Abstract
Long non-coding RNAs (lncRNAs) are involved in development of non-small cell lung cancer (NSCLC) by interacting with microRNAs (miRNAs) and/or mRNAs. However, the function of most lncRNAs in NSCLC remains unclear. Reverse transcription-quantitative PCR (RT-qPCR) and western blot were applied for detection of mRNA/miRNA and protein level. Interaction between LncRNA small nucleolar RNA host gene 14 (SNHG14) and miR-382-5p was validated by dual luciferase reporter assay. Cell proliferation and apoptosis was detected using Cell Counting Kit 8 (CCK8) and Flow cytometry, while cell migration and invasion was detected by scratch assay and Transwell assay. SNHG14 was upregulated in NSCLC tissues and cell lines. SNHG14 silencing reduced proliferation, migration and invasion, and induced apoptosis in A549 cells. SNHG14 was negatively correlated with miR-382-5p in NSCLC tissues, SNHG14 and miR-382-5p negatively regulated each other in A549 cells. SNHG14 promoted expression of miR-382-5p target genes, including LIM-only protein 3 (LMO3) and SET domain containing lysine methyltransferase 8 (SETD8), in A549 cells. miR-382-5p inhibition reversed SNHG14 knockdown-induced increase in apoptosis and decrease in proliferation, migration and invasion. SNHG14 was correlated with SETD8 and LMO3 in NSCLC tissues. Collectively, SNHG14 promoted NSCLC proliferation, migration and invasion, while inhibited apoptosis by sponging miR-382-5p in A549 cells.
Abbreviations: CCK8: Cell Counting Kit 8; LMO3: LIM-only protein 3; lncRNAs: long non-coding RNAs; miRNAs: microRNAs; NSCLC: non-small cell lung cancer; RT-qPCR: reverse transcription-quantitative PCR; SNHG14: LncRNA small nucleolar RNA host gene 14; SETD8: SET domain containing lysine methyltransferase 8.
Background
Lung cancer is the most commonly diagnosed cancer type and the leading cause of cancer related-mortalities worldwide (Bray et al. Citation2018). The poor prognosis of patients with lung cancer is partly due to the occurrence of metastasis before surgical removal of primary tumor (Klein Citation2009). As a major subtype, NSCLC accounts for >85% of lung cancer (Clinical Lung Cancer Genome P et al. Citation2013). Although numerous oncogenes and tumor suppressors have been identified in NSCLC, the molecular mechanisms underlying the development of NSCLC remains elusive and needs to be further investigated.
LncRNAs are non-coding, single-stranded transcripts with >200 nucleotides (Yang et al. Citation2013). Accumulating evidence suggests that dysregulation of lncRNAs contributes to the initiation and progression of cancer (Chen et al. Citation2016; Castro-Oropeza et al. Citation2018). Mechanistically, lncRNAs interact with miRNAs to regulate gene expression at the post-transcription level or to modify chromatin, controlling transcriptional gene silencing (Mercer et al. Citation2009). SNHG14 is associated with the development of breast cancer (Dong et al. Citation2018), glioma (Wang et al. Citation2018), and gastric cancer (Liu et al. Citation2018b). However, the role of SNHG14 in mediating metastasis of NSCLC has not been previously investigated.
miRNAs are small, non-coding, single-stranded RNAs with 18–22 nucleotides in length (Diederichs and Haber Citation2007). After interacting with the 3’-untranslated region (UTR) of their target mRNAs, miRNAs regulate the stability or translation rate of various transcripts (Ebert et al. Citation2007). The pivotal role of miRNAs during carcinogenesis has been well-characterized (Fang et al. Citation2012). Specific miRNAs act either as oncogenes or tumor suppressors depending on the cellular background (Gu et al. Citation2013). For instance, miR-382-5p serves a tumor suppressor in esophageal squamous cell carcinoma (Feng et al. Citation2017) and ovarian cancer (Tan et al. Citation2016). In particular, in NSCLC, miR-382-5p inhibits NSCLC progression by targeting SETD8 (Chen et al. Citation2017), or LMO3 (Chen et al. Citation2019). However, the precise mechanism underlying the interaction of SNHG14 and miR-382-5p in NSCLC remains unknown.
Results
SNHG14 is upregulated in NSCLC tissues and cell lines
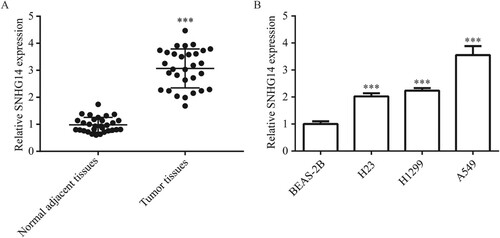
To investigate the role of SNHG14 in NSCLC, RT-qPCR was used to detect SNHG14 expression in NSCLC tissues and cell lines. SNHG14 expression was significantly increased in tumor tissues in comparison with normal lung tissues (Figure (A)), which was also significantly increased in NSCLC cell line (H23, H1299 and A549) compared with immortalized lung cell line (BEAS-2B) (Figure (B)). The highest expression of SNHG14 was observed in A549; therefore, this cell line was used for subsequent experiments.
Figure 1. Overexpression of SNHG14 mRNA in NSCLC tissues and cells. SNHG14 mRNA level was significantly overexpressed in NSCLC tissues compared to the adjacent normal tissues (A). SNHG14 mRNA level was significantly overexpressed in NSCLC cell lines (H23, H1299, A549) compared with BEAS-2B (B). ***p < 0.001.
SNHG14 is correlated with TNM of patients with NSCLC
The association between characteristics of patients with NSCLC and SNHG14 expression was exhibited in Table . SNHG14 is significantly correlated with TNM of patients with NSCLC, however, there was no significant correlation between SNHG14 and age, gender, smoking history or histological classification.
Table 1. The association between characteristics of patients with NSCLC and SNHG14 expression.
Downregulation of SNHG14 inhibits proliferation, migration and invasion, while promotes apoptosis in NSCLC cells
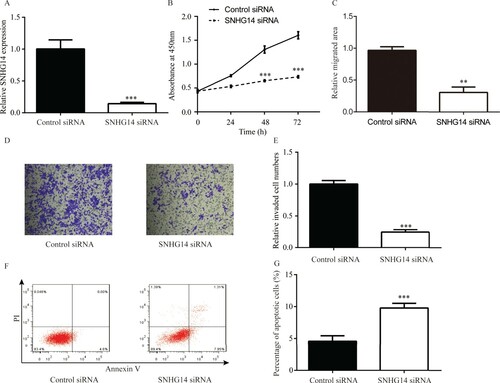
Compared with control siRNA, SNHG14 siRNA significantly decreased SNHG14 level in A549 cells (Figure (A)). Cell proliferation assay suggested that SNHG14 siRNA significantly inhibited proliferation of A549 cells compared with control siRNA (Figure (B)). Cell migration assay suggested that SNHG14 siRNA significantly reduced migrated area compared with control siRNA (Figure (C)). Cell invasion assay suggested that SNHG14 siRNA significantly reduced the number of invasive cells compared with control siRNA (Figure (D,E)). In the cell apoptosis assay, SNHG14 siRNA led to a small increase in apoptotic cells in comparison with control siRNA (Figure (F,G)).
Figure 2. SNHG14 regulated A549 cell proliferation/migration/invasion/apoptosis. Compared with control siRNA, SNHG14 siRNA significantly downregulated SNHG14 level in A549 cells (A). SNHG14 siRNA remarkably downregulated A549 cell proliferation (B), migration (C) and A549 cell invasion (D–E, ×100), while significantly upregulated A549 cell apoptosis (F,G). ***p < 0.001.
SNHG14 regulates miR-382-5p in NSCLC tissues and cells
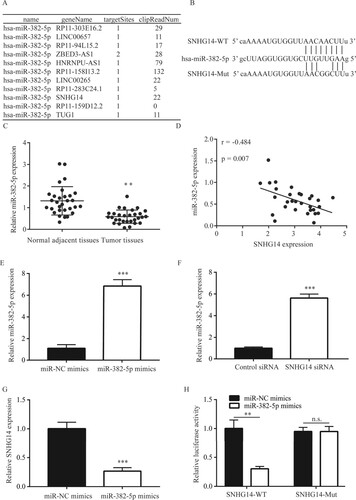
Following bioinformatic analysis, SNHG14 was found to potentially interact with miR-382-5p (Figure (A)). The potential binding site between miR-382-5p and SNHG14 was then identified (Figure (B)). RT-qPCR results suggested that, miR-382-5p was significantly lower in NSCLC tissues than in the adjacent normal tissues (Figure (C)), in addition, the expression of SNHG14 was significantly correlated (r = −0.484; P = 0.007) with miR-382-5p in 30 NSCLC tissues, and the 95% confidential interval was from −0.819 to −0.149 (Figure (D)). RT-qPCR results suggested that miR-382-5p level was significantly increased by miR-382-5p mimics (Figure (E)). In addition, SNHG14 siRNA significantly increased miR-382-5p level (Figure (F)). Conversely, miR-382-5p mimics significantly decreased SNHG14 level (Figure (G)). Moreover, dual luciferase assay suggested that miR-382-5p mimics decreased luciferase activity in A549 cells transfected with SNHG14-WT but not SNHG14-MT (Figure (H)).
Figure 3. SNHG14 regulated miR-382-5p expression in NSCLC tissues and cells. SNHG14 could potentially bind to miR-382-5p (A). The potential binding site between miR-382-5p and SNHG14 was exhibited (B). miR-382-5p was significantly lower in NSCLC tissues than in the adjacent normal tissues (C). There was a significant correlation between SNHG14 and miR-382-5p in NSCLC tissues (D). miR-382-5p mimic increased miR-382-5p levels in A549 cells (E). SNHG14 siRNA increased miR-382-5p levels in A549 cells (F). miR-382-5p mimic decreased SNHG14 levels in A549 cells (G). miR-382-5p mimic decreased luciferase activity of A549 cells transfected with SNHG14-WT (H). **p < 0.01; ***p < 0.001.
Downregulation of SNHG14 inhibits invasion, migration and proliferation, while induces apoptosis in NSCLC cells via miR-382-5p
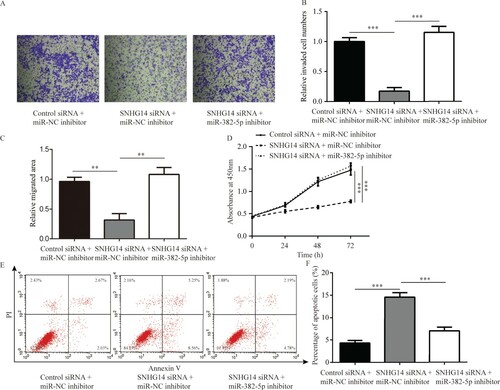
To investigate the function of SNHG14 in NSCLC cells, cell invasion, migration and proliferation were investigated in A549 cells following SNHG14 knockdown. SNHG14 siRNA significantly inhibited invasion of A549 cells, which was reversed following miR-382-5p inhibition (Figure (A,B)). Additionally, knockdown of SNHG14 decreased migration and proliferation of A549 cells, which was reversed following miR-382-5p inhibition (Figure (C,D)). Moreover, decreased expression of miR-382-5p attenuated cell apoptosis induced by SNHG14 knockdown in A549 cells (Figure (E,F)).
Figure 4. SNHG14 regulated cell apoptosis, invasion, migration and proliferation of NSCLC cells by miR-382-5p. SNHG14 siRNA significantly inhibited A549 cell invasion (A–B, ×100), migration (C) and proliferation (D), while induced apoptosis (E,F), which was reversed towards miR-382-5p inhibition. ***p < 0.001.
SNHG14 regulates LMO3 and SETD8 expression levels by downregulating miR-382-5p in NSCLC cells
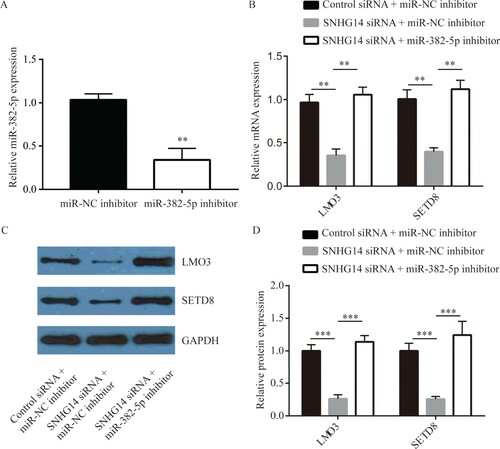
SETD8 is an oncogene targeted by miR-382-5p in NSCLC cells [17]. LMO3 was regulated by miR-382-5p [18]. RT-qPCR results suggested that miR-382-5p level was significantly decreased by miR-382-5p inhibitor (Figure (A)). SNHG14 knockdown significantly decreased mRNA and protein levels of SETD8 and LMO3 in A549 cells, which was reversed by miR-382-5p inhibitor (Figure (B–D)), suggesting that SNHG14 regulated these genes by sponging miR-382-5p.
Figure 5. SNHG14 regulated LMO3, SETD8, MMP2 and MMP9 expression by miR-382-5p in NSCLC cells. miR-382-5p inhibitor decreased miR-382-5p levels in A549 cells (A). SNHG14 siRNA decreased mRNA and protein levels of SETD8 and LMO3 in A549 cells, which were reversed by inhibition of miR-382-5p (B–D). **p < 0.01; ***p < 0.001.
SNHG14 regulates SETD8 and LMO3 mRNA level in NSCLC tissues
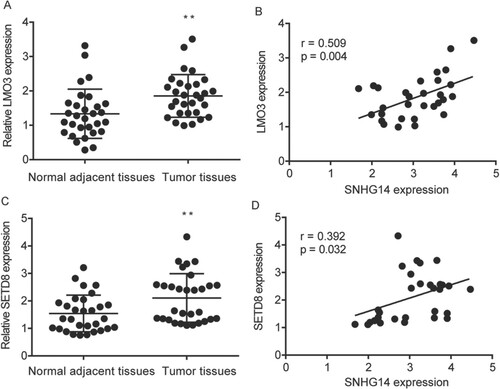
RT-qPCR results suggested that LMO3 was significantly increased in 30 NSCLC tissues compared to the adjacent normal tissues (Figure (A)), in addition, the expression of SNHG14 was significantly correlated (r = 0.509; P = 0.004) with LMO3 in 30 NSCLC tissues, and the 95% confidential interval was from 0.183 to 0.734 (Figure (B)). Meanwhile, RT-qPCR results suggested that SETD8 was significantly increased in 30 NSCLC tissues compared to the adjacent normal tissues (Figure (C)), in addition, SNHG14 was significantly correlated (r = 0.392; P = 0.032) with SETD8 in 30 NSCLC tissues, and the 95% confidential interval was from 0.037 to 0.659 (Figure (D)).
Figure 6. SNHG14 regulated SETD8 and LMO3 mRNA level in NSCLC tissues. LMO3 was significantly higher in NSCLC tissues than in the adjacent normal tissues (A), which was correlated with SNHG14 in NSCLC tissue (B). Meanwhile, SETD8 was significantly higher in NSCLC tissues than in the adjacent normal tissues (C), which was correlated with SNHG14 in NSCLC tissues (D). **p < 0.01.
Discussion
Current results suggested that SNHG14 was upregulated in NSCLC cells. Moreover, SNHG14 promoted proliferation, migration and invasion, while inhibited apoptosis in NSCLC cells by sponging miR-382-5p.
SNHG14 functions in the progression of several cancer types by sponging miRNAs, including NSCLC and colorectal cancer. For example, in NSCLC, SNHG14 was up-regulated and sponged miR-340 (Zhang et al. Citation2019), SNHG14 sponged miR-34a thus leading to cisplatin resistance (Jiao et al. Citation2019), and SNHG14 sponged miR-206-3p thus leading to gefitinib resistance (Wu et al. Citation2019); in colorectal cancer, SNHG14 sponged miR-32-5p, miR-3940-5p and miR-92b-3p, thus promoting cell proliferation, migration and invasion (Matboli et al. Citation2019; Ye et al. Citation2019; Zhang et al. Citation2020). In current study, SNHG14 was found to be increased in NCSLC tissues and cell line, and SNHG14 siRNA was discovered to inhibit proliferation and invasion, while promote apoptosis in NSCLC cell line. Whereas, in NSCLC, the miRNAs that could be sponged by SNHG14 were left to be explored.
miR-382-5p, a well-characterized tumor suppressor miRNA, interacted with oncogenes including NR2F2, RERG and SP1, inhibiting cell proliferation, metastasis and increasing chemotherapy sensitivity in colorectal cancer (Zhou et al. Citation2016; Ren et al. Citation2018; Yao et al. Citation2019). In addition, miR-382-5p could be sponged by lncRNAs TUG1 and NEAT1 in pancreatic cancer and ovarian cancer (Zhao et al. Citation2017; Liu et al. Citation2018a). In NSCLC, miR-382-5p level was decreased in NSCLC tissues and cell lines compared with normal tissues and cell lines (Chen et al. Citation2017). Consequently, the relation between miR-382-5p and SNHG14 in NSCLC attracted our attention. In present study, SNHG14 was found to be a ceRNA for miR-382-5p. Additionally, miR-382-5p was negatively correlated with SNHG14 in NSCLC tissues. Moreover, the effects of SNHG14 siRNA on proliferation, migration, invasion, and apoptosis of NSCLC cell line was rescued by miR-382-5p inhibitor. However, whether the molecules that were targeted by miR-382-5p were regulated by SNHG14 were left to be investigated.
SETD8 was a methyltransferase that specifically regulated the methylation levels of H4K20 and promoted cancer progression (Takawa et al. Citation2012) Inhibitors of SETD8 suppressed cancer cell proliferation (Veschi et al. Citation2017), moreover, miR-382-5p inhibits NSCLC progression by targeting SETD8 (Chen et al. Citation2017). LMO3, which was regulated by miR-382-5p, was a key regulator of metastasis in NSCLC cells (Chen et al. Citation2019). Currently, SNHG14 siRNA was found to inhibit the expression of SETD8 and LMO3 by sponging miR-382-5p.
Taken together, the present results suggested that SNHG14 acted as an oncogene in NSCLC, SNHG14/miR-382-5p/SETD8/LMO3 axis might play a crucial role in promoting NSCLC progression.
Material and methods
Patient samples
Paired tumor tissues and matched normal tissues were obtained from 30 patients with NSCLC who had undergone surgery to treat NSCLC at the Shenzhen People’s Hospital between January 2014 and January 2015. Diagnosis was confirmed by pathological analysis. No participant received radiotherapy or chemotherapy prior to surgical treatment. Written informed consent was provided by all patients enrolled in the present study. The use of human tissues for scientific research was approved by the Ethical Committee of Shenzhen People’s Hospital.
Cell culture
The normal human bronchial epithelial cell line BEAS-2B and human NSCLC cell lines (H1299, H23 and A549) were purchased from the American Type Culture Collection (ATCC). All cell lines were cultured in DMEM (Gibco) added with 10% FBS (HyClone) and 1% penicillin–streptomycin solution (Gibco) in a humidified incubator at 37˚C with 5% CO2.
Target prediction for miRNAs and lncRNAs
Bioinformatic prediction to identify the lncRNAs interacting with miR-328-5p and their putative binding sites were performed by online database StarBase (http://starbase.sysu.edu.cn/). In total, 34 lncRNAs were predicted to bind to miR-382-5p. Subsequently, miR-382-5p was validated as a target miRNA of SNHG14.
Plasmids construction
SNHG14 sequence containing the binding site for miR-382-5p was amplified from the cDNA of A549 cells by Taq PCR polymerase (New England Biolabs) and inserted into pGL3 (Invitrogen) to construct pGL3-SNHG14-wild-type (WT). A Stratagene mutation kit (Stratagene) was used to introduce point mutations into pGL3-SNHG14-Mutant (MT).
Small interfering (si)RNA transfection
siRNA targeting SNHG14 or control siRNA (GenePharma) was mixed with Lipofectamine RNAiMAX in OptiMEM (Invitrogen) and incubated for 15 min at room temperature. Subsequently, siRNA-containing solutions were added into each well in 6-well or 96-well plates. After incubation for 72 h, cells were harvested and subjected to reverse transcription-quantitative PCR to validate SNHG14 downregulation.
Transfection of miR-382-5p mimic and inhibitor
50 nM miR-382-5p mimics, miR-NC mimics, miR-382-5p inhibitor or miR-NC inhibitor (RiboBio) was transfected into the A549 cells by Lipofectamine 2000 (Invitrogen). After incubation for 48 h, cells were harvested for the following experiments.
Dual luciferase assay
A549 cells were cultured in 24-well plates and co-transfected with 50 nM miR-382-5p mimics or miR-NC mimics and pGL3-SNHG14-WT or pGL3-SNHG14-MT and Renilla luciferase vector by Lipofectamine 2000 (Invitrogen). After incubation for 48 h, the dual-luciferase activity in each well was examined by a dual-luciferase reporter assay system (Promega Corporation). Renilla luciferase activity was used as internal control.
Western blot
Cell lysates were prepared by RIPA lysis buffer (Beyotime). Protein concentration was determined using a bicinchoninic acid assay kit (Beyotime). Proteins were separated by SDS-PAGE using 8% gels and transferred on PVDF membranes. Membranes were subsequently blocked with 5% non-fat milk at room temperature for 1 h and incubated with the primary antibodies for LMO3, SETD8 and GAPDH overnight at 4°C. The next day, membranes were washed with TBS-Tween (0.2%) prior to incubation with the secondary antibody at room temperature for 1 h. Subsequently, the blots were developed using ECL (GE Healthcare Life Sciences). GAPDH served as the loading control. The quantification of band intensity was performed by ImageJ (version 1.50; NIH).
RNA extraction and reverse transcription-quantitative PCR
miRNeasy Mini kit (Qiagen) was used to extract total RNA from tissues and cells. RNA was reverse transcribed into cDNA using the TransScript First-Strand cDNA Synthesis SuperMix kit (Transgene SA). RT-qPCR was performed using the CFX96 real-time PCR system (Bio-Rad) with SYBR Premix Ex Taq (Takara). U6 and GAPDH were used as internal controls for miRNAs and mRNAs, respectively. The gene relative expression levels were quantified using the 2−ΔΔCq method (Livak and Schmittgen Citation2001).
Cell proliferation assay
Cell proliferation was detected using CCK8 (Dojindo Molecular Technologies). In total, 1,000 cells were seeded in each well of 96-well plates. On the following day, 10-μl CCK8 solution was added into each well and cells were incubated at 37°C for 2 h. After transfection with SNHG14 siRNA or control siRNA and/or miR-382 mimics or miR-NC mimics, the absorbance at 450 nm was detected at 0, 24, 48 and 72 h using a microplate reader (Bio-Rad).
Cell apoptosis assay
Cell apoptotic rate was analyzed by Annexin V–FITC (Sigma–Aldrich). Briefly, cells were treated by 1X binding buffer, added with Annexin V–FITC and incubated at room temperature for 15 min, followed with addition of propidium iodide (PI) and incubation for 5 min at room temperature. At last, cells were analyzed by FACSCalibur flow cytometer (BD Biosciences), data were analyzed by FlowJo.
Cell migration assay
Cell migration ability was measured by wound healing assay. A549 cells (1 × 105 cells/well) were seeded in 6-well plates and cultured for 6 h. Then, a wound was generated with a 10-µl Eppendorf pipette tip. After removement of the detached cells, A549 cells were incubated in DMEM for another 24 h. The distance between the edges of the gap was calculated by Image-Pro Plus v6.0.
Cell invasion assay
Cell invasion ability was measured using Matrigel invasion chambers (pore size, 8 µm; BD Biosciences). Briefly, 100-μl Matrigel (BD Biosciences) was plated onto the bottom of the upper Transwell chambers. Subsequently, 2 × 105 A549 cells in DMEM containing 1% FBS were added to the upper chamber, while DMEM containing 10% FBS was placed in the lower chamber. After 72 h, cells on the upper chamber were removed, while cells that invaded to the lower chamber were fixed using 4% formaldehyde at room temperature for 15 min. Cells were stained with crystal violet at room temperature for 20 min. The number of invaded cells was counted under a microscope. Representative images of invaded cells were obtained after staining.
Statistical analysis
Data were analyzed using GraphPad Prism 6.0 (GraphPad Software), presented as mean ± SD. Differences between two groups were analyzed using Students’ t-test and differences among multiple groups were analyzed by one-way ANOVA followed by Tukey’s test. The correlation between SNHG14 and miR-382-5p was analyzed by Pearson Correlation analysis. P < 0.05 indicated a statistically significant difference.
Author contributions
YZ, GX, MF, SW, ZT, and RQ carried out the experiments and analyzed the data. ZL and XL designed the experiments and wrote the manuscript.
Availability of data and materials
All data and materials were available from the corresponding author. Due to the nature of this research, participants of this study did not agree for their data to be shared publicly, so supporting data is not available.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. 2018. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 68:394–424. doi: https://doi.org/10.3322/caac.21492
- Castro-Oropeza R, Melendez-Zajgla J, Maldonado V, Vazquez-Santillan K. 2018. The emerging role of lncRNAs in the regulation of cancer stem cells. Cell Oncol. 41:585–603. doi: https://doi.org/10.1007/s13402-018-0406-4
- Chen DL, Ju HQ, Lu YX, Chen LZ, Zeng ZL, Zhang D-s, Luo H-y, Wang F, Qiu M-z, Wang D-s, et al. 2016. Long non-coding RNA XIST regulates gastric cancer progression by acting as a molecular sponge of miR-101 to modulate EZH2 expression. J Exp Clin Cancer Res. 35:142. doi: https://doi.org/10.1186/s13046-016-0420-1
- Chen T, Ren H, Thakur A, Yang T, Li Y, Zhang S, Wang T, Chen M. 2017. miR-382 inhibits tumor progression by targeting SETD8 in non-small cell lung cancer. Biomed Pharmacother. 86:248–253. doi: https://doi.org/10.1016/j.biopha.2016.12.007
- Chen D, Zhang Y, Lin Y, Shen F, Zhang Z, Zhou J. 2019. MicroRNA-382 inhibits cancer cell growth and metastasis in NSCLC via targeting LMO3. Exp Ther Med. 17:2417–2424.
- Clinical Lung Cancer Genome P, Network Genomic M. 2013. A genomics-based classification of human lung tumors. Sci Transl Med. 5:209ra153.
- Diederichs S, Haber DA. 2007. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell. 131:1097–1108. doi: https://doi.org/10.1016/j.cell.2007.10.032
- Dong H, Wang W, Mo S, Liu Q, Chen X, Chen R, Zhang Y, Zou K, Ye M, He X. 2018. Long non-coding RNA SNHG14 induces trastuzumab resistance of breast cancer via regulating PABPC1 expression through H3K27 acetylation. J Cell Mol Med. 22:4935–4947. doi: https://doi.org/10.1111/jcmm.13758
- Ebert MS, Neilson JR, Sharp PA. 2007. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 4:721–726. doi: https://doi.org/10.1038/nmeth1079
- Fang Y, Fang D, Hu J. 2012. MicroRNA and its roles in esophageal cancer. Med Sci Monit. 18:RA22–RA30.
- Feng J, Qi B, Guo L, Chen LY, Wei XF, Wei X-F, Liu Y-Z, Zhao B-S. 2017. miR-382 functions as a tumor suppressor against esophageal squamous cell carcinoma. World J Gastroenterol. 23:4243–4251. doi: https://doi.org/10.3748/wjg.v23.i23.4243
- Gu J, Wang Y, Wu X. 2013. MicroRNA in the pathogenesis and prognosis of esophageal cancer. Curr Pharm Des. 19:1292–1300. doi: https://doi.org/10.2174/1381612811319350009
- Jiao P, Hou J, Yao M, Wu J, Ren G. 2019. SNHG14 silencing suppresses the progression and promotes cisplatin sensitivity in non-small cell lung cancer. Biomed Pharmacother. 117:109164. doi: https://doi.org/10.1016/j.biopha.2019.109164
- Klein CA. 2009. Parallel progression of primary tumours and metastases. Nat Rev Cancer. 9:302–312. doi: https://doi.org/10.1038/nrc2627
- Liu Y, Wang Y, Fu X, Lu Z. 2018a. Long non-coding RNA NEAT1 promoted ovarian cancer cells’ metastasis through regulation of miR-382-3p/ROCK1 axial. Cancer Sci. 109:2188–2198. doi: https://doi.org/10.1111/cas.13647
- Liu Z, Yan Y, Cao S, Chen Y. 2018b. Long non-coding RNA SNHG14 contributes to gastric cancer development through targeting miR-145/SOX9 axis. J Cell Biochem. 119:6905–6913. doi: https://doi.org/10.1002/jcb.26889
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 25:402–408. doi: https://doi.org/10.1006/meth.2001.1262
- Matboli M, Shafei AE, Ali MA, El-Din Ahmed TS, Naser M, Abdel-Rahman T, Anber N, Ali M. 2019. Role of extracellular LncRNA-SNHG14/miRNA-3940-5p/NAP12 mRNA in colorectal cancer. Arch Physiol Biochem. 1–7. doi: https://doi.org/10.1080/13813455.2019.1650070
- Mercer TR, Dinger ME, Mattick JS. 2009. Long non-coding RNAs: insights into functions. Nat Rev Genet. 10:155–159. doi: https://doi.org/10.1038/nrg2521
- Ren Y, Zhang H, Jiang P. 2018. MicroRNA-382 inhibits cell growth and migration in colorectal cancer by targeting SP1. Biol Res. 51:51. doi: https://doi.org/10.1186/s40659-018-0200-9
- Takawa M, Cho HS, Hayami S, Toyokawa G, Kogure M, Yamane Y, Iwai Y, Maejima K, Ueda K, Masuda A, et al. 2012. Histone lysine methyltransferase SETD8 promotes carcinogenesis by deregulating PCNA expression. Cancer Res. 72:3217–3227. doi: https://doi.org/10.1158/0008-5472.CAN-11-3701
- Tan H, He Q, Gong G, Wang Y, Li J, Wang J, Zhu D, Wu X. 2016. miR-382 inhibits migration and invasion by targeting ROR1 through regulating EMT in ovarian cancer. Int J Oncol. 48:181–190. doi: https://doi.org/10.3892/ijo.2015.3241
- Veschi V, Liu Z, Voss TC, Ozbun L, Gryder B, Yan C, Hu Y, Ma A, Jin J, Mazur SJ. 2017. Epigenetic siRNA and chemical screens identify SETD8 inhibition as a therapeutic strategy for p53 activation in high-Risk neuroblastoma. Cancer Cell. 31:50–63. doi: https://doi.org/10.1016/j.ccell.2016.12.002
- Wang Q, Teng Y, Wang R, Deng D, You Y, Peng Y, Shao N, Zhi F. 2018. The long non-coding RNA SNHG14 inhibits cell proliferation and invasion and promotes apoptosis by sponging miR-92a-3p in glioma. Oncotarget. 9:12112–12124. doi: https://doi.org/10.18632/oncotarget.23960
- Wu K, Li J, Qi Y, Zhang C, Zhu D, Liu D, Zhao S. 2019. SNHG14 confers gefitinib resistance in non-small cell lung cancer by up-regulating ABCB1 via sponging miR-206-3p. Biomed Pharmacother. 116:108995. doi: https://doi.org/10.1016/j.biopha.2019.108995
- Yang L, Lin C, Jin C, Yang JC, Tanasa B, Li W, Merkurjev D, Ohgi KA, Meng D, Zhang J, et al. 2013. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature. 500:598–602. doi: https://doi.org/10.1038/nature12451
- Yao H, Xia D, Li ZL, Ren L, Wang MM, et al. 2019. MiR-382 functions as tumor suppressor and chemosensitizer in colorectal cancer. Biosci Rep. 39.
- Ye T, Zhang N, Wu W, Yang B, Wang J, Huang W, Tang D. 2019. SNHG14 promotes the tumorigenesis and metastasis of colorectal cancer through miR-32-5p/SKIL axis. In Vitro Cell Dev Biol Anim. 55:812–820. doi: https://doi.org/10.1007/s11626-019-00398-5
- Zhang W, Duan W, Mo Z, Wang J, Yang W, Wu W, Li X, Lin S, Tan Y, Wei W. 2020. Upregulation of SNHG14 suppresses cell proliferation and metastasis of colorectal cancer by targeting miR-92b-3p. J Cell Biochem. 121:1998–2008. doi: https://doi.org/10.1002/jcb.29434
- Zhang Z, Wang Y, Zhang W, Li J, Liu W, Lu W. 2019. Long non-coding RNA SNHG14 exerts oncogenic functions in non-small cell lung cancer through acting as an miR-340 sponge. Biosci Rep. 39:BSR20180941. doi: https://doi.org/10.1042/BSR20180941
- Zhao L, Sun H, Kong H, Chen Z, Chen B, Zhou M. 2017. The lncrna-TUG1/EZH2 axis promotes pancreatic cancer cell proliferation, migration and EMT phenotype formation through sponging mir-382. Cell Physiol Biochem. 42:2145–2158. doi: https://doi.org/10.1159/000479990
- Zhou B, Song J, Han T, Huang M, Jiang H, Qiao H, Shi J, Wang Y. 2016. MiR-382 inhibits cell growth and invasion by targeting NR2F2 in colorectal cancer. Mol Carcinog. 55:2260–2267. doi: https://doi.org/10.1002/mc.22466
