Abstract
The notable increase in drug-resistant infections and the failure of the most potent antibiotics to establish their curative effect without side effect have presented a serious need for the discovery of new therapeutic agent and the study of dietary implications on the mode of entry of these therapeutic agents in the human system. This review provides insight into the forms and modes of action, and roles of beneficial but limited and underutilized antimicrobial peptides for use in dietary formulations, with particular focus on the technologies employed for their discovery as well as their dietary efficacy. The wide spectrum of activities of these peptides will allow the opportunity to explore their benefits as dietary supplements and additives.
Introduction
Antibiotics have been widely used for the prevention of diseases and growth improvement in animal models (de la Fuente-núñez et al. Citation2017). The emergence and rapid dissemination of antibiotic-resistant microbes pose substantial risks for human health. It is predicted that by 2050, global human deaths due to antibiotic-resistant infections will reach 10 million more than the current death toll associated with different forms of cancer (Roy and Trinchieri Citation2017). This is exacerbated by the fact that the number of newly approved antibiotics has significantly and steadily decreased in the last 30 years (Martens and Demain Citation2017). In line with this, an urgent need to develop novel antimicrobial agents, including alternative drugs-based antimicrobial peptides (AMPs) and other compounds are imperative (Chandra et al. Citation2017).
AMPs, also known as host defense peptides (HDPs) are part of the innate immune response found among all classes of life and are mostly found in the tissues and organs that are exposed to airborne pathogens (Wang Citation2017). The uniqueness of AMPs is inherent in the fact that they target the cell membrane which is ubiquitous in microorganisms, unlike the conventional antibiotics which target specific cellular activities (e.g. synthesis of DNA, protein, or cell wall). Another vital role of AMPs is their rapid killing effect (within seconds) after initial contact with the cell membrane (Mai et al. Citation2017).
To date, more than 5,000 AMPs have been discovered in bacteria, protozoans, fungi, plants, insects, and animals or chemically synthesized (Wang Citation2017; Mishra et al. Citation2017). These peptides are generally between 5 and 100 amino acids in length which include two or more positively charged residues provided by arginine, lysine or, in acidic environments, histidine, and a large proportion (generally >50%) of hydrophobic residues (Jia et al. Citation2016). These peptides possess potent, broad-spectrum potential which demonstrates their capacity as novel therapeutic agents against bacteria, viruses, fungi and cancerous cells (Deslouches and Di Citation2017). This is due to their ability to rupture and destabilize biological membranes of pathogens, thereby forming transmembrane channels. They also have the tendency to enhance immunity by functioning as immune modulators which impact several advantages to these peptides over conventional antibiotics (Mishra et al. Citation2017).
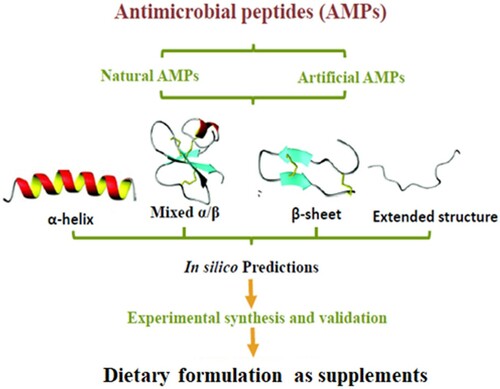
Furthermore, AMPs offer great promise because they combat low and high-affinity pathogen targets suggesting the development of very limited or no pathogen resistance towards them as opposed to conventional antibiotics. This property of AMPs makes it more difficult for target microbes to defend themselves by a single resistance mechanism (Stach et al. Citation2014; Haney et al. Citation2017; Mishra et al. Citation2017). Hence, the importance to study their modes of action and insights into their discovery processes and therapeutic relevance in diets. Functional foods fortified with AMPs can play a role in disease prevention and health promotion. Also, due to irritation and other side effects, it is easier and safer to consume drugs in form of foods than oral administration. This review addresses the need for AMPs development for use as dietary supplements for human for therapeutic purposes against pathogens (Figure ).
Antimicrobial peptides
AMPs are found in nearly all life forms and are produced by all organisms ranging from bacteria to plants, vertebrates, and invertebrates (Kumar et al. Citation2018). In bacteria, the AMPs benefit individual species by killing other bacterial species that may compete for nutrients and the same environmental niche. As a class of bacteria AMPs (class-1 bacteriocins), Lantibiotics are post-translationally modified AMPs (e.g. Nisin) containing non-proteinaceous amino acid lanthionine with a broad range of anti-bacterial activities. Other class II bacteriocins category includes non-lantibiotics (e.g bacteriocin-like inhibitory substance (BLIS)), which have bactericidal activities against limited range of bacteria (Alvarez-Sieiro et al. Citation2016). One of the first AMPs isolated and characterized was nisin from Lactococcus lactis in 1947 (Mattick and Hirsch Citation1947). With minimum inhibitory concentration in the range of nanomolar, nisin is effective against Gram-positive bacteria and has been used as a food preservative for 50 years with no significant development of resistance (Jenssen et al. Citation2006). Another bacteria AMPs studied for their possible use against antibiotic-resistant Gram-positive bacteria is mersacidin (Chatterjee et al. Citation1992). Plant, animal, and fungi have been reported as the origin of naturally discovered AMPs (Kumar et al. Citation2018). Generally, AMPs are cationic, amphipathic and have broad-spectrum activity (Table ) (Patterson-Delafield et al. Citation1981; Okada and Natori Citation1983; Zasloff Citation1987). Some of the roles of AMPs include innate immunity, immune modulation, and control of inflammation (Bowdish et al. Citation2005; Nijnik and Hancock Citation2009; Haney and Hancock Citation2013; Hilchie et al. Citation2013; Hancock et al. Citation2016). Eukaryotic AMP cecropins have been isolated in silk moth and fruit flies (Iwanaga and Kawabata Citation1998; Bachère et al. Citation2004). Tachyplesin was reported to have activities against both bacterial and fungal (Tincu and Taylor Citation2004) while polyphemusin is potent against viruses, bacterial and fungal (Masuda et al. Citation1992; Tincu and Taylor Citation2004).
Table 1. Broad-Spectrum Activity of Antimicrobial peptides from AMP database (APD).
Due to the increasing growth of antibiotics resistance, AMPs with multiple modes of action have been considered as alternatives to inhibit or combat pathogens. Two methods have been put into consideration prior to the development of AMPs. These methods include (1) the N-terminal modification with lipid acid which significantly improved the cationic AMPs’ antibacterial and antifungal activities with secondary structure changes. (2) The combination of in silico computational screening tool and experimental activity test for AMPs optimization. Both the chemical modification and in silico optimization would assist the potent antimicrobial agents’ development.
Structural categories of AMPs
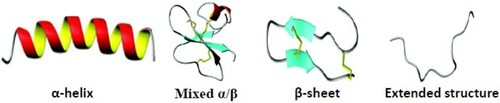
There are variety of approaches used for the classification of AMPs. AMPs are classified based on biosynthesis architecture (e.g. enzymatic, gene coded or synthetic) or molecular target (e.g. membrane targeting or cell penetrating target) or structure and activity-based classification. Here, AMPs have been categorized into subgroups on the basis of their amino acid composition and structures because AMPs structures have been said to be related to their functions. The secondary structural architecture of these vital molecules includes α-helical, mixed α/β, β-stranded, and extended structures with loops or hairpins (Figure ). Their β architecture is due to the presence of two or more disulfide bonds whilst the extended structure is contributed by a single disulfide bond and/or cyclization of the peptide chain (Kumar et al. Citation2018). However, some AMPs do not belong to any of these aforementioned groups (Gabere and Noble Citation2017) whilst some are composed of two different structural components (Jia et al. Citation2016). Certain AMPs form active structures only during interaction with membranes of target cells. An example of this is indolicin that shows a change in conformation during interaction with DNA observable with decreased fluorescence intensity and a slight shift in wavelength of maximum emission (Le et al. Citation2017), globular and amphipathic conformation in an aqueous solution and a wedge-shaped conformation in a lipid bilayer environment (Li et al. Citation2017). Generally, the hydrophilic amino acid residues of these peptides align along one side of their structure whilst their hydrophobic amino acid residues are formed along the opposite side of a helical molecule (Nordström and Malmsten Citation2017). These structural characteristics contribute to their amphipathicity that allows for partitioning into the membrane lipid bilayer, thereby enhancing their antimicrobial activities, thus impacting effective membrane permeability on a range of cytoplasmic targets (Bechinger and Gorr Citation2017).
Natural and artificial production of AMPs
Synthesis of most natural AMPs is carried out by specific cells within prokaryotic and eukaryotic organisms including viruses. These AMPs are produced either by ribosomal translation of mRNA through genetic coding in all life forms or by non-ribosomal peptide synthesis mainly in bacteria (Felgueiras and Amorim Citation2017). Lymph, epithelial cells in gastrointestinal and genitourinary systems (Zeth and Sancho-Vaello Citation2017), phagocytes (Ramesh et al. Citation2016) and lymphocytes (Haney et al. Citation2017) are some of the eukaryotic cells which are involved in the production of AMPs as ribosomal-gene encoded pre-peptides. Apart from the production in innate immune responses, AMPs can be produced by the influence of the host’s inflammatory responses during infection (Riool et al. Citation2017). This is evident from the treatment of mammals with bacterial lipopolysaccharide (LPS) antibiotics for host immunity where AMP production was induced, thus reducing the inflammatory response (Mangoni et al. Citation2016). Certain AMPs such as CAP18 (Kuroda et al. Citation2017), CAP35 (Moravej et al. Citation2018), and a lactoferrin-derivative (Lu et al. Citation2016) block LPS-induced cytokine release by macrophages. In contrast to this inflammatory regulation, antibiotics do not have this regulation on the inflammatory response of the host immune system; and LPS secretion results in an over-reaction of the host immune system, causing sepsis (Li et al. Citation2017).
It is relatively easy to modify the structure of AMPs because of their amino acid composition (Wang Citation2017). This is necessary for modification of existing AMPs and designing new synthetic ones for a potential change of targets and improved stability against proteases (Nordström and Malmsten Citation2017; Zorzi et al. Citation2017). Artificial synthesis is carried out by chemical synthesis using fully synthetic peptides (Zorzi et al. Citation2017) or recombinant expression systems (Haney et al. Citation2017). Despite these advantageous features of AMPs, certain expertise is required for the blueprint design or discovery of AMPs so that certain shortcomings are not introduced into their end products such as potential toxicity to humans (Deslouches and Di Citation2017), sensitivity to harsh environmental conditions such as proteases and extreme pH (Guidotti et al. Citation2017), lack of selectivity against specific strains (Wang et al. Citation2017), high production costs (Felício et al. Citation2017), folding issues of some large AMPs (Logashina et al. Citation2017), reduced activity when used for surface coating (Wang Citation2017), and bacterial resistance to some AMPs (Jia et al. Citation2016). Given the limitations associated with the chemical synthesis of AMPs, there is a need to explore more sensitive options including the use of in silico technologies for more sensitive, specific and accurate options towards their targets. These technologies consider the size, charge, hydrophobicity, amphipathicity, and solubility of AMPs to modify their activity and target spectrum for the production of novel AMPs and mutagenesis of existing ones.
Modes of action of AMPs
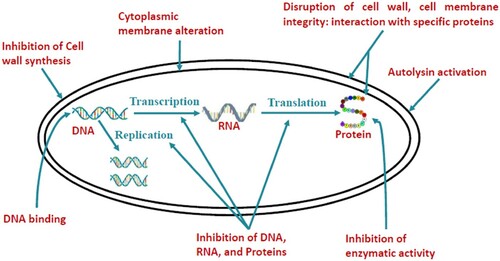
AMPs have varying modes of action by which they kill target microbes (Figure ) (Bechinger and Gorr Citation2017; Wang Citation2017). AMPs possess multiple activities including anti-Gram-positive bacterial, anti-Gram-negative bacterial, anti-fungal, anti-viral, anti-parasitic, and anti-cancer activities. These molecules interfere primarily with the cytoplasmic membrane of the target but can also interact with DNA, protein synthesis, protein folding, and cell wall synthesis (Kumar et al. Citation2018). For bacteria, the initial contact with the peptide is electrostatic, as most bacterial surfaces are anionic, or hydrophobic. AMPs attach to and insert into membrane bilayers forming pores by ‘barrel-stave’, ‘carpet’ or ‘toroidal-pore’ mechanisms due to their amino acid composition, amphipathicity, cationic charge and size. They may also bind intracellular molecules which are crucial to living cells causing inhibition of cell wall synthesis, alteration of the cytoplasmic membrane, activation of autolysin, inhibition of DNA, RNA, and protein synthesis, and inhibition of certain enzymes (Mishra et al. Citation2017). For instance, buforin II can diffuse into cells and bind to DNA and RNA without damaging the cell membrane (Haney et al. Citation2017). Drosocin, pyrrhocoricin, and apidaecin are other examples of such AMPs. These AMPs have 18–20 amino acid residues with an active site for their intracellular target (Felício et al. Citation2017). Other AMPs can block cell wall syntheses such as nicin, Gramicidin S, and PGLa (Mangoni et al. Citation2016; Mishra et al. Citation2017). Generally, their antimicrobial activity is determined by measuring the minimal inhibitory concentration (MIC), which is the lowest concentration of a drug that inhibits the growth of the pathogen (Haney et al. Citation2017). It is therefore not surprising that AMPs have been used for their therapeutic abilities against bacteria and other pathogens (Zhang et al. Citation2017) and diagnostic purposes for HIV (Tincho et al. Citation2016).
Techniques used to elucidate AMP function
The exact mechanism by which antimicrobial peptides establish their effect remains unclear. Three major mechanisms by which they establish their actions include the formation of toroidal pore, carpet, and barrel-stave (Figure ). Although the specificity of these mechanisms differs, they work by allowing peptide-induced membrane rupture and this gives rise to cytoplasmic leakage which ultimately leads to death of the target organisms. Several methods have been used to determine the mechanisms of antimicrobial peptide activity (O’Driscoll et al. Citation2013; Hein et al. Citation2015). These methods include dual polarization interferometry, solid-state NMR which has provided an atomic-level resolution explanation of membrane disruption by AMPs (Haney et al. Citation2017) (Schnabel Citation2017; Wei et al. Citation2017). X-ray crystallography has been employed to delineate how the family of plant defensins rupture membranes by identifying key phospholipids in the cell membranes of the pathogen (Poon et al. Citation2014; Järvå et al. Citation2018). Microscopy is used to visualize the effects of AMPs on microbial cells (Boge et al. Citation2017). Atomic emission spectroscopy is used to detect loss of intracellular potassium (an indication that bacterial membrane integrity has been compromised (Chen et al. Citation2015)). Fluorescent dyes are used to measure the ability of AMPs to transverse membrane vesicles (Figure ) (Scheinpflug et al. Citation2017). Ion channel formation is used to assess the formation and stability of an AMP-induced pore and circular dichroism and orientated circular dichroism is used to measure the orientation and secondary structure of an AMP bound to a lipid bilayer (Müller et al. Citation2017).
Figure 4. Mechanisms of linear AMPs interacting with lipid bilayers. Image modified from Giuliani et al. (Citation2008) (Copyright License No: 4686040911180).
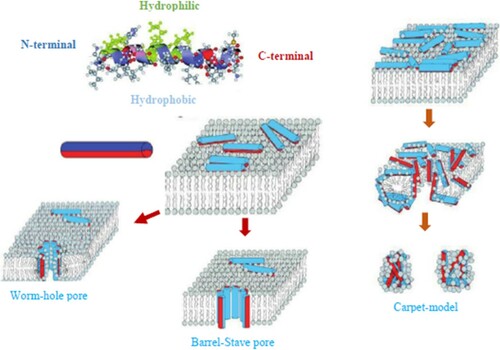
Many AMPs have the potential to fold into amphipathic a-helices with hydrophilic and hydrophobic sides (top, left). This conformation is schematically represented as an amphiphilic cylinder, with hydrophobic (red) and hydrophilic (blue) halves. AMPs bind to the membrane surface with the hydrophobic side groups anchored in the hydrophobic lipidic core of the bilayer, leading to different outcomes (center). The wormhole (or toroidal) pore model (bottom, left) as proposed for megainin (Bottom, center) Barrel-stave pore model, as proposed for alamethicin, a peptide antibiotic produced by the soil fungus Trichoderma viride. (Bottom, right) Carpet model: antimicrobial peptides crowd on the membrane’s surface and lead subsequently to micellation. Modified from Sani et al. (Citation2017).
In Silico predictions of AMPs
Several bioinformatics tools have been developed to generate new templates with appealing antimicrobial properties with the aim of discovering highly active peptide compounds with low cytotoxicity. It is imperative to explore these tools given to demonstrate the connection between the structural classes of AMPs, mode of actions, and the potency of activity among the series of host targets. Thus AMPs with similar structures can have a distinctly different mode of action and target cells. A structure-based precise prediction of activity, mode of action, and host range can only be possible if certain general design principles are considered. One of such is the length of an AMP which allows it to traverse the bilayer of the cell membrane in the barrel-slave model (Mishra et al. Citation2017) and also affect its cytotoxicity. The net charge is another main factor for the initial interaction with the cell membrane which when altered can abolish the selective killing of microbes (Bechinger and Gorr Citation2017). Helicity, although less important, when reduced by incorporating D-amino acids into the primary sequence has been shown to lower the hemolytic effect, while the antimicrobial activity was retained (Migoń et al. Citation2018). If more than 50% of amino acid residues of an AMP are hydrophobic, there is an increased positively charged side of the AMP below a threshold which increases its antimicrobial activity (Bechinger and Gorr Citation2017) and vice versa (Mishra et al. Citation2017). Amphipathicity is another property of AMPs that ensure their activity and interaction with microbial membranes and has been shown to be more important than hydrophobicity for binding to microbial membranes (Felício et al. Citation2017). This is because it confers a strong partition on AMP-membrane interaction for the target cells. In other words, if the AMP molecules aggregate, it will lose its ability to interact with the cell membrane.
Mutagenesis affects solubility in such a way that, if handled carelessly, can increase the dimerization of AMPs. For example, substituting a Lys residue on the non-polar face of a hybrid AMP prevents dimerization and leads to reduced hemolytic activity. Thus, loss of dimerization makes this AMP more effective for its incorporation into microbial membranes (Thamri et al. Citation2017). It is therefore imperative to predict and construct a functional peptide using in silico technologies, to achieve more accurate results. Application of these technologies to analyze the details of the folding of AMP molecules and interaction with target cells may be a promising approach to improve current trial and error methods. These types of artificial AMP design strategies hold the potential for developing new synthetic peptides against antibiotic-resistant pathogens (Riool et al. Citation2017).
Support Vector Machine (SVM)
SVM method is a polynomial kernel function which is regarded as the best model for classification of AMPs with excellent accuracy, sensitivity, specificity, and the Mathews Correlation Coefficient measure when used in a linear and nonlinear classification (Gabere and Noble Citation2017). It uses algorithms to analyze data used for classification and regression analysis. It constructs a hyperplane or set of hyperplanes in a high- or infinite-dimensional space, which can be used for classification, regression, or other tasks like outliers identification. The hyperplane that has the largest distance to the nearest training data point of any class is a good separation since the larger the margin, the lower the generalization error of the classifier (Hastie et al. Citation2015). SVM is a powerful tool for solving problems in classification, function estimation and density estimation with no prior assumptions about data and underlying distribution. Also, it does not necessitate a large number of training data to avoid overfitting. For this reason, it has been widely applied in biological and other sciences to classify proteins with up to 90% of the compounds classified correctly (Meher et al. Citation2017).
Hidden Markov Models (HMMER)
The Hidden Markov Models (HMMER) algorithm, used on Ubuntu LTS operating system which is based on the Linux kernel, has multiple modules to perform optimally using several command lines. HMMER only requires the sequences of experimentally validated biomolecules for the construction of models. These models will display their features based on the motifs of the input sequences and would have the desired activity against the specific target (Porto et al. Citation2017; Liu et al. Citation2017). The high sensitivity of the HMMER profiles is due to the combination of the scoring system and the low cut-off E-value calculated during the proteome sequences scanning steps. The E-value provides more information about the probability that predicted AMPs are true positives or false negatives (Hoppenbrouwers et al. Citation2017; Khater et al. Citation2017; Lopes et al. Citation2017). The use of the HMMER algorithm is deemed an appropriate tool which enables a more sophisticated search for novel peptides through proteome sequence scanning. Several AMPs discovered through this process, have been proven to detect certain diseases (Tincho et al. Citation2016).
Gap Local Alignment of Motifs 2 (GLAM2)
GLAM2 is designed for certain bio-sequence motifs that exhibit mutational changes either with insertion or deletion because it allows gaps within the AMPs. Discovery of gapped motifs is intrinsically more difficult than un-gapped motifs, therefore performing a simpler gapless motif analysis is recommended. Amino acid residues may be inserted between gaps and this assumes that their identity is irrelevant. GLAM2 reports a score for each motif that it discovers, with higher scores indicating stronger motifs and better matches to the overall motif. GLAM2 does not search for multiple distinct motifs but performs replicates in an attempt to discover motif with 10 times possibility strength, and displays the results in order of bit and E value scores. If the top few results are similar, this may be regarded as successful replication and a rerun is recommended to increase the ‘number of iterations’ parameters (Vannini et al. Citation2017). However, GLAM2 has difficulty adjusting the motif width when there are many short sequences. It should be noted that both protein and DNA motifs are often shorter than the defaults (50) used by GLAM2 for ‘maximum number of aligned columns’ and ‘maximum width’ (Brázda et al. Citation2018).
Quantitative Structure-Activity Relationship (QSAR)
QSAR plays a vital role in modern drug design since it is cost-effective, much easier to handle, faster and more amenable to automation. The QSAR model is built on the mathematical foundation of statistical and chemometric techniques. It is a computational screening model that is used whenever compounds with particular biological activity are known. It correlates macroscopic target properties with computed atom-based descriptors derived from the spatial representation of the molecular structures. Although QSAR performs a positive role in predicting and associating the biological features of molecules in many instances and conditions, it suffers from serious shortcomings given as follows: deficiency in sufficient number of training molecules, generation of only 2-D structure molecules, shortage of parameters for comparing drug-receptor relationships such as Hammett constant, lack specific physiochemical parameters, absence of representation of stereochemistry, unavailable unique dissolution with high risk of failure and chance correlations, requirement of substituent constants and chemistry utilized to design a molecule before action, and absence of suggestion to synthesize a new compound through classical QSAR equations with no graphical result (E Greber and Dawgul Citation2016). Except these shortcomings are given adequate consideration, QSAR will not design AMPs of desired specificity and accuracy.
Linear Discriminant analysis (LD)
Linear Discriminant Analysis (LDA), formulated by Ronald A. Fisher (Citation1938), is used to reduce dimensionality in the pre-processing step for pattern-classification and machine learning applications (Fisher Citation1938). The steps involved in performing the LDA include: compute the d-dimensional mean vectors for the different classes from the dataset, compute the scatter matrices (in-between-class and within-class scatter matrix), compute the eigenvectors (e1,e2, … ,ed1) and corresponding eigenvalues (λ1, λ2, … ,λd) for the scatter matrices, sort the eigenvectors by decreasing eigenvalues and choose k eigenvectors with the largest eigenvalues to form a d×k dimensional matrix W (where every column represents an eigenvector), and use this d×k eigenvector matrix to transform the samples onto the new subspace. This can be summarized by the matrix multiplication: Y = X×W (where X is an n×d-dimensional matrix representing then samples, and y are the transformed n×k-dimensional samples in the new subspace (Han et al. Citation2017)).
Random forest
Random forest, also known as random decision forest, is a method used for classification and regression by constructing a multitude of decision trees at training time, using the modal class (classification) or mean prediction (regression) of the individual trees (Manavalan et al. Citation2018). This algorithm contains the preliminary decision tree step, tree bagging, random forest step, and extra tree steps. The preliminary tree step involves an off-the-shelf procedure for data mining because it is invariant and robust but seldom inaccurate (Abdallh et al. Citation2016). Tree bagging decreases variance without leading to bias. In other words, the prediction of a single tree is sensitive to noise in its training set while training many trees in a single tree would give strongly correlated trees. Random forest step gives accuracy for the correlation of the trees in a bootstrap sample. Thus, creating extra trees is a randomized step yield extremely randomized trees. These are then trained using bagging and random subspace method with top-down splitting to select a random value feature’s empirical range. Random forest predictors naturally lead to a dissimilarity measure among the observations which are then measured between unlabelled data with the idea to construct a random forest predictor that distinguishes the ‘observed’ data from suitably generated synthetic data (Meher et al. Citation2017).
Sliding Window (SW)
Sliding Window (SW) is created to solve inherent problems such as high memory bandwidth requirements, segregation of the memory space and centralized control of switching functions which are associated with present methods used in handling packet biological data traffic efficiently. This method introduces a switch architecture to overcome these problems facing the scalability of shared-memory switches (Liu et al. Citation2018). It makes use of a wide range of coincidental memory modules that are logically divided by all input and output lines to store and execute data packets. The special ability to accomplish parallel operation and the simplicity of the control functions make this technique a better method for prediction of AMPs (Pane et al. Citation2017).
Sequence alignment and feature selection methods
Sequence Alignment and Feature Selection methods were developed since most tools in Bioinformatics do not have the function to identify which features are optimal for accurately predicting and meaningfully interpreting their biological implications. In the feature selection method, each peptide will be coded with several features, including amino acid composition (Meher et al. Citation2017) and pseudo amino acid composition (Gabere and Noble Citation2017) that incorporated electrostatic charge, codon diversity, molecular volume, polarity, and secondary structure (W. Chen et al. Citation2016) while the sequence alignment method uses the BLASTP function. BLASTP (Bhadra et al. Citation2018) was used to predict AMPs, which can be described with a query peptide P and the training set, then the high-scoring segment pairs (HSPs) score between the query peptide and each peptide in the training set are calculated by BLASTP with default parameters. Sequence Alignment would incorporate sensitivity but cannot deal with much data accurately and this is complemented by the Feature Selection method (Liu et al. Citation2017).
Dietary consequences of AMPs as therapeutic agents
The widespread distribution and abundance of AMPs in living organisms underscore their critical role in innate immunity as antimicrobial and immunomodulatory agents (Wang Citation2017). There are numerous AMPs currently under clinical development for therapeutic indications besides antimicrobials or antifungal agents. To mention a few, LL-37 has recently been evaluated in a clinical trial as a local treatment to enhance healing of venous leg ulcers (Grönberg et al. Citation2014). Although the mechanisms by which LL-37 promotes wound healing are not fully understood but may involve several wound repair components such as re-epithelialization, angiogenesis, and inflammation. Apart from this, PXL01 has been explored for its prevention of post-surgical adhesion formation in connection to hand surgery, besides its anti-infection roles (Wiig et al. Citation2014).
Studies are ongoing to use AMPs to increase their endogenous production by the body in order to boost the innate immune responses and thereby combating infections. For instance, vitamin D has been evaluated for its modulatory expression on several AMPs applicability for the treatment of infections in several ongoing trials (Wang Citation2017). However, the anti-infection activity of AMPs is highly sensitive to environmental conditions, which results in discrepancies between in vitro versus in vivo efficacy and makes the accurate prediction of anti-infection properties in a clinical situation very difficult. This has been described in several reports where the in vivo antimicrobial properties of AMPs in experimental animal models appear inactive or minimally active in the presence of physiological salt concentrations evaluated in minimum inhibitory concentration (MIC) (Radzishevsky et al. Citation2007; Jennings et al. Citation2014; Krizsan et al. Citation2014; Ageitos et al. Citation2017; Haney et al. Citation2017). For example, the presence of carbonates dramatically increases the sensitivity of AMPs in both Gram-positive and Gram-negative bacteria (Gong et al. Citation2017).
Furthermore, certain AMPs derived from food proteins (e.g milk) which are harmless substances have been discovered to have immense advantage for use in medicine and food industry (Sah et al. Citation2018). These advantages transcend the nutritive value to be derived from them with possible resource to improve the innate defence of the organisms against invading pathogens. Examples of these proteins are lactoferrin (Liu et al. Citation2018) and lysozyme (Wu et al. Citation2018) which are bactericidal and have been modelled through the process of ‘tailoring and modelling’ as food supplements. Other examples include ovotransferrin, alpha-lactalbumin and beta-lactoglobulin which are products of proteolytic digestion of food proteins which are multifunctional molecules with physiological roles (Chalamaiah et al. Citation2018). Also, pyroglutamyl leucine, a peptide present in protein hydrolysates and fermented foods, has been isolated for oral administration at 0.1 mg/kg body weight in different animal models (Shirako et al. Citation2019). It has been said to function in the attenuation of hepatitis, colitis and dysbiosis, a term used for microbial imbalance or maladaptation on or inside the body (Sato Citation2018).
Moreover, the low metabolic stability of AMPs, which is an inherent risk of therapeutic peptides in general, is considered another key factor limiting their clinical application. Peptide drugs are subjected to pre-systemic enzymatic degradation and poor penetration in intestinal mucosa thereby limiting their bioavailability during oral administration. Also, the rapid degradation by proteolytic enzymes in blood plasma and rapid removal of AMPs from the circulation by the liver and kidneys are negatively affected by their short half-life during intravenous injection (Zhang et al. Citation2017). It is believed that their use as food supplements would enhance their therapeutic efficacy. Yoon et al. (Citation2013)investigated the effect of dietary supplementation of basal diet with antimicrobial peptide-P5 (AMP-P5) on weanling pig nutrition; this enhanced the peptide with improved growth performance and total tract nutrients assimilation potential with reduced coliforms in weaning pigs. This result is in line with the work of Xiong et al. (Citation2014) which reported that the addition of AMPs to the basal diet had positive effects on growth action, ameliorative occurrence of diarrhea, and a large survival rate of weaned pigs.
Furthermore, Gadde et al. (Citation2017), investigated the effects of antimicrobial peptide-A3 (AMP-A3) at varying concentration to the basal diet on the growth performance, nutrient retention, intestinal microflora and intestinal morphology of broilers fed with basal diet. The result showed that 90 mg/kg AMP-A3 has the best performance which can be used as a potential antimicrobial growth promoter. In addition, Liu et al. (Citation2017) reported the effects of the mixture of feed additives such as recombinant swine defensin and a fly antibacterial peptide in the ratio 1:1 in juvenile goats which lead to increased body weight, average daily weight gain, improved enzymatic activity (pectinase, xylanase, and lipase) and microorganism diversity indices. However, further experimental research is required to establish whether these effects are the same for the human model. Two AMPs which belong to the bacteriocins were reported, namely nisin and pediocin PA-1/AcH, for their firm presence in line with other natural food preservatives, either claimed as an active substance in the formulation or being present in a fermented active extract (Barbosa et al. Citation2017). This has the potential to reduce the carcinogenic and other side effects of some preservatives used in foods and antibiotics.
AMPs are prone to contain certain amino acid types, such as cysteine, proline, arginine, lysine, tryptophan, and histidine which are rich in antimicrobial activities (Jia et al. Citation2016). There is sufficient evidence that cysteine, tryptophan, lysine, and histidine are essential amino acids that could be used to supplement diet in humans. Cysteine-rich peptides are particularly typical of plants (Ageitos et al. Citation2017) and animals (Wang et al. Citation2017). Pairs of cysteines forming intramolecular disulfide bridge are common in AMPs, thus allowing a complex three-dimensional structure, such as beta-sheets (Haney et al. Citation2017) and beta-turn (Patel and Akhtar Citation2017).
The discovery and commercial development of these peptides and the understanding of their mechanism of action, resistance patterns, and smart formulation strategies is an active area of research. Formation of AMPs cyclization, inclusion of certain amino acids and artificial analogs of amino acids, and peptide mimetics with distinct structural backbones have also been reported to solve related problems of local delivery through degradation by proteolytic enzymes (Bechinger and Gorr Citation2017). Furthermore, blockage of N- or C-terminal ends of the AMPs by modifications such as N-pyroglutamate, N-acetylation, or C-amidation is frequently being used to promote resistance toward peptidases (Kang et al. Citation2017). These are some of the possible modifications through their use as food supplements. While AMPs will probably not replace conventional antibiotics (Li et al. Citation2017), they may serve as a valuable asset to already marketed drugs, especially in considering safety features of these peptides such as biodegradability and lack of immunogenicity when added to diets ().
Table 2. Some antimicrobial peptides (AMPs) used as food supplements, their biological roles and mechanism of action.
Challenges of the use of AMPs as dietary supplements
In spite of the numerous advantages of AMPs, certain challenges can hinder their applications in dietary formulation which include high costs of production, potential toxicity to humans (Wang Citation2017) and sensitivity to proteases and extreme pH (Wong et al. Citation2017). Their lack of selectivity against specific strains and folding issues of high molecular weight ones may reduce their antimicrobial properties when AMPs are used for surface coating (Baumann et al. Citation2017; Zhu et al. Citation2017). This is why many AMPs including the natural ones (such as magainins which are active in vitro) have high activities in animal models of infection only at very high doses.
Conclusion
Antimicrobial peptides are invaluable molecules whose potency suffers serious compromise by the incidence and expansion of antimicrobial resistance. This challenges their translation from non-clinical candidates into successful clinical products and could be enhanced by dietary influences as described by in vivo studies. AMPs have properties which make them promising alternatives to conventional antibiotics as outlined in this review. The provision of AMPs in animal foods strengthens a wide variety of therapeutic purposes that include growth promotion. Therefore, the discovery of more AMPs with their use as AMP-diet supplement is imperative for improved sensitivity, accuracy, and specificity to target pathogens as it would enhance their therapeutic effect in overcoming the shortcomings such as untimely degradation and reduced efficacy for effective combat against resistant pathogens. With the aid of evolving technology such as in silico technology and advancement in knowledge, effective use of AMPs has the potential to improve the efficiency of medical practitioners in treating their patients through dietary means to reduce side effect caused by antibiotics. However, there is a need for research to evaluate the pharmacokinetics of these AMPs due to the fact that the fate of AMPs in vivo is still poorly understood.
Future perspectives
Natural and artificial sources of AMPs, including their modes of actions, should be investigated more deeply. More tools that can elucidate the structure–function relationship of AMPs should be developed for efficient synthesis and modification of AMPs. Although overexposure of microbes to high level of AMPs may bring about resistance, this may serve as a good challenge to randomize their use.
Acknowledgments
The authors would like to acknowledge funding from the National Research Foundation (NRF) which helped to support our research described herein.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Abdallh MMA, Bilal KH, Babiker A. 2016. Machine learning algorithms. Int J Eng Appl Manag Sci Paradigms. 36(01):17–27.
- Ageitos J, Sánchez-Pérez A, Calo-Mata P, Villa T. 2017. Antimicrobial Peptides (AMPs): ancient compounds that represent novel weapons in the fight against bacteria. Biochem Pharmacol. 133:117–138. doi: https://doi.org/10.1016/j.bcp.2016.09.018
- Alvarez-Sieiro P, Montalbán-López M, Mu D, Kuipers OP. 2016. Bacteriocins of lactic acid bacteria: extending the family. Appl Microbiol Biotechnol. 100(7):2939–2951. doi: https://doi.org/10.1007/s00253-016-7343-9
- Bachère E, Gueguen Y, Gonzalez M, De Lorgeril J, Garnier J, Romestand B. 2004. Insights into the anti-microbial defense of marine invertebrates: the penaeid shrimps and the oyster Crassostrea gigas. Immunol Rev. 198(1):149–168. doi: https://doi.org/10.1111/j.0105-2896.2004.00115.x
- Barbosa AAT, Mantovani HC, Jain S. 2017. Bacteriocins from lactic acid bacteria and their potential in the preservation of fruit products. Crit Rev Biotechnol. 37(7):852–864. doi: https://doi.org/10.1080/07388551.2016.1262323
- Baumann T, Nickling JH, Bartholomae M, Buivydas A, Kuipers OP, Budisa N. 2017. Prospects of in vivo incorporation of non-canonical amino acids for the chemical diversification of antimicrobial peptides. Front Microbiol. 8:124. doi: https://doi.org/10.3389/fmicb.2017.00124
- Bechinger B, Gorr S-U. 2017. Antimicrobial peptides: mechanisms of action and resistance. J Dent Res. 96(3):254–260. doi: https://doi.org/10.1177/0022034516679973
- Bhadra P, Yan J, Li J, Fong S, Siu SW. 2018. AmPEP: sequence-based prediction of antimicrobial peptides using distribution patterns of amino acid properties and random forest. Sci Rep. 8(1):1697. doi: https://doi.org/10.1038/s41598-018-19752-w
- Boge L, Umerska A, Matougui N, Bysell H, Ringstad L, Davoudi M, Andersson M. 2017. Cubosomes post-loaded with antimicrobial peptides: characterization, bactericidal effect and proteolytic stability. Int J Pharm. 526(1-2):400–412. doi: https://doi.org/10.1016/j.ijpharm.2017.04.082
- Bowdish DM, Davidson DJ, Scott MG, Hancock RE. 2005. Immunomodulatory activities of small host defense peptides. Antimicrob Agents Chemother. 49(5):1727–1732. doi: https://doi.org/10.1128/AAC.49.5.1727-1732.2005
- Brázda V, Červeň J, Bartas M, Mikysková N, Coufal J, Pečinka P. 2018. The amino acid composition of quadruplex binding proteins reveals a shared motif and predicts new potential quadruplex interactors. Molecules. 23(9):2341. doi: https://doi.org/10.3390/molecules23092341
- Chalamaiah M, Yu W, Wu J. 2018. Immunomodulatory and anticancer protein hydrolysates (peptides) from food proteins: A review. Food Chem. 245:205–222. doi: https://doi.org/10.1016/j.foodchem.2017.10.087
- Chandra H, Bishnoi P, Yadav A, Patni B, Mishra A, Nautiyal A. 2017. Antimicrobial resistance and the alternative resources with special emphasis on plant-based antimicrobials—a review. Plants. 6(2):16. doi: https://doi.org/10.3390/plants6020016
- Chatterjee S, Chatterjee S, Lad SJ, Phansalkar MS, Rupp R, Ganguli B, Kogler H. 1992. Mersacidin, a new antibiotic from Bacillus Fermentation, isolation, purification and chemical characterization. J Antibiot. 45(6):832–838. doi: https://doi.org/10.7164/antibiotics.45.832
- Chen WY, Chang HY, Lu JK, Huang YC, Harroun SG, Tseng YT, Chang HT. 2015. Self-assembly of antimicrobial peptides on gold nanodots: against multidrug-resistant bacteria and wound-healing application. Adv Funct Mater. 25(46):7189–7199. doi: https://doi.org/10.1002/adfm.201503248
- Chen W, Ding H, Feng P, Lin H, Chou K-C. 2016. iACP: a sequence-based tool for identifying anticancer peptides. Oncotarget. 7(13):16895.
- de la Fuente-núñez C, Silva ON, Lu TK, Franco OL. 2017. Antimicrobial peptides: role in human disease and potential as immunotherapies. Pharmacol Ther. 178:132–140. doi: https://doi.org/10.1016/j.pharmthera.2017.04.002
- Deslouches B, Di YP. 2017. Antimicrobial peptides with selective antitumor mechanisms: prospect for anticancer applications. Oncotarget. 8(28):46635. doi: https://doi.org/10.18632/oncotarget.16743
- E Greber K, Dawgul M. 2016. Antimicrobial peptides under clinical trials. Curr Top Med Chem. 17(5):620–628. doi: https://doi.org/10.2174/1568026616666160713143331
- Felgueiras HP, Amorim MTP. 2017. Functionalization of electrospun polymeric wound dressings with antimicrobial peptides. Colloids Surf B. 156:133–148. doi: https://doi.org/10.1016/j.colsurfb.2017.05.001
- Felício MR, Silva ON, Gonçalves S, Santos NC, Franco OL. 2017. Peptides with dual antimicrobial and anticancer activities. Front Chem. 5:5. doi: https://doi.org/10.3389/fchem.2017.00005
- Fisher RA. 1938. The statistical utilization of multiple measurements. Ann Eugen. 8(4):376–386. doi: https://doi.org/10.1111/j.1469-1809.1938.tb02189.x
- Gabere MN, Noble WS. 2017. Empirical comparison of web-based antimicrobial peptide prediction tools. Bioinformatics. 33(13):1921–1929. doi: https://doi.org/10.1093/bioinformatics/btx081
- Gadde U, Kim W, Oh S, Lillehoj HS. 2017. Alternatives to antibiotics for maximizing growth performance and feed efficiency in poultry: a review. Anim Health Res Rev. 18(1):26–45. doi: https://doi.org/10.1017/S1466252316000207
- Giuliani A, Pirri G, Bozzi A, Di Giulio A, Aschi M, Rinaldi A. 2008. Antimicrobial peptides: natural templates for synthetic membrane-active compounds. Cell Mol Life Sci. 65(16):2450–2460. doi: https://doi.org/10.1007/s00018-008-8188-x
- Gong Y, Andina D, Nahar S, Leroux J-C, Gauthier MA. 2017. Releasable and traceless PEGylation of arginine-rich antimicrobial peptides. Chem Sci. 8(5):4082–4086. doi: https://doi.org/10.1039/C7SC00770A
- Grönberg A, Mahlapuu M, Ståhle M, Whately-Smith C, Rollman O. 2014. Treatment with LL-37 is safe and effective in enhancing healing of hard-to-heal venous leg ulcers: a randomized, placebo-controlled clinical trial. Wound Repair Regen. 22(5):613–621. doi: https://doi.org/10.1111/wrr.12211
- Guidotti G, Brambilla L, Rossi D. 2017. Cell-penetrating peptides: from basic research to clinics. Trends Pharmacol Sci. 38(4):406–424. doi: https://doi.org/10.1016/j.tips.2017.01.003
- Han J, Cheng H, Wang B, Braun MS, Fan X, Bender M, Lindner T. 2017. A Polymer/peptide complex-based Sensor Array that Discriminates bacteria in Urine. Angew Chem Int Ed. 56(48):15246–15251. doi: https://doi.org/10.1002/anie.201706101
- Hancock RE, Haney EF, Gill EE. 2016. The immunology of host defence peptides: beyond antimicrobial activity. Nat Rev Immunol. 16(5):321–334. doi: https://doi.org/10.1038/nri.2016.29
- Haney EF, Hancock RE. 2013. Peptide design for antimicrobial and immunomodulatory applications. Pept Sci. 100(6):572–583. doi: https://doi.org/10.1002/bip.22250
- Haney EF, Mansour SC, Hancock REW. 2017. Antimicrobial peptides: an introduction. In: Hansen P., editor. Antimicrobial peptides. Methods in molecular biology. Vol. 1548. New York, NY: Humana Press.
- Hastie T, Tibshirani R, Wainwright M. 2015. Statistical learning with sparsity: the lasso and generalizations. London: CRC press.
- Hein KZ, Takahashi H, Tsumori T, Yasui Y, Nanjoh Y, Toga T, Wehkamp J. 2015. Disulphide-reduced psoriasin is a human apoptosis-inducing broad-spectrum fungicide. Proc Natl Acad Sci U S A. 112(42):13039–13044. doi: https://doi.org/10.1073/pnas.1511197112
- Hilchie AL, Wuerth K, Hancock RE. 2013. Immune modulation by multifaceted cationic host defense (antimicrobial) peptides. Nat Chem Biol. 9(12):761–768. doi: https://doi.org/10.1038/nchembio.1393
- Hoppenbrouwers T, Autar AS, Sultan AR, Abraham TE, van Cappellen WA, Houtsmuller AB, de Maat MP. 2017. In vitro induction of NETosis: Comprehensive live imaging comparison and systematic review. PloS one. 12(5):e0176472. doi: https://doi.org/10.1371/journal.pone.0176472
- Iwanaga S, Kawabata S-I. 1998. Evolution and phylogeny of defense molecules associated with innate immunity in horseshoe crab. Front Biosci. 3:D973–D984. doi: https://doi.org/10.2741/A337
- Järvå M, Lay FT, Phan TK, Humble C, Poon IK, Bleackley MR, Kvansakul M. 2018. X-ray structure of a carpet-like antimicrobial defensin–phospholipid membrane disruption complex. Nat Commun. 9(1):1962. doi: https://doi.org/10.1038/s41467-018-04434-y
- Jennings MC, Ator LE, Paniak TJ, Minbiole KP, Wuest WM. 2014. Biofilm-eradicating properties of Quaternary Ammonium Amphiphiles: Simple Mimics of antimicrobial peptides. ChemBioChem. 15(15):2211–2215. doi: https://doi.org/10.1002/cbic.201402254
- Jenssen H, Hamill P, Hancock RE. 2006. Peptide antimicrobial agents. Clin Microbiol Rev. 19(3):491–511. doi: https://doi.org/10.1128/CMR.00056-05
- Jia B, Raphenya AR, Alcock B, Waglechner N, Guo P, Tsang KK, Sharma AN. 2016. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. gkw1004:D566–D573.
- Kang H-K, Kim C, Seo CH, Park Y. 2017. The therapeutic applications of antimicrobial peptides (AMPs): a patent review. J Microbiol. 55(1):1–12. doi: https://doi.org/10.1007/s12275-017-6452-1
- Khater S, Gupta M, Agrawal P, Sain N, Prava J, Gupta P, Mohanty D. 2017. SBSPKSv2: structure-based sequence analysis of polyketide synthases and non-ribosomal peptide synthetases. Nucleic Acids Res. 45(W1):W72–W79. doi: https://doi.org/10.1093/nar/gkx344
- Krizsan A, Volke D, Weinert S, Sträter N, Knappe D, Hoffmann R. 2014. Insect-derived proline-rich antimicrobial peptides kill bacteria by Inhibiting bacterial protein translation at the 70 S Ribosome. Angew Chem Int Ed. 53(45):12236–12239. doi: https://doi.org/10.1002/anie.201407145
- Kumar P, Kizhakkedathu J, Straus S. 2018. Antimicrobial peptides: diversity, mechanism of action and strategies to improve the activity and biocompatibility in vivo. Biomolecules. 8(1):4. doi: https://doi.org/10.3390/biom8010004
- Kuroda K, Fukuda T, Krstic-Demonacos M, Demonacos C, Okumura K, Isogai H, Isogai E. 2017. miR-663a regulates growth of colon cancer cells, after administration of antimicrobial peptides, by targeting CXCR4-p21 pathway. BMC Cancer. 17(1):33. doi: https://doi.org/10.1186/s12885-016-3003-9
- Le C-F, Fang C-M, Sekaran SD. 2017. Intracellular targeting mechanisms by antimicrobial peptides. Antimicrob Agents Chemother. 61(4):e02340–e02316.
- Li J, Koh J-J, Liu S, Lakshminarayanan R, Verma CS, Beuerman RW. 2017. Membrane active antimicrobial peptides: translating mechanistic insights to design. Front Neurosci. 11:73.
- Liu S, Fan L, Sun J, Lao X, Zheng H. 2017. Computational resources and tools for antimicrobial peptides. J Pept Sci. 23(1):4–12. doi: https://doi.org/10.1002/psc.2947
- Liu B, Yang F, Huang D-S, Chou K-C. 2018. iPromoter-2L: a two-layer predictor for identifying promoters and their types by multi-window-based PseKNC. Bioinformatics. 34(1):33–40. doi: https://doi.org/10.1093/bioinformatics/btx579
- Liu Q, Yao S, Chen Y, Gao S, Yang Y, Deng J, Hu Y. 2017. Use of antimicrobial peptides as a feed additive for juvenile goats. Sci Rep. 7(1):12254. doi: https://doi.org/10.1038/s41598-017-12394-4
- Liu F, Zhang S, Li J, McClements DJ, Liu X. 2018. Recent development of lactoferrin-based vehicles for the delivery of bioactive compounds: Complexes, emulsions, and nanoparticles. Trends Food Sci Technol. 79:67–77. doi: https://doi.org/10.1016/j.tifs.2018.06.013
- Logashina YA, Solstad RG, Mineev KS, Korolkova YV, Mosharova IV, Dyachenko IA, Arseniev AS. 2017. New disulfide-stabilized fold provides sea anemone peptide to exhibit both antimicrobial and TRPA1 potentiating properties. Toxins (Basel). 9(5):154. doi: https://doi.org/10.3390/toxins9050154
- Lopes NA, Pinilla CMB, Brandelli A. 2017. Pectin and polygalacturonic acid-coated liposomes as novel delivery system for nisin: Preparation, characterization and release behavior. Food Hydrocolloids. 70:1–7. doi: https://doi.org/10.1016/j.foodhyd.2017.03.016
- Lu Y, Zhang T-F, Shi Y, Zhou H-W, Chen Q, Wei B-Y, Kang J. 2016. PFR peptide, one of the antimicrobial peptides identified from the derivatives of lactoferrin, induces necrosis in leukemia cells. Sci Rep. 6:20823. doi: https://doi.org/10.1038/srep20823
- Mai S, Mauger MT, Niu L-N, Barnes JB, Kao S, Bergeron BE, Tay FR. 2017. Potential applications of antimicrobial peptides and their mimics in combating caries and pulpal infections. Acta Biomater. 49:16–35. doi: https://doi.org/10.1016/j.actbio.2016.11.026
- Manavalan B, Shin TH, Kim MO, Lee G. 2018. AIPpred: sequence-based prediction of anti-inflammatory peptides using random forest. Front Pharmacol. 9:276. doi: https://doi.org/10.3389/fphar.2018.00276
- Mangoni ML, McDermott AM, Zasloff M. 2016. Antimicrobial peptides and wound healing: biological and therapeutic considerations. Exp Dermatol. 25(3):167–173. doi: https://doi.org/10.1111/exd.12929
- Martens E, Demain AL. 2017. The antibiotic resistance crisis, with a focus on the United States. J Antibiot. 70(5):520–526. doi: https://doi.org/10.1038/ja.2017.30
- Masuda M, Nakashima H, Ueda T, Naba H, Ikoma R, Otaka A, Murakami T. 1992. A novel anti-HIV synthetic peptide, T-22 ([Tyr5, 12, Lys7]-polyphemusin II). Biochem Biophys Res Commun. 189(2):845–850. doi: https://doi.org/10.1016/0006-291X(92)92280-B
- Mattick A, Hirsch A. 1947. Further observations on an inhibitory substance (nisin) from lactic streptococci. Lancet. 250:5–8. doi: https://doi.org/10.1016/S0140-6736(47)90004-4
- Meher PK, Sahu TK, Saini V, Rao AR. 2017. Predicting antimicrobial peptides with improved accuracy by incorporating the compositional, physico-chemical and structural features into Chou’s general PseAAC. Sci Rep. 7:42362. doi: https://doi.org/10.1038/srep42362
- Migoń D, Neubauer D, Kamysz W. 2018. Hydrocarbon stapled antimicrobial peptides. Protein J. 37(1):2–12. doi: https://doi.org/10.1007/s10930-018-9755-0
- Mishra B, Reiling S, Zarena D, Wang G. 2017. Host defense antimicrobial peptides as antibiotics: design and application strategies. Curr Opin Chem Biol. 38:87–96. doi: https://doi.org/10.1016/j.cbpa.2017.03.014
- Moravej H, Moravej Z, Yazdanparast M, Heiat M, Mirhosseini A, Moosazadeh Moghaddam M, Mirnejad R. 2018. Antimicrobial peptides: features, action, and their resistance mechanisms in bacteria. Microb Drug Resist. 24(6):747–767. doi: https://doi.org/10.1089/mdr.2017.0392
- Müller AT, Gabernet G, Hiss JA, Schneider G. 2017. modlAMP: Python for antimicrobial peptides. Bioinformatics. 33(17):2753–2755. doi: https://doi.org/10.1093/bioinformatics/btx285
- Nijnik A, Hancock R. 2009. Host defence peptides: antimicrobial and immunomodulatory activity and potential applications for tackling antibiotic-resistant infections. Emerg Health Threats J. 2(1):7078. doi: https://doi.org/10.3402/ehtj.v2i0.7078
- Nordström R, Malmsten M. 2017. Delivery systems for antimicrobial peptides. Adv Colloid Interface Sci. 242:17–34. doi: https://doi.org/10.1016/j.cis.2017.01.005
- O’Driscoll NH, Labovitiadi O, Cushnie TT, Matthews KH, Mercer DK, Lamb AJ. 2013. Production and evaluation of an antimicrobial peptide-containing wafer formulation for topical application. Curr Microbiol. 66(3):271–278. doi: https://doi.org/10.1007/s00284-012-0268-3
- Okada M, Natori S. 1983. Purification and characterization of an antibacterial protein from haemolymph of Sarcophaga peregrina (flesh-fly) larvae. Biochem J. 211(3):727–734. doi: https://doi.org/10.1042/bj2110727
- Pane K, Durante L, Crescenzi O, Cafaro V, Pizzo E, Varcamonti M, Notomista E. 2017. Antimicrobial potency of cationic antimicrobial peptides can be predicted from their amino acid composition: application to the detection of “cryptic” antimicrobial peptides. J Theor Biol. 419:254–265. doi: https://doi.org/10.1016/j.jtbi.2017.02.012
- Patel S, Akhtar N. 2017. Antimicrobial peptides (AMPs): The quintessential ‘offense and defense’molecules are more than antimicrobials. Biomed Pharmacother. 95:1276–1283. doi: https://doi.org/10.1016/j.biopha.2017.09.042
- Patterson-Delafield J, Szklarek D, Martinez R, Lehrer R. 1981. Microbicidal cationic proteins of rabbit alveolar macrophages: amino acid composition and functional attributes. Infect Immun. 31(2):723–731. doi: https://doi.org/10.1128/IAI.31.2.723-731.1981
- Poon IK, Baxter AA, Lay FT, Mills GD, Adda CG, Payne JA, Veneer PK. 2014. Phosphoinositide-mediated oligomerization of a defensin induces cell lysis. Elife. 3:e01808. doi: https://doi.org/10.7554/eLife.01808
- Porto W, Pires A, Franco O. 2017. Computational tools for exploring sequence databases as a resource for antimicrobial peptides. Biotechnol Adv. 35(3):337–349. doi: https://doi.org/10.1016/j.biotechadv.2017.02.001
- Radzishevsky IS, Rotem S, Bourdetsky D, Navon-Venezia S, Carmeli Y, Mor A. 2007. Improved antimicrobial peptides based on acyl-lysine oligomers. Nat Biotechnol. 25(6):657–659. doi: https://doi.org/10.1038/nbt1309
- Ramesh S, Govender T, Kruger HG, de la Torre BG, Albericio F. 2016. Short AntiMicrobial peptides (SAMPs) as a class of extraordinary promising therapeutic agents. J Pept Sci. 22(7):438–451. doi: https://doi.org/10.1002/psc.2894
- Riool M, de Breij A, Drijfhout JW, Nibbering PH, Zaat SA. 2017. Antimicrobial peptides in biomedical device manufacturing. Front Chem. 5:63. doi: https://doi.org/10.3389/fchem.2017.00063
- Roy S, Trinchieri G. 2017. Microbiota: a key orchestrator of cancer therapy. Nat Rev Cancer. 17(5):271–285. doi: https://doi.org/10.1038/nrc.2017.13
- Sah B, Vasiljevic T, McKechnie S, Donkor O. 2018. Antioxidative and antibacterial peptides derived from bovine milk proteins. Crit Rev Food Sci Nutr. 58(5):726–740. doi: https://doi.org/10.1080/10408398.2016.1217825
- Sani M-A, Carne S, Overall SA, Poulhazan A, Separovic F. 2017. One pathogen two stones: are Australian tree frog antimicrobial peptides synergistic against human pathogens? Eur Biophys J. 46(7):639–646. doi: https://doi.org/10.1007/s00249-017-1215-9
- Sato K. 2018. Structure, content, and bioactivity of food-derived peptides in the body. J Agric Food Chem. 66(12):3082–3085. doi: https://doi.org/10.1021/acs.jafc.8b00390
- Scheinpflug K, Wenzel M, Krylova O, Bandow JE, Dathe M, Strahl H. 2017. Antimicrobial peptide cWFW kills by combining lipid phase separation with autolysis. Sci Rep. 7:44332. doi: https://doi.org/10.1038/srep44332
- Schnabel R. 2017. Squeezed states of light and their applications in laser interferometers. Phys Rep. 684:1–51. doi: https://doi.org/10.1016/j.physrep.2017.04.001
- Shirako S, Kojima Y, Tomari N, Nakamura Y, Matsumura Y, Ikeda K, Sato K. 2019. Pyroglutamyl leucine, a peptide in fermented foods, attenuates dysbiosis by increasing host antimicrobial peptide. NPJ Science of Food. 3(1):1–9. doi: https://doi.org/10.1038/s41538-019-0050-z
- Stach M, Siriwardena TN, Köhler T, Van Delden C, Darbre T, Reymond JL. 2014. Combining Topology and sequence design for the discovery of potent antimicrobial peptide Dendrimers against Multidrug-resistant Pseudomonas aeruginosa. Angew Chem Int Ed. 53(47):12827–12831. doi: https://doi.org/10.1002/anie.201409270
- Thamri A, Létourneau M, Djoboulian A, Chatenet D, Déziel E, Castonguay A, Perreault J. 2017. Peptide modification results in the formation of a dimer with a 60-fold enhanced antimicrobial activity. PloS one. 12(3):e0173783. doi: https://doi.org/10.1371/journal.pone.0173783
- Tincho M, Gabere M, Pretorius A. 2016. In silico identification and molecular validation of putative antimicrobial peptides for HIV therapy. J AIDS Clinical Res. 7(9):1–11. doi: https://doi.org/10.4172/2155-6113.1000606
- Tincu JA, Taylor SW. 2004. Antimicrobial peptides from marine invertebrates. Antimicrob Agents Chemother. 48(10):3645–3654. doi: https://doi.org/10.1128/AAC.48.10.3645-3654.2004
- Tucker AT, Leonard SP, DuBois CD, Knauf GA, Cunningham AL, Wilke CO, Davies BW. 2018. Discovery of next-generation antimicrobials through bacterial self-screening of surface-displayed peptide libraries. Cell. 172(3):618–628.e13. doi: https://doi.org/10.1016/j.cell.2017.12.009
- Vannini A, Pinatel E, Costantini PE, Pelliciari S, Roncarati D, Puccio S, Danielli A. 2017. Comprehensive mapping of the Helicobacter pylori NikR regulon provides new insights in bacterial nickel responses. Sci Rep. 7:45458. doi: https://doi.org/10.1038/srep45458
- Wang G. 2017. Antimicrobial peptides: discovery, design and novel therapeutic strategies. Cabi.
- Wang Q, Yang S, Liu J, Terecskei K, Ábrahám E, Gombár A, Wang T. 2017. Host-secreted antimicrobial peptide enforces symbiotic selectivity in Medicago truncatula. Proc Natl Acad Sci U S A. 114(26):6854–6859.
- Wei DS, van der Sar T, Sanchez-Yamagishi JD, Watanabe K, Taniguchi T, Jarillo-Herrero P, Yacoby A. 2017. Mach-Zehnder interferometry using spin-and valley-polarized quantum Hall edge states in graphene. Sci Adv. 3(8):e1700600. doi: https://doi.org/10.1126/sciadv.1700600
- Wiig ME, Dahlin LB, Friden J, Hagberg L, Larsen SE, Wiklund K, Mahlapuu M. 2014. PXL01 in sodium hyaluronate for improvement of hand recovery after flexor tendon repair surgery: randomized controlled trial. PloS one. 9(10):e110735. doi: https://doi.org/10.1371/journal.pone.0110735
- Wong D, Nielsen TB, Bonomo RA, Pantapalangkoor P, Luna B, Spellberg B. 2017. Clinical and pathophysiological overview of Acinetobacter infections: a century of challenges. Clin Microbiol Rev. 30(1):409–447.
- Wu T, Huang J, Jiang Y, Hu Y, Ye X, Liu D, Chen J. 2018. Formation of hydrogels based on chitosan/alginate for the delivery of lysozyme and their antibacterial activity. Food Chem. 240:361–369. doi: https://doi.org/10.1016/j.foodchem.2017.07.052
- Xiong X, Yang H, Li L, Wang Y, Huang R, Li F, Qiu W. 2014. Effects of antimicrobial peptides in nursery diets on growth performance of pigs reared on five different farms. Livest Sci. 167:206–210. doi: https://doi.org/10.1016/j.livsci.2014.04.024
- Yamamoto Y, Mizushige T, Mori Y, Shimmura Y, Fukutomi R, Kanamoto R, Ohinata K. 2015. Antidepressant-like effect of food-derived pyroglutamyl peptides in mice. Neuropeptides. 51:25–29. doi: https://doi.org/10.1016/j.npep.2015.04.002
- Yoon JH, Ingale SL, Kim JS, Kim KH, Lohakare J, Park YK, Chae BJ. 2013. Effects of dietary supplementation with antimicrobial peptide-P5 on growth performance, apparent total tract digestibility, faecal and intestinal microflora and intestinal morphology of weanling pigs. J Sci Food Agric. 93(3):587–592. doi: https://doi.org/10.1002/jsfa.5840
- Zasloff M. 1987. Magainins, a class of antimicrobial peptides from Xenopus skin: isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proc Natl Acad Sci U S A. 84(15):5449–5453. doi: https://doi.org/10.1073/pnas.84.15.5449
- Zeth K, Sancho-Vaello E. 2017. The human antimicrobial peptides dermcidin and LL-37 show novel distinct pathways in membrane interactions. Front Chem. 5:86. doi: https://doi.org/10.3389/fchem.2017.00086
- Zhang Y, Algburi A, Wang N, Kholodovych V, Oh DO, Chikindas M, Uhrich KE. 2017. Self-assembled cationic amphiphiles as antimicrobial peptides mimics: role of hydrophobicity, linkage type, and assembly state. Nanomed Nanotechnol Biol Med. 13(2):343–352. doi: https://doi.org/10.1016/j.nano.2016.07.018
- Zhu M, Liu P, Niu Z-W. 2017. A perspective on general direction and challenges facing antimicrobial peptides. Chin Chem Lett. 28(4):703–708. doi: https://doi.org/10.1016/j.cclet.2016.10.001
- Zorzi A, Deyle K, Heinis C. 2017. Cyclic peptide therapeutics: past, present and future. Curr Opin Chem Biol. 38:24–29. doi: https://doi.org/10.1016/j.cbpa.2017.02.006