Abstract
Abnormal long noncoding RNAs (lncRNAs) participate in the generation and progression of malignancies. We aimed to determine the expression and clinical importance of lncRNA ENST433110 in thyroid cancer (TC). We thus investigated the role of ENST433110 in TC cell invasion, migration, and proliferation. ENST433110 expression was assessed using RT-PCR, cell migration was evaluated using a Transwell assay, and cell apoptosis was analyzed by flow cytometry. Our findings revealed that ENST433110 expression was dramatically reduced in TC sample tissues and TC cells (B-CPAP and TPC-1). Moreover, we showed that lncRNA ENST433110 overexpression inhibited cell migration and proliferation in TC cells. Furthermore, lncRNA ENST433110 overexpression enhanced cell apoptosis and induced a noticeable reduction in Bcl-2 expression and an increase in BAX expression in both TPC-1 and B-CPAP cells compared with that in control groups. In summary, our results indicate that lncRNA ENST433110 is important in the development of thyroid carcinoma and could thus be a promising target in thyroid carcinoma treatment.
Introduction
Thyroid carcinoma (TC) is the most prevalent cancer of this endocrinal gland, and its worldwide incidence continues to increase (Kwon et al. Citation2018; Weeks et al. Citation2018; Xhaard et al. Citation2018). TC affects follicular and parafollicular cells and connective tissues, and can be divided into papillary thyroid cancer (PTC), follicular TC (FTC), medullary TC (MTC), anaplastic TC (ATC), primary thyroid lymphomas, and primary thyroid sarcomas (Takahashi et al. Citation2017; Giusti et al. Citation2018). PTC is the predominant type of TC, representing over 80% of all thyroid malignancies (Stojanovic et al. Citation2017; Wang et al. Citation2018). Despite its inertness, a favorable clinical outcome, and low mortality rate, some clinicopathological features of TC can be lethal, including large primary cancer and metastasis (Beltrami et al. Citation2017; Kwon et al. Citation2017; Tavarelli et al. Citation2017). Thus, a better understanding of TC etiology is essential if TC treatments are to be improved.
Long noncoding RNAs (lncRNAs) are endogenous transcripts that consist of at least 200 nucleotides (Zhou et al. Citation2018) that are present in the transcriptome of mammals (Balcin et al. Citation2018). Previous research has focused mainly on gene regulation by lncRNAs at the transcriptional or post-transcriptional level (Zhang et al. Citation2018). As lncRNAs do not encode proteins, they were once considered to be ‘noise’ in the genome. However, several studies have since suggested that lncRNAs participate in various processes such as viability, differentiation, proliferation, and chromatin remodeling (Li et al. Citation2017; Shi et al. Citation2017). It has been shown that lncRNAs are involved in the pathophysiology of many disorders and that alterations in lncRNA expression induce diverse illnesses and malignancies(Liao et al. Citation2018). For example, lncRNA SAMMSON upregulation leads to melanoma, which is inhibited by silencing of lncRNA SAMMSON (Citation2016; Goding Citation2016). Overexpression of lncRNA SChLAP1 has been linked to a poor prognosis for prostate cancer (Mehra et al. Citation2014). Moreover, lncRNA SChLAP1 is necessary for cell invasion and metastasis in prostate cancer. These findings suggest lncRNA play important roles in cancer. Therefore, we first detected some lncRNA expression in thyroid cancer cell lines and found that lncRNA ENST433110 is significantly decreased in thyroid cancer cell lines. Thus, the present study examined lncRNA ENST433110 expression in TC cells and evaluated its influence on cell migration and proliferation.
Materials and methods
Cell lines and patient specimens
Human thyroid cancer cell lines B-CPAP and TPC-1, and immortalized follicular epithelial thyroid cells (Nthy-ori 3-1) were obtained from the Chinese Academy of Sciences (Shanghai, China) and cultured in RPMI-1640 medium (Thermo Fisher Scientific, Waltham, MA, USA) containing 10% fetal bovine serum (Thermo Fisher Scientific) in 5% CO2 at 37°C.
A total of seventeen tissue samples from patients with TC clinically and histologically diagnosed at the Third Xiangya Hospital of Central South University between 2011 and 2015 were obtained with previous patient consent and approval from the Institutional Research Ethics Committee. Seven normal tissue samples were obtained from individuals who suffered from simple goiter but were free of any detectable pathological conditions. All the patients signed written informed consent. This study was approved by the Ethical Committee of the Third Xiangya Hospital of Central South University.
Transfection
A lentivirus-lncRNA ENST433110 plasmid (pLv-ENST433110, 1 × 108 Fu/mL) was used to overexpress lncRNA ENST433110 in TPC-1 and B-CPAP cells. A lentivirus carrying an empty vector (pLv-NC; Invitrogen, Carlsbad, CA, USA) was used as a negative control. The sequence of the lncRNA ENST433110 is as follows:
TTTTGAAAACTTCTGGGTTTCTGAAATGAA GGTTCTTTGCAGAGGACAATCTTGGAAAAAA AGATGAAGTTTGATTTCCCTTTTTTCTTTGC ACTATAGGGGTGTTCACTGAGGTTGCAGAGC ACCAACCATGTCTGCAGAGGAGTTCACACCT CACTGGTGTTCATTGTATTGCTTCACCGTTG AGAGCCTGAATGTGAACCTTGTTGTATTAAG GTGATATTCTTGTCCTTCATGTGTTAGAATA TCTATTTGCTTTGAAGAGAGAGTCAGACTCC CTGCCTTTTGGGACATGGCTTTGAGATCTGC CTTTCAGTAAGTAAGACGCTATCCTCAAATT GAGCTGGGCTACCTCTCTACATGGGAAAACC TAAGTACTTCACAGAAAGCTCGGTGACGGTT TGAGTTTCCCAGAATGCCGCACTCATCCCCA AGGATAAGTCACGTCTTTGGGGGATGATGGG GCTTCGACACTGGACACGTGGACACTGGACC GGAAGTTAGAATCCAGTCTCCCAGCCCTCAA CCCCATGCTCCTTTCCATGGCGAAGGAGGAG AAGCTCGCCGCCACCCCACTGGCCTGCTCTG CTGAGGTTGTGGAGCACCGGGAAGCAGGGGC CGAGCCTTCCCCGCTCTGTGCCTGGGGGCTC TGGGCACTGTCTACACCTGGGAGGTGGTCAG CATATGTGCGTGGGAACTTGCTGGAAGGAAG GAATGACCCTTCTAGCCATGGAACGGTGGTC ATCCGTGAGCCACGGCCTCAGGAGGTGGGTT CCCTTAAGGCTCCGGCTCTGGGGGCAGCGCC CCCTGGGAGCGTCCATTTTTAAGCCTGTGAA GAAAGGGCTGCTGACGTTTGGGGCCCCCGCT ATGAGCATAGCCTCCACTCCTCCCCAATCCC TGTCCGTGCCTCTACATACCTCTGTGGCATT TGCAAACCCTTTTCTTAGCAGGTGCTCCCAT CTGTGTTTGCCCTTCTCTCCTCCCGTTTCCA TGGAGAGGCTGGCACCAGGACGCGTGACTGA CAGTGCTGTCTCCCCCAGCAGTCCCCTCTGA GTCTGTCGGGTGTGGGCTCCGGGCTCCACCC TGCTTCTTCCTTAGCCCGCTCTCTGCCCCTC CCACTGCCCAGGCTGCCGGGGCTTCCAGCTC CCGTGGTGCAAACGTGGCCTTTGCTGGCGGC AGGATGCGGTTGCGTCAGGAGTAAGCAGAGC TTTTGTGACCAAAATCAACCCCGGGGCATCT CAGGCTCTAGCGCTGGTGGGCGTGTGAGATC ATTTCAGCCCAACCCACATCCCAGGCTTGAA TCCCATTACCAGGGGCAACCGATCCTTCTTG TCCTTTCCCAGATTGGTGATGGATGTCCACT GTGTCTCCATGCAGCTGGCCACTCTGTGGAC AGCTGTTTGTGACTCAGCTCCTCCCAGCCTG AGGGCATCTGGGCCCTGCAACTCCATCCTTT GCTCCCCTTTCTTCCCGCGGGGACCCTTTCA TCTCCCTGGCAGGACCCTGTCTCCACCCGAG GGCACGTTCTCTGACCTCTTCTGCGGAGTAC CCTGGCTTGATGACGCTTCCTGTTCCTTCCC AGGGTTGTGCTGGGGGCATCAGTCTTTGAAC AGTCCCTCCCTGAGGGCTGCCGTGTCCCCTC AGTGCCCAGGCACATCAAGCACTGCTACAAA CCCAGACATTCACAGGGACCCCTCCACCTGA AGTCCACCATAAGGTAGAAGCTTCCGGAAGG GGTGATGCAAAGGGGTCAAACATTTAGACTA CGCCTCAGAGCTGCATGATTCCTTCCTCTAA CTGTGAAGCAAATACAGCGATTTCTCTGGGT CTGCCATCGAGGCCTTCTTCCCTCTGCTCTC ACCTCAGGTGCCTTCCTAATGTGCAGGTGAC GATTGCCTCTGAGCTCAGGCATGCAGCTTCT GGGATGCACTCTCCTTTCACGGCAAAGCATT CTGCATAGGAAAAACCGTCTCTGCGTCTCCA AGCTGTGGCCTAGCCCTTCATTCATCAAATG GGATACAACTTTCATTTCTTATTAAAATCTGGAA.
RNA isolation and RT-PCR
Total RNA was extracted from cells using TRIzol (Life Technologies) and was purified using an RNeasy Mini kit (Qiagen, Hilden, Germany). A Superscript III kit (Life Technologies) was used for reverse transcription (RT), and the resulting cDNA was used for qRT-PCR. Primers were: lncRNA ENST433110 F: 5′-ACCACTGCACTACCTGACTC-3′; R: 5′-CCGCAGGGGGTGGCCATGAA-3′. ROR F: CTCAGTGGGGAAGACTCCAG, R: AGGAAGCCTGAGAGTTGGC. LINC00961 F: CTGTTCTGG ATGGGAGCGAA; R: ACAGTCACCACGAAC AGCAC. Transcription was quantified and evaluated using an SYBR Green PCR Supermix kit (Bio-Rad Laboratories, Hercules, CA, USA). All procedures were performed three times for each specimen, with a minimum of three independent runs. Results were evaluated using Real-Time StatMiner software (Integromics, Madrid, Spain).
Cell survival assay
Cell survival was evaluated using an MTT assay. Cells were plated onto 96-well plates (5 × 103 cells/well) and cultured at 37°C in 5% CO2. The MTT assay was carried out 48 h after transfection, and the cells were incubated for 4 h at 37°C. A microplate reader (Thermo Fisher Scientific) was used to measure absorbance at 570 nm. All procedures were performed three times.
Cell cycle assay
Cells were starved for 12 h for synchronization followed by restimulation with 10% fetal bovine serum for 24 h. Cells were fixed with 75% ethanol and treated with the Cell Cycle Detection Kit (BD Biosciences, Bedford, MA, USA) according to the manufacturer’s instructions. Cells were then sorted by a FACS Calibur flow cytometer (BD Biosciences, San Jose, CA, USA). The cell phase distribution was analyzed with the FlowJo software (Treestar Inc., USA).
Apoptosis assay
Cell apoptosis was evaluated using an annexin V-FITC/PI apoptosis detection kit (Shanghai Kaifeng Biotechnology, Shanghai, China). Annexin V (100 µl) was added to cells and, after a 15-min incubation at room temperature, 4 µl of PI diluted at 1:10 in annexin V binding buffer was added to each well and incubated for a further 15 min. Flow cytometry (BD Biosciences) was used to evaluate cell apoptosis.
Cell migration assay
Cell migration was evaluated using a Transwell assay. Cells were cultured in DMEM containing 1 μg/mL mitomycin C without serum to create a cell suspension. Cells (5 × 105/well) were seeded onto polycarbonate Transwell filters in a 24-well plate (Millipore, Bedford, MA, USA). DMEM containing 10% serum was added to the lower well. After 48 h, cells in the bottom chamber were mixed, stained, and quantified. The remaining cells in the top chamber were eliminated.
Cell invasion test
Twenty-four-well transwell chambers with 8-µm pore size polycarbonate membranes were used for the invasion test. Transwell chambers initially coated with Matrigel were seeded with cells. Cells suspended in 200 µL of DMEM without serum were seeded on the top chamber, while those in 8 µL of DMEM with 10% FBS were seeded on the bottom chamber. Cells that did not undergo invasion were eliminated from top chamber using a cotton swab after 24 h of incubation, and those that invaded the bottom were quantified in 6 random fields for every well.
Western blot analysis
Lysates were obtained by treating cells with a lysis buffer (Beyotime, China). A Bradford assay (Bio-Rad Laboratories) was used for protein quantification. Samples were subjected to SDS-PAGE using 8–15% Tris-HCl polyacrylamide gels (Bio-Rad Laboratories) and then transferred onto a PVDF membrane (Millipore). Membranes were blocked and then incubated with specific primary antibodies diluted in tris-buffered saline and Tween-20 overnight at 4°C. The primary antibodies used were anti-β-actin, anti-Bcl-2, and anti-BAX (Cell Signaling Technology, Beverly, MA, USA). After washing in tris-buffered saline and Tween-20, membranes were incubated with secondary antibodies. Bands were detected using ECL Plus detection reagent (Pierce, Rockford, IL, USA) and evaluated using an Omega 16ic chemiluminescence imaging system (Ultra-Lum, Claremont, CA, USA).
Statistical analysis
Results are represented as mean ± SEM. Two-tailed, unequal-variance Student’s t-test and ANOVA before Tukey’s post hoc analysis were used to determine differences among groups. P < 0.05 was regarded as significant.
Results
LncRNA ENST433110 expression is decreased in TC tissue and TC cells
To explore the role of lncRNA ENST433110 in TC, we examined some lncRNA expression in TC tissue and TC cells, including B-CPAP, TPC-1, and Nthy-ori 3-1 cells. The expression of lncRNA ENST433110 was significantly lower in TC cells than in normal cells (Figure A-1E). In addition, the expression of lncRNA ENST433110 was dramatically reduced in TC tissue samples (Figure C). These data suggest that lncRNA ENST433110 participates in TC development.
Figure 1. LncRNA ENST433110 expression is decreased in TC cells and tissues. A, RT-PCR was used to evaluate lncRNA ENST433110 expression. B, Results are represented as the mean ± SEM of three independent experiments. **P < 0.01 vs. Nthy-ori 3-1. C, lncRNA ENST433110 expression was detected by quantitative real-time RT-PCR in TC and in normal tissues (normal). D-E, lncRNA-ROR expression and lncRNA00961 expression were detected by quantitative real-time RT-PCR in TC cells. Data are represented as the mean ± SEM, n = 7, **P < 0.01 vs. normal group.
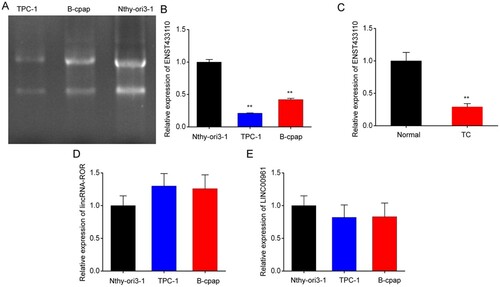
Overexpression of lncRNA ENST433110 inhibits TC cell proliferation
To determine the role of lncRNA ENST433110 in TC cell proliferation, TPC-1 and B-CPAP cells were transfected with lncRNA ENST433110, and RT-PCR was used to evaluate lncRNA ENST433110 expression. The results showed that lncRNA ENST433110 expression was increased in both TPC-1 (Figure A) and B-CPAP cells (Figure C) after transfection compared with that in controls transfected with the empty vector. Furthermore, MTT assay showed that lncRNA ENST433110 overexpression significantly decreased the proliferation rate of TPC-1 and B-CPAP cells (Figure B and D). In addition, the percentages of TPC-1 and B-CPAP cells in S phase were found to be significantly lower in the lncRNA ENST433110 overexpression group than in the empty vector group by flow cytometry (Figure E and 2F). These results suggest that lncRNA ENST433110 suppresses the proliferation of TC cells.
Figure 2. Overexpression of lncRNA ENST433110 suppresses TC cell proliferation. A and C, Expression of lncRNA ENST433110 in TPC-1 (A) and B-CPAP (C) cells after transient transfection with a lentivirus with the lncRNA ENST433110 plasmid (pLv-ENST433110) or lentivirus with an empty vector (pLv-NC). B and D, MTT assay to evaluate the effect of lncRNA ENST433110 overexpression on TPC-1 (B) and B-CPAP (D) cell proliferation. E and F, Cell cycle phase distribution was analyzed by flow cytometry in TPC-1 (E) and B-CPAP (F) cells. Data are represented as the mean ± SEM of three independent experiments. *P < 0.05, **P < 0.01 vs. pLv-NC.
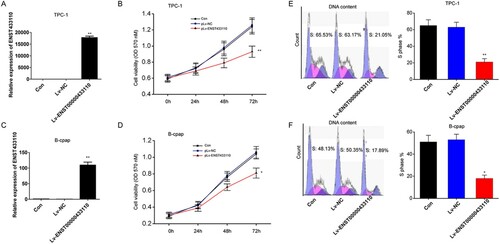
Overexpression of lncRNA ENST433110 inhibits TC cell migration
To determine the influence of lncRNA ENST433110 on TC cell migration, we performed Transwell assays. Our results showed that overexpression of lncRNA ENST433110 significantly inhibited the migration of TPC-1 (Figure A and 3B) and B-CPAP (Figure C) cells. Thus, lncRNA ENST433110 may play a role in the migration and proliferation of TC cells.
Figure 3. Overexpression of lncRNA ENST433110 suppresses TC cell migration. A–C, (A) Transwell assay was used to evaluate the effect of lncRNA ENST433110 overexpression on TPC-1 (B) and B-CPAP (C) cell migration. Representative images are shown. Scale bars = 100 µm. Data are represented as the mean ± SEM of three independent experiments. **P < 0.01 vs. pLv-NC.
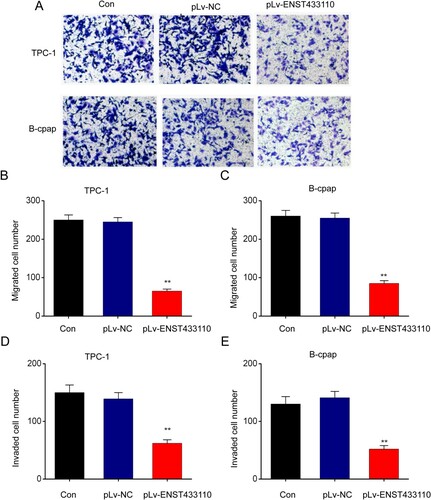
Overexpression of lncRNA ENST433110 enhances TC cell apoptosis
Next, to examine the role of lncRNA ENST433110 in PTC cell apoptosis, we used flow cytometry to evaluate Annexin V-FITC/PI double staining. Figure shows that cell apoptosis was increased in the pLv-ENST433110 group compared with the pLv-NC control group. These results suggest that lncRNA ENST433110 enhances cell apoptosis in TC cells.
Figure 4. Overexpression of lncRNA ENST433110 enhances TC cell apoptosis. A–D, Flow cytometry was used to determine the effect of lncRNA ENST433110 overexpression on TPC-1 (A, B) and B-CPAP (C, D) migration. Representative images are shown. Scale bars = 100 µm. Data are represented as the mean ± SEM of three independent experiments. **P < 0.01 vs. pLv-NC.
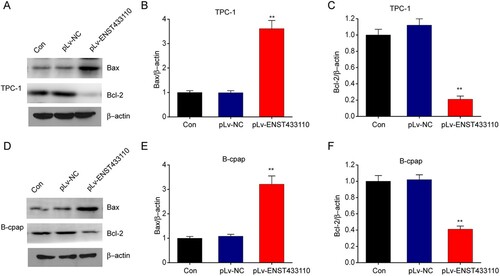
Overexpression of lncRNA ENST433110 regulates Bcl-2 and BAX expression in TC cells
The relationship between lncRNA ENST433110 and the expression of Bcl-2 and BAX was explored by using western blotting. Our results revealed that lncRNA ENST433110 overexpression noticeably reduced Bcl-2 expression and increased BAX expression in both TPC-1 (Figure A-5C) and B-CPAP (Figure D-5F) cells compared with the control groups. These results show that the expression of cell apoptosis-related proteins is enhanced by lncRNA ENST433110 in TC cells.
Figure 5. Overexpression of lncRNA ENST433110 regulates Bcl-2 and BAX expression in TC cells. A–F, Representative immunoblots and quantification of Bcl-2 and BAX expression in TPC-1 (A–C) and B-CPAP (D–F) cells. Data are represented as the mean ± SEM of three independent experiments. **P < 0.01 vs. pLv-NC.
Discussion
The major finding of the current study is that lncRNA ENST433110 expression was downregulated in TC cells compared with normal thyroid cells. Furthermore, lncRNA ENST433110 overexpression induced significant inhibition of cell migration and proliferation, while enhancing cell apoptosis. These findings suggest that lncRNA ENST433110 is a potential target for the premalignancy detection of TC.
Some patients with TC undergo surgery to remove the tumor, whereas a larger number of patients receive treatment with radioactive iodine to eliminate malignant cells (2002). However, developing innovative strategies is necessary to prevent an increase in the incidence of TC, as well as to improve the clinical outcome (Staunton and Skeet Citation1979; Ladurner et al. Citation1984; Liu et al. Citation2017). The molecular etiology of TC has attracted much attention recently (Williard et al. Citation1992). As a result, it has become increasingly desirable to discover biomarkers that can predict TC in patients, especially those with metastasis. Previous research has shown that various lncRNAs participate in diverse malignancies (Chen et al. Citation2018; Zhai and Xu Citation2018; Zhang et al. Citation2018), but an understanding of the role of lncRNA ENST433110 in TC is lacking. In this study, we explored the role and expression of lncRNA ENST433110 in the development of TC. Our findings revealed that lncRNA ENST433110 expression was downregulated in TC cells, and we identified a role for lncRNA ENST433110 in PC cell migration and proliferation. We found that lncRNA ENST433110 overexpression remarkably reduced TC cell migration and proliferation, indicating that restoration of the lncRNA ENST433110 physiological level could be an innovative approach for the treatment of TC. Previous studies have shown that lncRNAs modulate migration and proliferation inPTC cells (Huang et al. Citation2017; Wang et al. Citation2017; Ding et al. Citation2018; Zhang et al. Citation2018); however, the current study is the first to report an essential role for lncRNA in TC and demonstrate its participation in malignancy. The exact mechanism underlying the promotion of migration and proliferation by ENST433110 still requires further investigation.
In addition, we showed that lncRNA ENST433110 participated in TC cell apoptosis. Overexpression of lncRNA ENST433110 induced a remarkable increase in the number of dead cells. Located in the cytosol, BAX promote cell apoptosis and Bcl-2 suppress it. Bcl-2 relocates to the surface of mitochondria to regulate cell apoptosis. Thus, the ratio of BAX to Bcl-2 is a crucial factor in the regulation of cell apoptosis (Hao et al. Citation2003). We found that ENST433110 overexpression reduced Bcl-2 expression and promoted BAX expression, which is crucial for the promotion of cell apoptosis.
Based on these results, we will continue to further study whether overexpression of the lncRNA could sensitize the two tumor cell lines to therapeutic agents (radiation or a type of chemotherapy) in vivo and in vitro to further validate the relevance of this work in clinical practice. For example, suppression of lncRNA SChLAP1 inhibits cell invasion and metastasis of prostate cancer.
In summary, we showed that lncRNA ENST433110 overexpression can suppress migration and proliferation and enhance cell apoptosis in TC cells in vitro. This work could provide an innovative strategy in which a network of ENST433110 activity is constructed with regard to TC. lncRNA ENST433110 could be a promising target for the premalignant detection of TC.
Data availability statement
Due to the nature of this research, participants of this study did not agree for their data to be shared publicly, so supporting data is not available.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 2016. SAMMSON long noncoding RNA Is essential for melanoma cell viability. Cancer Discov.
- Balcin O, Ak Aksoy S, Tunca B, Kaya E, Egeli U, Tezcan G, Ugras N, Cecener G, Isik O, Dundar HZ, Yerci O. 2018. Overexpression of the long noncoding RNA homeoboxa transcript at the distal tip predicts poor prognosis in a KRAS-independent manner in periampullary Region Tumors. Pancreas. 47(2):213–220. doi: https://doi.org/10.1097/MPA.0000000000000984
- Beltrami CM, Dos Reis MB, Barros-Filho MC, Marchi FA, Kuasne H, Pinto CAL, Ambatipudi S, Herceg Z, Kowalski LP, Rogatto SR. 2017. Integrated data analysis reveals potential drivers and pathways disrupted by DNA methylation in papillary thyroid carcinomas. Clin Epigenetics. 9:45. doi: https://doi.org/10.1186/s13148-017-0346-2
- Chen JF, Wu P, Xia R, Yang J, Huo XY, Gu DY, Tang CJ, De W, Yang F. 2018. STAT3-induced lncRNA HAGLROS overexpression contributes to the malignant progression of gastric cancer cells via mTOR signal-mediated inhibition of autophagy. Mol Cancer. 17(1):6. doi: https://doi.org/10.1186/s12943-017-0756-y
- Ding J, Wang F, Xiang T, Qiao M. 2018. Expression and function of long noncoding RNA NONHSAT129183 in papillary thyroid cancer. Oncol Res. 26(7):1047–1053. doi: https://doi.org/10.3727/096504018X15152037713570
- Giusti M, Mittica M, Comite P, Campana C, Gay S, Mussap M. 2018. Anti-mullerian hormone in pre-menopausal females after ablative radioiodine treatment for differentiated thyroid cancer. Endocrine. 60(3):516–523. doi: https://doi.org/10.1007/s12020-017-1510-3
- Goding CR. 2016. Targeting the lncRNA SAMMSON reveals metabolic vulnerability in melanoma. Cancer Cell. 29(5):619–621. doi: https://doi.org/10.1016/j.ccell.2016.04.010
- Hao XS, Hao JH, Liu FT, Newland AC, Jia L. 2003. Potential mechanisms of leukemia cell resistance to TRAIL-induced apopotosis. Apoptosis. 8(6):601–607. doi: https://doi.org/10.1023/A:1026131425204
- Huang JK, Ma L, Song WH, Lu BY, Huang YB, Dong HM, Ma XK, Zhu ZZ, Zhou R. 2017. LncRNA-MALAT1 promotes angiogenesis of thyroid cancer by modulating tumor-associated macrophage FGF2 protein secretion. J Cell Biochem. 118(12):4821–4830. doi: https://doi.org/10.1002/jcb.26153
- Kwon H, Jeon MJ, Yoon JH, Hong SJ, Lee JH, Kim TY, Shong YK, Kim WB, Kim WG, Song DE. 2017. Preoperative clinicopathological characteristics of patients with solitary encapsulated follicular variants of papillary thyroid carcinomas. J Surg Oncol. 116(6):746–755. doi: https://doi.org/10.1002/jso.24700
- Kwon MR, Shin JH, Hahn SY, Oh YL, Kwak JY, Lee E, Lim Y. 2018. Histogram analysis of greyscale sonograms to differentiate between the subtypes of follicular variant of papillary thyroid cancer. Clin Radiol. 73(6):591 e1-591 e7. doi: https://doi.org/10.1016/j.crad.2017.12.008
- Ladurner D, Seeber G, Hofstadter F. 1984. [Papillary thyroid cancer--prognosis and prognostic factors]. Langenbecks Arch Chir. 363(1):43–55. doi: https://doi.org/10.1007/BF01255776
- Li Q, Pan X, Wang X, Jiao X, Zheng J, Li Z, Huo Y. 2017. Long noncoding RNA MALAT1 promotes cell proliferation through suppressing miR-205 and promoting SMAD4 expression in osteosarcoma. Oncotarget. 8(63):106648–106660. doi: https://doi.org/10.18632/oncotarget.20678
- Liao X, Chen J, Liu Y, He A, Wu J, Cheng J, Zhang X, Lv Z, Wang F, Mei H. 2018. Knockdown of long noncoding RNA FGFR3- AS1 induces cell proliferation inhibition, apoptosis and motility reduction in bladder cancer. Cancer Biomark. 21(2):277–285. doi: https://doi.org/10.3233/CBM-170354
- Liu N, Zhou Q, Qi YH, Wang H, Yang L, Fan QY. 2017. Effects of long non-coding RNA H19 and microRNA let7a expression on thyroid cancer prognosis. Exp Mol Pathol. 103(1):71–77. doi: https://doi.org/10.1016/j.yexmp.2017.06.004
- Mehra R, Shi Y, Udager AM, Prensner JR, Sahu A, Iyer MK, Siddiqui J, Cao X, Wei J, Jiang H, et al. 2014. A novel RNA in situ hybridization assay for the long noncoding RNA SChLAP1 predicts poor clinical outcome after radical prostatectomy in clinically localized prostate cancer. Neoplasia. 16(12):1121–1127. doi: https://doi.org/10.1016/j.neo.2014.11.006
- Shi J, Wang YJ, Sun CR, Qin B, Zhang Y, Chen G. 2017. Long noncoding RNA lncHERG promotes cell proliferation, migration and invasion in glioblastoma. Oncotarget. 8(64):108031–108041. doi: https://doi.org/10.18632/oncotarget.22446
- Staunton MD, Skeet RG. 1979. Thyroid cancer: prognosis in 469 patients. Br J Surg. 66(9):643–647. doi: https://doi.org/10.1002/bjs.1800660915
- Stojanovic M, Stojanovic D, Rancic N, Ignjatovic A, Antic Z, Miljkovic S, Rajovic T. 2017. Trends in thyroid cancer incidence and mortality in Central Serbia, 1999-2014. Ann Ist Super Sanita. 53(4):299–304.
- Takahashi H, Takahashi K, Shimura H, Yasumura S, Suzuki S, Ohtsuru A, Midorikawa S, Ohira T, Ohto H, Yamashita S, Kamiya K. 2017. Simulation of expected childhood and adolescent thyroid cancer cases in Japan using a cancer-progression model based on the national cancer registry: application to the first-round thyroid examination of the Fukushima Health Management Survey. Medicine (Baltimore). 96(48):e8631. doi: https://doi.org/10.1097/MD.0000000000008631
- Tavarelli M, Sarfati J, Chereau N, Tissier F, Golmard JL, Ghander C, Lussey-Lepoutre C, Trésallet C, Menegaux F, Leenhardt L, Buffet C. 2017. Heterogeneous prognoses for pT3 papillary thyroid carcinomas and impact of delayed risk stratification. Thyroid. 27(6):778–786. doi: https://doi.org/10.1089/thy.2016.0512
- Wang CW, Lee YC, Calista E, Zhou F, Zhu H, Suzuki R, Komura D, Ishikawa S, Cheng SP. 2018. A benchmark for comparing precision medicine methods in thyroid cancer diagnosis using tissue microarrays. Bioinformatics. 34(10):1767–1773. doi: https://doi.org/10.1093/bioinformatics/btx838
- Wang P, Liu G, Xu W, Liu H, Bu Q, Sun D. 2017. Long noncoding RNA H19 inhibits cell viability, migration, and invasion via downregulation of IRS-1 in thyroid cancer cells. Technol Cancer Res Treat. 16(6):1102–1112. doi: https://doi.org/10.1177/1533034617733904
- Weeks KS, Kahl AR, Lynch CF, Charlton ME. 2018. Racial/ethnic differences in thyroid cancer incidence in the United States, 2007-2014. Cancer. 124(7):1483–1491. doi: https://doi.org/10.1002/cncr.31229
- Williard W, Borgen P, Bol R, Tiwari R, Osborne M. 1992. Cowden's disease. A case report with analyses at the molecular level. Cancer. 69(12):2969–2974. doi: https://doi.org/10.1002/1097-0142(19920615)69:12<2969::AID-CNCR2820691217>3.0.CO;2-2
- Xhaard C, Rubino C, Souchard V, Maillard S, Ren Y, Borson-Chazot F, Sassolas G, Schvartz C, Colonna M, Lacour B, et al. 2018. Dietary habits during the 2 months following the Chernobyl accident and differentiated thyroid cancer risk in a population-based case-control study. Cancer Epidemiol. 52:142–147. doi: https://doi.org/10.1016/j.canep.2017.12.015
- Zhai X, Xu W. 2018. Long noncoding RNA ATB Promotes proliferation, migration, and invasion in bladder cancer by suppressing MicroRNA-126. Oncol Res. 26(7):1063–1072. doi: https://doi.org/10.3727/096504018X15152072098476
- Zhang H, Cai Y, Zheng L, Zhang Z, Lin X, Jiang N. 2018. Long noncoding RNA NEAT1 regulate papillary thyroid cancer progression by modulating miR-129-5p/KLK7 expression. J Cell Physiol. 233(10):6638–6648. doi: https://doi.org/10.1002/jcp.26425
- Zhou J, Xiao H, Yang X, Tian H, Xu Z, Zhong Y, Ma L, Zhang W, Qiao G, Liang J. 2018. Long noncoding RNA CASC9.5 promotes the proliferation and metastasis of lung adenocarcinoma. Sci Rep. 8(1):37. doi: https://doi.org/10.1038/s41598-017-18280-3
