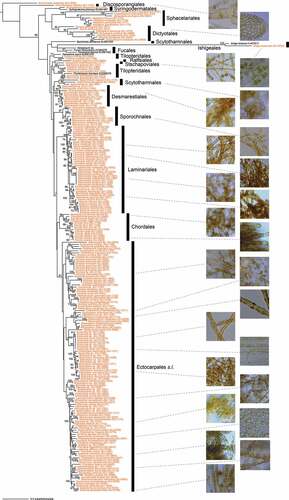
The molecular phylogeny of Phaeophyceae (brown algae) was examined using concatenated DNA sequences of seven chloroplast and mitochondrial genes (atpB, psaA, psaB, psbA, psbC, rbcL and cox1) using 36 KU-MACC unialgal cultures, and the origin and divergence times for brown algae were investigated. These were the first comprehensive analyses based on culture strains (Kawai et al., Citation2015). The molecular phylogeny based only on the cox3 gene () showed a tree topology similar to the multigene molecular phylogeny in Kawai et al. (Citation2015) except for the position of Ishige okamurae, although the bootstrap supports of most nodes were low (< 50%). In the tree, orders with isomorphic life history and apical growth such as Discosporangiales, Sphacelariales and Dictyotales were basal. Syringodermatales were also basal as in the multigene tree although they have heteromorphic life histories. Ishigeales branched near the base of Phaeophyceae next to Discosporangiales in the multigene molecular phylogeny, but in the cox3 tree, it showed a long branch and was sister to Fucales. The position of Fucales was also remarkably different from that in the multigene molecular phylogeny, but this may due to sampling bias; only three species were included in the analysis because the members of Fucales are difficult to maintain in unialgal cultures and are not included in the culture collection.
Figure 4. Maximum likelihood (ML) molecular phylogenetic tree of Phaeophyceae based on mitochondrial cox3 gene sequences of KU-MACC culture strains (indicated by brown letters) and published sequences (black letters).
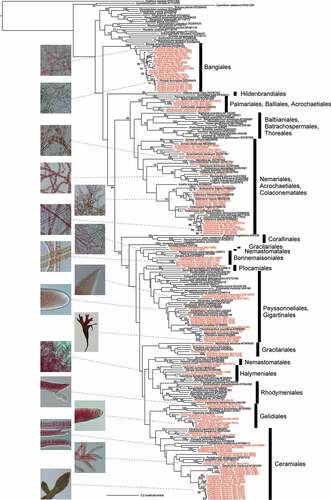
In the molecular phylogeny of Rhodophyta (), 14 orders are represented by KU-MACC strains covering about half of currently recognized Bangiophycean and Florideophycean orders (Guiry & Guiry, Citation2020). As to the coverage of the taxa, in KU-MACC, basal lineages as well as freshwater taxa of Rhodophyta such as Cyanidiophyceae, Stylonematophyceae, Compsopogonophyceae, Rhodellophyceae and Batrachospermales of Florideophyceae are not covered. Also, coralline algae are not covered because of the technical problems in maintaining the cultures.
Figure 5. Maximum likelihood (ML) molecular phylogenetic tree of Rhodophyta based on chloroplast rbcL gene sequences of KU-MACC culture strains (indicated by red letters) and published sequences (black letters).
KU-MACC holds an international workshop for students every year at the marine station of the KURCIS and KU-MACC facilities. This workshop teaches basic techniques for collecting and identifying macroalgae, establishing unialgal cultures, preparing culture media, maintaining cultures in culture chambers, and cryopreservation.
Kawai_et_al_supplementary_information.pdf
Download PDF (469.2 KB)Acknowledgments
We are grateful to Eric Henry for his valuable comments on the article.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Cock, J. M., Sterck, L., Rouzé, P., Scornet, D., Allen, A. E., Amoutzias, G., … Wincker, P. (2010). The Ectocarpus genome and the independent evolution of multicellularity in the brown algae. Nature, 465, 617–621.
- Collén, J., Porcel, B., Carré, W., Ball, S. G., Chaparro, C., Tonon, T., … Boyen, C. (2013). Genome structure and metabolic features in the red seaweed Chondrus crispus shed light on evolution of the Archaeplastida. Proceedings of the National Academy of Sciences of the United States of America, 110, 5247–5252.
- Guiry, M. D., & Guiry, G. M. (2020). AlgaeBase. Galway: World-wide electronic publication, National University of Ireland. Retrieved from http://www.algaebase.org
- Heesch, H., Day, J. G., Yagagishi, T., Kawai, H., Müller, D. G., & Küpper, F. C. (2012). Cryopreservation of the model alga Ectocarpus (Phaeophyceae). CryoLetters, 33, 327–336.
- Katoh, K., & Toh, H. (2008). Recent developments in the MAFFT multiple sequence alignment program. Briefings in Bioinformatics, 9, 286–298.
- Kawai, H., Hanyuda, T., Draisma, S. G., Wilce, R. T., Andersen, R. A., & Gabrielson, P. (2015). Molecular phylogeny of two unusual brown algae, Phaeostrophion irregulare and Platysiphon glacialis, proposal of the Stschapoviales ord. nov. and Platysiphonaceae fam. nov., and a re-examination of divergence times for brown algal orders. Journal of Phycology, 51, 918–928.
- Kawai, H., Motomura, T., & Okuda, K. (2005). Isolation and purification techniques for macroalgae. In R. A. Andersen (Ed.), Algal culturing techniques (pp. 133–143). Burlington, MA: Elsevier Academic Press.
- Lis, J. T. (1980). Fractionation of DNA fragments by polyethylene glycol induced precipitation. Methods in Enzymology, 65, 347–353.
- Nishiyama, T., Sakayama, H., de Vries, J., Bushmannn, H., Sant-Marcoux, D., Ullrich, K. K., … Rensing, S. A. (2018). The Chara genome: Secondary complexity and implications for plant terrestrialization. Cell, 174, 448–464.
- Silvestro, D., & Michalak, I. (2012). raxmlGUI: A graphical front-end for RAxML. Organisms Diversity & Evolution, 12, 335–337.
- Starr, R. C., & Zeikus, J. (1993). UTEX - The culture of algae at the University of Texas at Austin. Journal of Phycology, 29, 1–106.
- Tatewaki, M. (1966). Formation of a crustaceous sporophyte with unilocular sporangia in Scytosiphon lomentaria. Phycologia, 6, 62–66.
