ABSTRACT
Pneumonia remains the leading infectious cause of death among children under five years of age, and the elderly. Several biomarkers, which have been identified for its diagnosis lack specificity, as they could not differentiate viral from bacterial pathogens of the disease; these biomarkers also fail to establish a distinction between pneumonia and other associated diseases such as pulmonary tuberculosis and Human Immunodeficiency Virus (HIV). This review outlined the menace of pneumonia disease from the statistical prevalence, clinical and immunological view, challenges with the methods used in diagnosis, and more useful information about methods of diagnosis of pneumonia with their limitations as well. Additionally, the use of aptamers and antimicrobial peptides (AMPs) rather than antibodies to bind and recognize receptors for diagnostics, offers several advantages over other biomarkers shortcomings such as non-specificity.
Introduction
There is nothing more worrisome to researchers and the international health community than the rising incidence of mortality and major economic losses resulting from pneumonia. Approximately 2.56 million people die from pneumonia annually (Lin et al. Citation2020). In 2017, for instance, 808,694 children under the age of five died of pneumonia, accounting for fifteen percent of all deaths of children in that year (Shen et al. Citation2020a). There are over 1400 cases of pneumonia out of every 100,000 children worldwide with the highest incidence occurring in South Asia, West and Central Africa (Zhu et al. Citation2020). Recently, the world health organization (WHO) Child Health Epidemiology Reference Group gathered that the estimated median global cases of pneumonia is 0.28 episodes per child year, equating to an annual of 150.7 million in which 11–20 million of these cases are severe enough to require hospitalization (Li et al. Citation2020). There is very little progress in reducing death due to pneumonia in children. The yearly incidence of death cases in the elderly is also estimated at 18.2 per 1000 persons within age 65–69 years and 52.3 cases in 1000 persons within age 85 years and above. Huge amount is budgeted in billions of dollars for diseases treatment support from government and international community, in which pneumonia expenses accounted for eighty-one percent of annual budget (Tong et al. Citation2018).
Pneumonia is caused by either a virus, bacterium or fungus and it is related with a huge morbidity and mortality in older patients compared with more youthful adults (Chen et al. Citation2020). Those most in danger are individuals who are under 5 years or 65 years or more; smoke tobacco, devour a lot of liquor, or both; have fundamental conditions, for example, cystic fibrosis, chronic obstructive pulmonary disorder (COPD), asthma, or conditions that influence the kidneys, heart, or liver; have a debilitating or impaired immune system, for instance, because of AIDS, HIV, or malignant growth; take prescriptions for gastroesophageal reflux disease (GERD); have as of late recouped from a cold or influenza infection; experience malnutrition; have been as of late hospitalized in an emergency unit; have been presented to specific synthetic substances or toxins (Bai et al. Citation2020a). A few groups are more inclined than others to pneumonia, including Native Alaskan or certain Native American ethnicities (Kim et al. Citation2017). Despite considerable effort by health agencies and organizations, mortality rates due to pneumonia in most developing countries are growing at an alarming increase (Liu et al. Citation2018).
In the last years, the methods of detection of these diagnostic biomarkers have been constantly advanced, ranging from blood cultures, polymerase chain reaction, matrix assisted laser desorption or ionization time of flight, immunofiltration, turbidimetric immunoassay based on latex agglutination, just to mention a few (Eddabra and Benhassou Citation2018). With these methods, it is possible to detect locally confined bacterial infections like pneumonia. However, these methods show false negative results for viral pneumonia and patients with high titres of rheumatoid factors may also give false results (Al-Ali and Indorf Citation2018). It is therefore imperative to explore other more reliable methods with improved sensitivity and accuracy towards diagnosis of pneumonia.
Due to this, efforts have been made by researchers to fill the remaining gaps in understanding this intense respiratory infection in children and the elderly. Almeida and Boattini (Citation2017) revised national-level estimates of the morbidity, extreme morbidity, mortality, etiology and hazard factors for acute lower respiratory infections. This gave an evaluation of comorbidity between childhood pneumonia and diarrhoea with the utilization of reviews to consider the impression of individuals towards oral rehydration sachets, antibiotics, and different treatments for diarrhoea and pneumonia in India and Kenya; Momosaki et al. (Citation2016) analyzed scaling up access to oral rehydration solution for diarrhoea and pneumonia, with reference from verifiable involvement with low–and high–performing nations; Wazny et al. (Citation2013) added to this work by surveying caregivers in Kenya to evaluate efficacy of zinc as a treatment for diarrhoea and pneumonia. Finally, Bhutta et al. (Citation2013) reported on the global action plan for childhood diarrhoea and pneumonia and showed how research priorities were developed resulting from this effort, among others. Despite all these, experts have called for more diagnostic research on the pathogens causing pneumonia and the routes through which it is transmitted, as this is of critical importance for treatment and prevention (Rajpurkar et al. Citation2017).
Furthermore, there is continuous need to improve the research into pneumonia diagnostics in light of the fact that the Alexandre Project, an overall surveillance study found that the worldwide pace of pneumococcal macrolide resistance had ascended from 16.5_% to 21.9_% in 1996–1997 to 29_% in 1998–2000 (Rudan et al. Citation2005). Resistance to macrolides and fluoroquinolones which were originally endorsed with the approval of levofloxacin for the treatment of pneumococcal infection in 1996 have been reported, with global pace of resistance from S. pneumonia strains running from 4_% to 100_% (Zilberberg et al. Citation2017). In the face of growing bacterial resistance, the utility of conventional antibiotics for the treatment of pneumonia is strained and development of new biomarkers for accurate and sensitive diagnosis to aid proper treatment and understanding of the pathogens is essential (Fernández-Barat et al. Citation2017).
Also, antigenic shift and drift are the major mechanisms by which viruses mutate to escape being discovered (Ramirez et al. Citation2017). As a result of this, many global pandemics have resulted in which a particular known virus would mutate to evade detection. Of particular interest is the advent of COVID-19 from SARS-CoV-2 that presents with a flu-like symptoms causing a global pandemic which calls for more serious attention and shows how serious the menace of pneumonia infection could be (Bai et al. Citation2020b).
The aim of this review is to give an overview of bacterial and viral pneumonia, stating the difficulty in the current diagnostic methods and biomarkers whilst giving an insight into the discovery of new biomarkers using bioinformatics for instance, for more accurate search. This will allow sensitive and accurate screening procedure for differential diagnostics of the specific pneumonia pathogens in patients for early treatment before or after onsets of symptoms, such that they can develop an accommodating lifestyles.
Pneumonia and pneumonitis
Pneumonia is an inflammation of the lung parenchyma (Curbelo et al. Citation2017). It is derived from a Greek word, Pneuma, which means ‘breath’. Parenchyma is the functional part of any organ whereas stroma means the structural part made of connective tissue. This disease begins when the lungs parenchyma gets inflamed, and the tiny air sacs, or alveoli, inside them, fills up with fluid. At this point, pneumonia-causing germs then settle in the alveoli and multiply, causing the lung sacs to be filled with pus (Ngari et al. Citation2014). This contagious disease is then spread through contact with or the touch of shared objects, sneezing, and coughing or even inhaled. Pneumonia is generally risky for older adults, babies, individuals with underlying illnesses, and those with an impaired immune system. Pneumonia influences kids and families everywhere in the world however it is generally predominant in Africa (Kalil et al. Citation2016) (Figure ).
Figure 1. The lungs and chest X-rays showing inflammation leading to pneumonia. (A) The lung presented with inflammation (left) and normal right lung. (B) The chest X-rays showing the normal chest with clear lungs without unusual opacification, Bacterial pneumonia displaying focal lobar consolidation in the right upper lobe, and Viral pneumonia showing diffuse interstitial patterns in the two lungs.
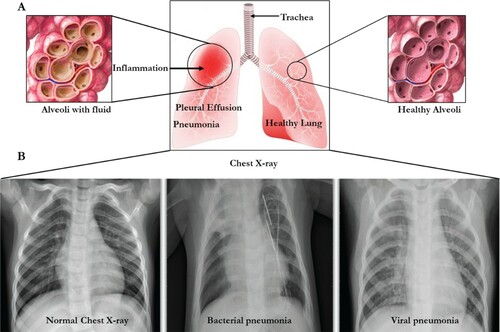
Pneumonitis is a term used to refer to a non-infectious causes of lung inflammation as a result of drugs, exposure to birds’ excrement or feather, and radiation/chemotherapy treatment from cancer (Salisbury et al. Citation2019). In other words, pneumonitis is used to describe sudden inflammation of the lung tissues without an infection whereas pneumonia results into a gradual inflammation caused by infection either from bacteria, viruses or fungi (Son et al. Citation2017). Pneumonitis is caused when an irritant, as aforementioned, causes the alveoli of the lungs to become inflamed, making oxygen difficult to cross into the bloodstream, in which in some people, the irritant is never detected (Bowman et al. Citation2018). Pneumonitis is common among farmers who use aerosols, pesticides and during harvest of grains and hay; among poultry workers as a result of exposure to droppings, feathers and other materials; during cancer treatment using chemotherapy and among hot tub and humidifier users (Tjalvin et al. Citation2018). The most common symptoms of pneumonitis is shortness of breath, fatigue, weight loss, loss of appetite and dry cough which may become chronic if undetected, resulting in fibrosis, heart failure, respiratory failure and death (Sommerfeld et al. Citation2018). Diagnosis of pneumonitis is carried out using imaging teats such as chest X ray test and computerized tomography (CT) scan, blood test, pulmonary function test using oximeter or spirometer, bronchoscopy and surgical lung biopsy (Morisset et al. Citation2018). Corticosteroids and oxygen therapy are procedures used for treating chemical and hypersensitivity from pneumonitis (Helber et al. Citation2018).
Pathogenesis and symptoms of pneumonia
Pathogenesis of pneumonia
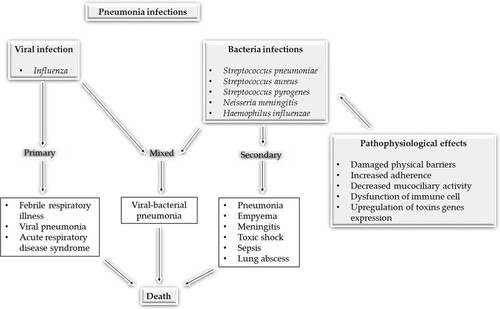
Pneumonia, in clinical term, is caused by either bacteria, viruses or fungi (Baral et al. Citation2018). Based on this, the aetiology of pneumonia can be divided into (i) community-acquired acute pneumonia which can be caused by Streptococcus pneumoniae, Haemophilus influenzae, Moraxella catarrhalis, Staphylococcus aureus, Legionella pneumophila, Enterobacteriaceae (Klebsiella pneumoniae and Pseudomonas spp.), (ii) community-acquired atypical pneumonia which is caused by Mycoplasma pneumonia, Chlymadia spp., Coxiella burnetti and viruses (Respiratory Syncytial Virus, Influenzae types A and B, Adenovirus known as SARS virus), (iii) hospital-acquired pneumonia which is caused by Gram-negative rod, Enterobacteriaceae spp., Staphylococcus aureus, and Pseudomonas spp., (iv) aspiration pneumonia (means loss of cough reflex occurring in alcoholics) which is caused by anaerobic oral flora such as (Bacteroides, prevotella, Fusobacterium, Pepto-streptococcus, admixed with aerobic bacteria), (v) chronic pneumonia which is caused by Nocardia, Actinomyces, granulomatous, Mycobacterium tuberculosis, atypical mycobacteria, Histoplasma capsulatum, Coccidiodes immitis, Blastomyces, Dermatidis and (vi) necrotizing pneumonia and lung abscesses caused by anaerobic bacteria with/or without mixed infection that occurs in immune-compromised patients as a result of Cytomegalovirus, Pneumocystis jiroveci, Mycobacterium avium-intracellulare, invasive Aspergillosis, invasive candidiasis infection (Heron Citation2018).
Pneumonia can result from one or more pathogens as highlighted above but some are common than others. The most common bacterial cause of community-acquired pneumonia (CAP) is S. pneumoniae (DeMuri et al. Citation2018) whilst influenzae viruses, followed by Respiratory syncytial virus are the commonest viral causes of pneumonia which are mostly severe and often, fatal. Fungal pneumonia can result from a condition, for example, a valley fever, brought about by the Coccidioides fungus (Viriyakosol et al. Citation2019). Aspiration pneumonia which is not infectious can show when an individual inhales food, fluids, or stomach substance into the lungs (Dragan et al. Citation2018). Hospital-acquired pneumonia can happen in patients being treated for other conditions, for instance, those connected to a respirator, or a breathing machine (Giuliano et al. Citation2018).
This can be strengthened by a recent state-of-the-art diagnostic techniques study for bacterial, viral, and fungal infections in which 38% of a specific pathogen was detected for community-acquired pneumonia (CAP) cases. The detection of one or more viruses was seen in 23% of cases and 11% of other bacteria. Bacterial and viral pathogen combinations were seen in 3% of cases with fungal and mycobacterial organisms accounting for 1% of cases. Human rhinoviruses were isolated in 9% of cases and influenza virus in 6% of cases (Jain et al. Citation2015). However, regardless of the cause, the signs and symptoms will be the same (Figures and ).
Figure 2. Pathophysiology of pneumonia adopted from flash share at https://www.slideshare.net/hasnahnoi/pneumonia-38313892.
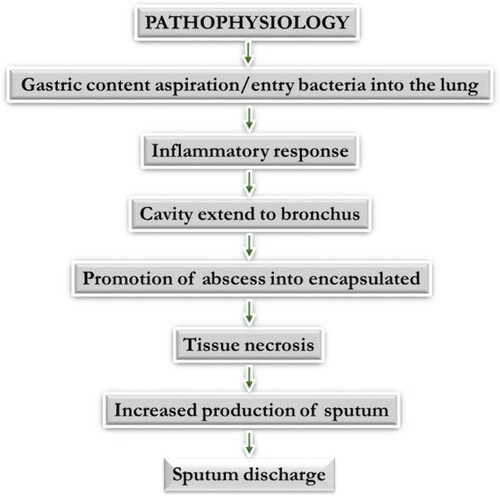
Figure 3. Pathogenesis of pneumonia disease using some bacterial pathogens and influenza as representative of bacterial and viral pneumonia pathogenesis. Adapted from Brundage (Citation2006).
However, only pneumonia caused by bacteria have definitive diagnosis, necessitating differentiation of bacterial from viral causative agents of the disease, with the effectiveness of the drugs and vaccines against the disease being controversial (Liu et al. Citation2015).
In addition, pneumonia is more severe in an immunocompromised individual with an underlying sickness (Messiaen et al. Citation2017). This is because the immune system is a host’s defense system comprising numerous biological structures and processes within an organism that protects against diseases. Pathogens can rapidly develop and adjust, and thereby avoid detection and neutralization by the immune system (Lee Citation2017). The capacity of the immune system to respond to pathogens is reduced in both the young and the elderly, with immune responses starting to decrease at around 50 years of age due to immuno-senescence (Rajpurkar et al. Citation2017). Underlying factors such as obesity, alcoholism, drug abuse, and malnutrition are common causes of poor immune function that can make an individual vulnerable to pneumonia infection (Nguyen et al. Citation2017).
Symptoms of pneumonia
The manifestations of pneumonia can shift from gentle to serious which may require hospitalization, contingent upon the type of germ causing the disease, population type, age and general well-being (Zhu et al. Citation2020). The first symptoms of pneumonia for the most part look like that of a cold or flu. The individual at that point builds up a high fever, chills, and cough with sputum. Other symptoms include phlegm/sputum coughed up from lungs, quick and difficult breathing, chest pain that generally declines when taking a full breath, known as pleuritic pain, quick heartbeat, fatigue and weakness, nausea and vomiting, diarrhea, sweating, headache, muscle pain, confusion or delirium, particularly in older adults, dusky or purplish skin colour, or cyanosis, from ineffectively oxygenated blood (Torres et al. Citation2018). These symptoms can differ contingent upon other fundamental conditions, the type of pneumonia and clinical history (Gu et al. Citation2018). An X-ray can show if there is any harm to the lungs. It is regular to speculate pneumonia if coarse breathing, wheezing, popping, or diminished breath sounds is noticed from listening to the chest through a stethoscope. The oxygen levels in the blood, with an painless monitor on the finger, called a heartbeat oximeter (Baral et al. Citation2018). However, it is always advised to carry out a diagnostic test to establish the status of a patient rather than a suspicion-based judgment.
Treatment and vaccination of pneumonia
The treatment of pneumonia relies upon the type and seriousness of pneumonia. Bacterial types of pneumonia are typically treated with antibiotics, contingent upon the kind of bacteria, while viral sorts have no conclusive treatment yet are normally treated with consumption of a lot of fluids, in any case, antiviral drugs can be utilized in influenza (Shields et al. Citation2018). Fungal types of pneumonia are typically treated with antifungal drugs (Schwartz et al. Citation2018). Specialists normally recommend over-the-counter (OTC) drugs to help deal with the symptoms of pneumonia, for example, fever, aches, pains, and coughs (Chen et al. Citation2018b). It is prescribed to rest and drink a lot of liquids to remain hydrated so as to disperse thick mucus and phlegm, making it simpler to cough up. Hospitalization for pneumonia might be required with the utilization of a supplemental oxygen if symptoms are particularly awful or if an individual has a debilitated immune system or different chronic ailments (Abbott et al. Citation2018). However, effective treatment is limited by sensitive and accurate diagnosis.
Two different vaccines exist to forestall bacterial pneumonia which covers a wide assortment of pneumococcal infections and is recommended for both children and adults, contingent upon their immune system capacity. Pneumococcal conjugate antibody (otherwise called Prevnar) and pneumococcal polysaccharide vaccine (known as Pneumovax) are utilized for prevention of bacterial pneumonia (Ngari et al. Citation2014; Andrews et al. Citation2019). Prevnar (PCV13) is ordinarily included as a major aspect of a newborn child’s normal vaccinations (Reglinski et al. Citation2018). It is recommended for kids under 2 years, adults more than 65 years, and those between the ages of 2 and 64 years with certain ailments. Pneumovax (PPSV23) is recommended for kids and adults who are at expanded danger of creating pneumococcal diseases (Parker et al. Citation2018). This incorporates adults aged 65 years or more, individuals with diabetes, those with terminal heart, lung, or kidney disease, individuals who devour a lot of alcohol or who smoke, and those without a spleen. Those within the range of 2 and 64 years with certain other ailments might be encouraged to have this immunization (McLaughlin et al. Citation2018). Also, there is a limitation to the use of vaccines as their efficacy is concentrated on pneumococcal pneumonia but not viral pneumonia (Demicheli et al. Citation2018). Along with vaccinations against bacterial pneumonia, physicians suggest frequent hand washing, covering of mouth and nose whilst coughing and sneezing, refraining from smoking, eating wholesomely, normal exercise and avoiding the sputum or cough particles of pneumonia patients. This goes to say that vaccination might not fully protect an individual from the disease.
Available methods for pneumonia diagnosis with their shortcomings
The core component of pneumonia etiology is diagnostics which largely relies on medical history, physical examination before an order for diagnostic test is established (Murdoch et al. Citation2012). The routine evaluation of patients for pneumonia diagnosis relies on certain methods used for decades which are based on microscopy and culture of respiratory tract specimens, blood cultures, and discovery of antigens in urine and respiratory specimens, and serological test (detection of specific antibodies in blood). Nucleic acid detection methods such as Polymerase chain reaction (PCR) are now established tools in diagnostic laboratories (Douglas Citation2016). The detection of a potential pathogen from upper and lower respiratory tract of a patient does not interpret pneumonia (Zar et al. Citation2017). This is because some pneumonia pathogens may colonize the upper airways of healthy individuals, thus a distinguishing feature of colonization from infection is very imperative, most especially from sputum sample (Iosifidis et al. Citation2018). A good method of diagnosis is very important for early onset of treatment, decreased resistance to antibiotics and reduced mortality. Such method should be cheap, sensitive, and accurate and give result in a short time (Marik and Kaplan Citation2003). This section therefore attempts to introduce some of these methods used in the diagnosis of bacterial and viral pneumonia, giving precise account of their use and stating their challenges which necessitate the quest for more sensitive options.
Old methods used for pneumonia diagnosis
The use of microscopy and culture of sputum or respiratory tract specimens (e.g. Broncho alveolar lavage fluid) and blood cultures have been used for the diagnosis of pneumonia (Kitsios et al. Citation2018). The identification of respiratory pathogens in these specimens collected from the lungs provide good evidence of a likely microorganism causing the pneumonia. Laboratory culture of specimens can be done on standard microbiological media such as combination of blood, chocolate and MacConkey agars (Moreno et al. Citation2018). Specimen collection should be checked for quality before diagnosis; for instance a high-quality expectorated sputum specimen in adult contains <10 squamous epithelial cells (SECs) and >25 polymorphonuclear cells (PMNs) per low power field (magnification *100) or >10 for each squamous epithelial cells (SECs) (Para et al. Citation2018). Sputum analysis is a phenotypic method that can determine which organism is causing pneumonia. A bronchoscopy is sometimes used for this investigation (Ngari et al. Citation2014). A thin, flexible, and lighted tube called a bronchoscope is passed down into the lungs. This enables the doctor to examine directly the infected parts of the airways and lungs.
Direct immunofluorescence microscopy and isolation in cell cultures are regarded as standard diagnostic approach for viral pneumonia. Blood culture is a phenotypic method that has been used for the detection of bacterial and viral pneumonia (Kermany et al. Citation2018). It is noteworthy to mention that blood tests can help to determine locally confined bacterium and virus causing pneumonia infection (Davis et al. Citation2017). Blood cultures may also reveal whether the microorganism from the lungs has spread into the bloodstream (Kalil et al. Citation2016).
Antigen detection assays has been used as point-of-care tests for pneumonia which relies on the discovery of suitable antigens in clinical specimens (Wunderink et al. Citation2018). However, commercial assays have only been developed for selected bacterial pneumonia pathogens such as Streptococcus pneumonia and Legionella pneumophila using immunochromatographic test of C-polysaccharide cell wall antigen in urine and viral pneumonia such influenza and Respiratory syncytial virus in respiratory specimens in which results depend on the viral types or subtypes, timing of specimen collection, specimen type, patient age and test comparator (Suzuki et al. Citation2017).
Chest X-rays are a phenotypic method that can be used for pneumonia diagnosis and will also show which areas of (Shen et al. Citation2020b) the lungs are affected (Figure (b)). A computerized tomography (CT) scan of the chest may provide more detailed information (Amatya et al. Citation2018).
Recent methods for pneumonia diagnosis
Nucleic acid detection can be used for the diagnosis of all pneumonia types with very low levels of nucleic acids (Shen et al. Citation2020b). It does not depend on the viability of the pathogens and can provide result within a clinically relevant time which is not affected by a previous antibiotic resistance genes. Commercial assays are now available with nucleic acid detection and have contributed to the understanding of the role of pathogens in the causation and microbial etiology of pneumonia (Wagner et al. Citation2018). PCR is a molecular method uses nucleic acid detection and it includes quantitative (qPCR) and reverse transcriptase (RT-PCR) assays which have been developed to diagnose the majority of bacterial and viral pneumonia pathogens such as Mycobacteria Influenzae viruses, Streptococcus pneumoniae and Chlamydia trachomatis in blood samples (Wagner et al. Citation2018; Freeman et al. Citation2019); and it is also replacing viral culture and serial viral antigens titre which are deemed problematic because of delay in result outcome (Freeman et al. Citation2019).
Apart from this, there is also a wide and useful epidemiological data for the use of antibody detection for pneumonia diagnosis (Cassirer et al. Citation2018). Serology has advantage over other methods such as PCR and cultures because it is more sensitive such as the case of a bacterial pneumonia subsequently triggered by a viral one because the viral pathogen may no longer be detectable in respiratory specimens by these methods (Nascimento-Carvalho et al. Citation2018). Immunoassay such as enzyme-linked immunosorbent assay (ELISA) is carried out for the diagnosis of bacterial and viral pneumonia using a receptor protein, that is highly purified as an antigen. For instance, the use of pneumolysin has been used as an antigen for Streptococcus pneumonia for its serological diagnosis using antibodies (Blanco-Covián et al. Citation2017).
Biomarkers used in assessment of pneumonia severity
Measurement of biomarkers such as procalcitonin (PCT), C-reactive protein (CRP), Soluble Triggering Receptor Expressed in Myeloid Cell-1 (STREM-1), copeptin, pro-ANP (atrial natriuretic peptide), adrenomedullin, cortisol and D-dimers is carried out using particular enhanced turbidimetric assay to monitor pneumonia severity index (severity scores) as well as CURB-65 (confusion, urea, respiratory rate scores, arterial blood pressure and age) can help decision making processes in hospitalization referral and ICU (Intensive Care Unit) for diagnosis, prognosis and follow-up of treatment of pneumonia. These biomarkers have proven useful in diagnosis as they are produced in considerably high concentration but there is ambiguity in their specificity towards pneumonia as they can be produced in response to other inflammatory stimuli in the neuron, atherosclerotic plaques, myocytes, and lymphocytes (Yang et al. Citation2018); whilst, the mechanism regulating their syntheses at these sites is not clearly understood (Tekerek et al. Citation2018). CRP, even though being nonspecific, has proven to help establish the etiology of some infections. A high CRP value (100 mg/L) can indicate a severe bacterial infection (Rawson et al. Citation2018). Streptococcus pneumoniae, viruses, Chlamydia pneumoniae, Mycoplasma pneumoniae, Legionella pneumophila, and Coxiella burnetii were detected from bronchoalveolar lavage (BAL) culture of the patients and the serum CRP values revealed: (1) high CRP values in pneumonia patients with S. pneumoniae or L. pneumophila; (2) increased CRP values with the severity of disease showing the use of CRP in pneumonia prognosis; (3) the use of CRP for the commencement of treatment procedure (Mendelson et al. Citation2018). CRP values are higher in certain pathogens than others and cannot differentiate viral from bacterial pneumonia (Chen et al. Citation2018a). Also, CRP does not allow for the distinguishing between all the types of pneumonia, its sensitivity is questionable. In a systematic review published in 2005, it was stated that ‘testing for C reactive protein is neither sufficiently sensitive to rule out nor sufficiently specific to rule in an infiltrate on chest radiograph and bacterial etiology of lower respiratory tract infection’ (Beltempo et al. Citation2018; Keramat et al. Citation2018;). However, the advantage of CRP is that its levels can be used for the follow-up of pneumonia as well as to evaluate patient management and response to antibiotic therapy (Beltempo et al. Citation2018).
Apart from this, PCT has been used as biomarker for sepsis and other infection since it satisfies all the required criteria such as (1) high specificity and sensitivity; (2) ready measurability; (3) less expensive and ready availability; (4) reproducibility; and (5) half-life of 24 hours (Stockmann et al. Citation2018). However, reduced sensitivity of PCT is observed due to its elevated concentration during non-infectious conditions such as trauma, surgery, cardiogenic shock, burns, heat, stroke, acute respiratory distress syndrome, infectious necrosis after acute pancreatitis, and rejection after transplantation (Jung et al. Citation2008). PCT sensitivity has been compared with CRP, interleukin-6 and interferon-alpha to detect possible bacterial CAP infection with PCT, showing high reliability more than others in the detection of CAP (Sager et al. Citation2017). However, the use of procalcitonin is difficulty in the diagnosis of pneumonia due to its nonspecific symptoms and may cause delay in treatment which may have fatal outcomes in these patients (Shehabi and Seppelt Citation2008).
The concentration of TREM-1 increases during inflammation such as sepsis and ventilator acquired pneumonia (VAP) but not in noninfectious inflammatory conditions like psoriasis, ulcerative colitis, and vasculitis (Kumar and Lodha Citation2018). Tejera et al. (Citation2007) found that serum STREM-1 is high in patients with CAP and that the prognostic value of STREM-1 is independent of age, other inflammatory markers such as IL-6, pneumonia severity index, sepsis, and nutritional status. The study found that patients who had increased STREM-1 levels on admission had the worst prognostic values.
Many biomarkers such as endotoxin, proadrenomedullin, natriuretic peptides, endothelin-1 precursor peptides, copeptin, and cortisol have been linked with the diagnosis and prognosis of pneumonia (Christ-Crain et al. Citation2008). These biomarkers are useful in guiding culture sampling, empirical antibiotics prescription, following the clinical course of the condition and identify those who do not respond to therapy (Ito et al. Citation2017). It is good to point out that while endotoxin is a diagnostic marker just like CRP, PCT, and sTREM-1, the other biomarkers mentioned (i.e. proadrenomedullin, natriuretic peptides, endothelin-1 precursor peptides, as well as copeptin and cortisol levels) have, up till now, only been found to be useful as prognostic markers and may be of great help in the risk stratification of patients.
Moreover, genetic markers such as single nucleotide polymorphs (SNP) and haplotypes such as CD143 rs4340 have been linked to increased risk of community-acquired pneumonia (CAP) (Schürch et al. Citation2018). Genetic factor outcomes related to cytokines expression may be influenced where patients with pulmonary sepsis were found to have specific IL-10 haplotype production with significantly higher mortality than patients who had alternative haplotypes (Wattanathum et al. Citation2005). Gene polymorphisms in human during regulated signal transduction events of the innate immune response, including Mal (Von Bernuth et al. Citation2008), IRAK-4 (Ku et al. Citation2007) and MyD88 (Khor et al. Citation2007), can increase the risk of invasive bacterial pneumonia. The non-specific nature of these human-mediated biomarkers in the diagnosis and prognosis of the severity and etiology of pneumonia necessitates the quest for new biomarkers which are cheaper, more sensitive, accurate and specific in nature.
Use of aptamers and antimicrobial peptides as biomarkers for potential use in pneumonia diagnosis
Aptamers have been designed in recent years for the detection of many difficult-to-treat cancer types against cancer specific proteins or whole cells (Molefe et al. Citation2018) and viral or bacterial pneumonia proteins in the serum and plasma of infected cells (Banerjee and Jaiswal Citation2018). Aptamers are short sequences of either DNA or RNA or peptide molecules that have been identified in vitro through several methods such as systematic evolution of ligands by exponential enrichment (SELEX) process (Park Citation2018). For instance, DNA aptamers probes have been used to detect S. pneumonia as low as 15 CFU/ml with the aid of SELEX which shows high specificity result against closely related streptococci and other bacteria (Bayrac and Donmez Citation2018). Tat and Rev are other aptamer examples used to recognize HIV proteins using Surface Plasmon Resonance (SPR), quartz crystal microbalance (QCM) and diamond field effect transistor (FET) techniques as biosensors whilst over 40 DNA and RNA aptamers kits have been developed for influenza viruses (González et al. Citation2016).
Furthermore, the use of antimicrobial peptides (AMPs), which was originally known for therapeutics, is also becoming increasingly popular in the field diagnostics (Torres et al. Citation2019). These peptides, once in a target microbial membrane, binds target cells through diverse mechanisms (Demaria et al. Citation2017). Several research work have been carried out on the diverse use of these molecules in diagnostics. For instance, the diagnostic use of cathelicidin AMP, LL-37, was carried out in congenital pneumonia utilizing its relationship with 25-hydroxycholeciferol where the result came out positive with a 93_% sensitivity and 86_% specificity (Gad et al. Citation2015). Another research where antimicrobial peptides were utilized for p24 detection in HIV, using in silico tools such as HMMER was the work of Tincho et al. (Citation2016) and were validated for use in a lateral flow device in a specific and sensitive manner (Williams et al. Citation2016) (Table ).
Table 1. Summary of some biomarkers used in pneumonia diagnosis.
In silico methods for prediction of aptamers and antimicrobial peptides for use in pneumonia diagnosis
It is imperative to explore other methods for the discovery of AMPs and optimization of the aptamer sequences obtained from SELEX because they might not give the required length during their in vitro generation, thus straining their specificity and sensitivity towards the receptors or manipulation during molecular conjugation on the matrix surfaces. Many computational methods have been introduced to predict AMPs and aptamers based on the different features such as the binary pattern method using AntiBP server, for prediction of antibacterial peptides (Veltri et al. Citation2018), the CAMP methods (Tucker et al. Citation2018), the combination of sequence alignment and the feature selection method (Beltran et al. Citation2018), and the pseudo amino acid composition method (Bhadra et al. Citation2018). Other methods for predicting peptides include Support Vector Machine (SVM), quantitative matrices (QM), and artificial neural network (ANN).
Unfortunately, AMPs have many variations in size with the aforementioned machine learning methods only working well on AMPs of fixed lengths (Lee et al. Citation2018). For the CAMP methods (Tucker et al. Citation2018), the AMPs prediction is performed using Random Forests (RF), SVM, and Discriminant Analysis (DA) based on all classes of full AMPs sequences while in silico tools like molecular docking and Molecular dynamics (MD) simulations are used for aptamers (Sabri et al. Citation2019a). The sequence alignment method enjoys high prediction accuracy but it is not able to predict all sequences (Yoshida et al. Citation2018). This is because the classification concept used in the sequence alignment relies on High Scoring Segment Pairs (HSPs) scores which represent the similarity scores between two sequences using BLASTP. If the test sequence has no relationship with any training sequence, an HSP score cannot be generated; thus the classification concept cannot be performed on that particular sequence. Incorporation of high through-put bioinformatics sequencing techniques (HTS), in silico aptamer and AMPs optimization, simulation, molecular dynamics and patterning of libraries would help to reduce the time for aptamer and AMP design, including their specificity (Kinghorn et al. Citation2017).
Following the discovery of aptamers and AMPs using in silico tools, molecular validation using in vitro study of these biomolecules for the selective index of the optimized forms for complex formation with the pathogen surface receptors in kits is important for diagnostics. Several biomarkers have been used for the development of diagnostic kits in pharmaceutical and biotechnology industries because they have advantages such as ease of operation, ready availability, portability and cost-effectiveness (Cho et al. Citation2013). The use of commercial immunoassay kits has been used to support drug and diagnostic development but their validation can be laborious (Nerenberg et al. Citation2018). Several studies have developed lateral flow devices for the diagnosis of pathogens such as invasive Aspergillus (Thornton Citation2008), malaria parasites (Tahar et al. Citation2013) just to mention a few. Six commercially available immuno-chromatographic lateral flow device (LFD) kits were recently tested for rabies of the brain (Eggerbauer et al. Citation2016). These developments can be useful in pneumonia diagnosis, if incorporated with in silico design of aptamers and AMPs, for accurate, sensitive and specific viral and bacterial pneumonia pathogens diagnostics.
Shortcomings and benefits of the methods of pneumonia diagnosis
Most developmental work is ongoing for the improvement of the nucleic acid and antigen detection to minimize the manual steps towards development of point-of-care test for pneumonia diagnosis. Several challenges such as specimen collection and contamination from the upper respiratory tract secretions, most especially sputum, have huge implication on the result outcome of microscopy and cultures which can lead to an incorrect conclusion that an upper airway colonizer is a pneumonia pathogen. A sputum with relatively low PMNs and high SECs indicates oropharyngeal contamination. Another shortcoming of blood culture for pneumonia diagnosis is that some patients with pneumonia have documented bloodstream infections.
In general, blood cultures have shortcomings such as poor sensitivity with further limitations observed in the strength of the agglutination, which is not indicative of the pathogen causing the pneumonia and patients have to wait for days before getting a result. A blood culture may also produce false negative and false positive results as seen in patients with high titres of rheumatoid factors which may give false-positive results (Pulido et al. Citation2018). However, chest X-ray test cannot tell what kind of germ is causing pneumonia (Rajpurkar et al. Citation2017). Bronchoscopy sputum analysis has shortcoming such as poor sensitivity and is time-consuming (Cilloniz et al. Citation2016).
One of the shortcomings of nucleic acid detection in pneumonia diagnosis is lack of suitable comparator gold standard, however, positive nucleic acid detection cannot be dismissed because of negative result from a sensitive comparator. The role of quantitative PCR needs to be clarified by using indices of certain clinical significance to distinguish carriage and disease as well as false result from upper respiratory tract infection and pneumonia. Another challenge is the detection of multiple pathogens using multiple sensitive testing methods such as multiplex PCR assays in a single patient which can present causality problem that requires further investigation into which one is responsible for pneumonia or whether all pathogens have a role in the pneumonia pathogenesis. PCR is prone to carryover contamination during manipulation and amplification of its products and it cannot provide anything other than supplemental data for the clinician; and there is an overwhelming lack of data on the efficacy of this method as a diagnostic method for other bacterial and viral pneumonia (Ahn et al. Citation2016). Immunoassay method is time-consuming, expensive and lacks sensitivity towards the diagnosis of certain bacterial and viral pathogens such as Haemophilus influenza, Legionella pneumophila, and Influenza A virus (Schiffner-Rohe et al. Citation2016). Analysis of the alveolar breath using non-invasive and easily repeatable biomarkers with minimal specimens from blood through passive diffusion may prove pivotal in mitigating the shortcomings of pneumonia diagnosis.
The use bioinformatics approaches offer new insights into designing alternative diagnostic kits for commercial use using aptamers and AMPs because a large number of bacterial strains and viruses escape detection with the aforementioned methods contributing to their shortcomings. Aptamer and AMP molecules are less expensive compared to antibodies and nucleic acid detection and can be modified chemically for optimum function (Jo et al. Citation2018). Other benefits of AMPs and aptamers such as small size, homogenous structure, high density and increased binding capacity make them potential candidates for sensitive and accurate diagnosis of disease pathogens, including pneumonia (Kaur Citation2018). The use of bioinformatics approach saves time and offers accuracy for the potential discovery of more biomarkers for the sensitive diagnosis of pneumonia and the severity of its conditions. However, the choice and combination of in silico methods for the prediction and design of aptamers and AMPs remain a bottleneck in achieving expectedly high sensitivity, accuracy and specificity results to effectively utilize them for pneumonia diagnostics.
Conclusion
This review gives insight into the diagnosis of bacterial and viral pneumonia with emphasis on methods and biomarkers used which would aid medical practitioners to carry out their work effectively and allow/enable patients to develop an accommodating lifestyles. The incorporation of in silico design and modelling of aptamers, as well as AMPs, as biomarkers would facilitate pneumonia diagnosis in an easy, fast, and sensitive manner, due to their advantages over the years in the field of diagnostics. This review would also provide information for improved awareness and understanding of pneumonia infection from clinical and immunological point of view that would eventually reduce the incidence of this disease by optimizing the use and discovery of these biomarkers for sensitive diagnosis. There is a promising perspective towards facilitation of collaborations with industries for sustainable investment to develop diagnostic kits from these biomarkers which offer the prospect of job creation and economic growth through the technologies developed from scarce-skilled fields such as Bioinformatics and Biomedical sciences.
Acknowledgements
Special appreciation to National Research Foundation (NRF) (grant number 112332), South Africa and South African National Zakat Fund (SANZAF). Conceived and designed the concept: AP, OOB; and wrote the paper: AOF, OOB; read and review the paper: MK, AOF, AK.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Abbott T, Fowler A, Pelosi P, De Abreu MG, Møller A, Canet J, Creagh-Brown B, Mythen M, Gin T, Lalu M. 2018. A systematic review and consensus definitions for standardised end-points in perioperative medicine: pulmonary complications. Br J Anaesth. 120:1066–1079. doi: https://doi.org/10.1016/j.bja.2018.02.007
- Ahn IE, Jerussi T, Farooqui M, Tian X, Wiestner A, Gea-Banacloche J. 2016. Atypical Pneumocystis jirovecii pneumonia in previously untreated patients with CLL on single-agent ibrutinib. Blood. 128:1940–1943. doi: https://doi.org/10.1182/blood-2016-06-722991
- Al-Ali A, Indorf KW. 2018. Pneumonia screener. Google Patents.
- Almeida A, Boattini M. 2017. Community-acquired pneumonia in HIV-positive patients: an update on etiologies, epidemiology and management. Curr Infect Dis Rep. 19:2. doi: https://doi.org/10.1007/s11908-017-0559-8
- Amatya Y, Rupp J, Russell FM, Saunders J, Bales B, House DR. 2018. Diagnostic use of lung ultrasound compared to chest radiograph for suspected pneumonia in a resource-limited setting. Int J Emerg Med. 11:8. doi: https://doi.org/10.1186/s12245-018-0170-2
- Andrews N, Kent A, Amin-Chowdhury Z, Sheppard C, Fry N, Ramsay M, Ladhani SN. 2019. Effectiveness of the seven-valent and thirteen-valent pneumococcal conjugate vaccines in England: the indirect cohort design, 2006–2018. Vaccine. 37:4491–4498. doi: https://doi.org/10.1016/j.vaccine.2019.06.071
- Bai HX, Hsieh B, Xiong Z, Halsey K, Choi JW, Tran TML, Pan I, Shi L-B, Wang D-C, Mei J. 2020a. Performance of radiologists in differentiating COVID-19 from viral pneumonia on chest CT. Radiology. 200823.
- Bai Y, Yao L, Wei T, Tian F, Jin D-Y, Chen L, Wang M. 2020b. Presumed asymptomatic carrier transmission of COVID-19. Jama.
- Banerjee R, Jaiswal A. 2018. Recent advances in nanoparticle-based lateral flow immunoassay as a point-of-care diagnostic tool for infectious agents and diseases. Analyst. 143:1970–1996. doi: https://doi.org/10.1039/C8AN00307F
- Baral P, Umans BD, Li L, Wallrapp A, Bist M, Kirschbaum T, Wei Y, Zhou Y, Kuchroo VK, Burkett PR. 2018. Nociceptor sensory neurons suppress neutrophil and γδ T cell responses in bacterial lung infections and lethal pneumonia. Nat Med. 24:417. doi: https://doi.org/10.1038/nm.4501
- Bayrac AT, Donmez SI. 2018. Selection of DNA aptamers to Streptococcus pneumonia and fabrication of graphene oxide based fluorescent assay. Anal Biochem. 556:91–98. doi: https://doi.org/10.1016/j.ab.2018.06.024
- Beltempo M, Viel-Thériault I, Thibeault R, Julien A-S, Piedboeuf B. 2018. C-reactive protein for late-onset sepsis diagnosis in very low birth weight infants. BMC Pediatr. 18:16. doi: https://doi.org/10.1186/s12887-018-1002-5
- Beltran JA, Aguilera-Mendoza L, Brizuela CA. 2018. Optimal selection of molecular descriptors for antimicrobial peptides classification: an evolutionary feature weighting approach. BMC Genomics. 19:672. doi: https://doi.org/10.1186/s12864-018-5030-1
- Bhadra P, Yan J, Li J, Fong S, Siu SW. 2018. AmPEP: sequence-based prediction of antimicrobial peptides using distribution patterns of amino acid properties and random forest. Sci Rep. 8:1–10. doi: https://doi.org/10.1038/s41598-018-19752-w
- Bhutta ZA, Das JK, Walker N, Rizvi A, Campbell H, Rudan I, Black RE. 2013. Interventions to address deaths from childhood pneumonia and diarrhoea equitably: what works and at what cost? The Lancet. 381:1417–1429. doi: https://doi.org/10.1016/S0140-6736(13)60648-0
- Blanco-Covián L, Montes-García V, Girard A, Fernández-Abedul MT, Pérez-Juste J, Pastoriza-Santos I, Faulds K, Graham D, Blanco-López MC. 2017. Au@ Ag SERRS tags coupled to a lateral flow immunoassay for the sensitive detection of pneumolysin. Nanoscale. 9:2051–2058. doi: https://doi.org/10.1039/C6NR08432J
- Bowman N, Caravati EM, Horowitz BZ, Crouch BI. 2018. Acute pneumonitis associated with nickel carbonyl exposure in the workplace. Clin Toxicol. 56:223–225. doi: https://doi.org/10.1080/15563650.2017.1355057
- Brundage JF. 2006. Interactions between influenza and bacterial respiratory pathogens: implications for pandemic preparedness. Lancet Infect Dis. 6:303–312. doi: https://doi.org/10.1016/S1473-3099(06)70466-2
- Cassirer EF, Manlove KR, Almberg ES, Kamath PL, Cox M, Wolff P, Roug A, Shannon J, Robinson R, Harris RB. 2018. Pneumonia in bighorn sheep: risk and resilience. J Wildl Manage. 82:32–45. doi: https://doi.org/10.1002/jwmg.21309
- Chen Y, Wu Q, Zhou C, Jin Q. 2018b. Facile preparation of Ce-doped TiO2/diatomite granular composite with enhanced photocatalytic activity. Adv Powder Technol. 29:106–116. doi: https://doi.org/10.1016/j.apt.2017.10.017
- Chen C, Yan M, Hu C, Lv X, Zhang H, Chen S. 2018a. Diagnostic efficacy of serum procalcitonin, C-reactive protein concentration and clinical pulmonary infection score in ventilator-associated pneumonia. Médecine/Sciences. 34:26–32. doi: https://doi.org/10.1051/medsci/201834f105
- Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, Qiu Y, Wang J, Liu Y, Wei Y. 2020. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. The Lancet. 395:507–513. doi: https://doi.org/10.1016/S0140-6736(20)30211-7
- Cho CH, Woo MK, Kim JY, Cheong S, Lee C-K, An SA, Lim CS, Kim WJ. 2013. Evaluation of five rapid diagnostic kits for influenza A/B virus. J Virol Methods. 187:51–56. doi: https://doi.org/10.1016/j.jviromet.2012.09.003
- Christ-Crain M, Breidthardt T, Stolz D, Zobrist K, Bingisser R, Miedinger D, Leuppi J, Tamm M, Mueller B, Mueller C. 2008. Use of B-type natriuretic peptide in the risk stratification of community-acquired pneumonia. J Intern Med. 264:166–176. doi: https://doi.org/10.1111/j.1365-2796.2008.01934.x
- Cilloniz C, Martin-Loeches I, Garcia-Vidal C, San Jose A, Torres A. 2016. Microbial etiology of pneumonia: Epidemiology, diagnosis and resistance patterns. Int J Mol Sci. 17:2120. doi: https://doi.org/10.3390/ijms17122120
- Curbelo J, Bueno SL, Galván-Román JM, Ortega-Gómez M, Rajas O, Fernández-Jiménez G, Vega-Piris L, Rodríguez-Salvanes F, Arnalich B, Díaz A. 2017. Inflammation biomarkers in blood as mortality predictors in community-acquired pneumonia admitted patients: importance of comparison with neutrophil count percentage or neutrophil-lymphocyte ratio. PLoS One. 12:e0173947. doi: https://doi.org/10.1371/journal.pone.0173947
- Davis TR, Evans HR, Murtas J, Weisman A, Francis JL, Khan A. 2017. Utility of blood cultures in children admitted to hospital with community-acquired pneumonia. J Paediatr Child Health. 53:232–236. doi: https://doi.org/10.1111/jpc.13376
- Demaria M, O’Leary MN, Chang J, Shao L, Liu S, Alimirah F, Koenig K, Le C, Mitin N, Deal AM. 2017. Cellular senescence promotes adverse effects of chemotherapy and cancer relapse. Cancer Discov. 7:165–176. doi: https://doi.org/10.1158/2159-8290.CD-16-0241
- Demicheli V, Jefferson T, Ferroni E, Rivetti A, Di Pietrantonj C. 2018. Vaccines for preventing influenza in healthy adults. Cochrane Database Syst Rev. 3:1–219.
- DeMuri GP, Gern JE, Eickhoff JC, Lynch SV, Wald ER. 2018. Dynamics of bacterial colonization with Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis during symptomatic and asymptomatic viral upper respiratory tract infection. Clin Infect Dis. 66:1045–1053. doi: https://doi.org/10.1093/cid/cix941
- Douglas IS. 2016. New diagnostic methods for pneumonia in the ICU. Curr Opin Infect Dis. 29(3):197–204. doi: https://doi.org/10.1097/QCO.0000000000000249
- Dragan V, Wei Y, Elligsen M, Kiss A, Walker SA, Leis JA. 2018. Prophylactic antimicrobial therapy for acute aspiration pneumonitis. Clin Infect Dis. 67:513–518. doi: https://doi.org/10.1093/cid/ciy120
- Eddabra R, Benhassou HA. 2018. Rapid molecular assays for detection of tuberculosis. Pneumonia. 10:4. doi: https://doi.org/10.1186/s41479-018-0049-2
- Eggerbauer E, de Benedictis P, Hoffmann B, Mettenleiter TC, Schlottau K, Ngoepe EC, Sabeta CT, Freuling CM, Müller T. 2016. Evaluation of six commercially available rapid immunochromatographic tests for the diagnosis of rabies in brain material. PLoS Negl Trop Dis. 10:1–16. doi: https://doi.org/10.1371/journal.pntd.0004776
- Fernández-Barat L, Ferrer M, De Rosa F, Gabarrús A, Esperatti M, Terraneo S, Rinaudo M, Bassi GL, Torres A. 2017. Intensive care unit-acquired pneumonia due to Pseudomonas aeruginosa with and without multidrug resistance. J Infect. 74:142–152. doi: https://doi.org/10.1016/j.jinf.2016.11.008
- Freeman AM, Soman-Faulkner K, Leigh Jr TR. 2019. Viral pneumonia. StatPearls [Internet]. StatPearls Publishing.
- Gad GI, Abushady NM, Fathi MS, Elsaadany W. 2015. Diagnostic value of anti-microbial peptide, cathelicidin in congenital pneumonia. J Matern Fetal Neonatal Med. 28:2197–2200. doi: https://doi.org/10.3109/14767058.2014.981806
- Giuliano KK, Baker D, Quinn B. 2018. The epidemiology of nonventilator hospital-acquired pneumonia in the United States. Am J Infect Contr. 46:322–327. doi: https://doi.org/10.1016/j.ajic.2017.09.005
- González VM, Martín ME, Fernández G, García-Sacristán A. 2016. Use of aptamers as diagnostics tools and antiviral agents for human viruses. Pharmaceuticals. 9:78. doi: https://doi.org/10.3390/ph9040078
- Gu D, Dong N, Zheng Z, Lin D, Huang M, Wang L, Chan EW-C, Shu L, Yu J, Zhang R. 2018. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis. 18:37–46. doi: https://doi.org/10.1016/S1473-3099(17)30489-9
- Helber HA, Hada AL, Pio RB, Moraes PHZD, Gomes DBD. 2018. Immunotherapy-induced pneumonitis: cases report. Einstein (Sao Paulo). 16:1–5. doi: https://doi.org/10.1590/s1679-45082018rc4030
- Heron MP. 2018. Deaths: leading causes for 2016.
- Hu Z, Tan J, Lai Z, Zheng R, Zhong J, Wang Y, Li X, Yang N, Li J, Yang W. 2017. Aptamer combined with fluorescent silica nanoparticles for detection of hepatoma cells. Nanoscale Res Lett. 12:96. doi: https://doi.org/10.1186/s11671-017-1890-6
- Iosifidis E, Pitsava G, Roilides E. 2018. Ventilator-associated pneumonia in neonates and children: a systematic analysis of diagnostic methods and prevention. Future Microbiol. 13:1431–1446. doi: https://doi.org/10.2217/fmb-2018-0108
- Ito A, Ishida T, Tokumasu H, Washio Y, Yamazaki A, Ito Y, Tachibana H. 2017. Prognostic factors in hospitalized community-acquired pneumonia: a retrospective study of a prospective observational cohort. BMC Pulm Med. 17:78. doi: https://doi.org/10.1186/s12890-017-0424-4
- Jain S, Self WH, Wunderink RG, Fakhran S, Balk R, Bramley AM, Reed C, Grijalva CG, Anderson EJ, Courtney DM. 2015. Community-acquired pneumonia requiring hospitalization among US adults. N Engl J Med. 373:415–427. doi: https://doi.org/10.1056/NEJMoa1500245
- Jo N, Kim B, Lee S-M, Oh J, Park IH, Lim KJ, Shin J-S, Yoo K-H. 2018. Aptamer-functionalized capacitance sensors for real-time monitoring of bacterial growth and antibiotic susceptibility. Biosens Bioelectron. 102:164–170. doi: https://doi.org/10.1016/j.bios.2017.11.010
- Jung D-Y, Park JB, Lee E-N, Lee H-A, Joh J-W, Kwon CH, Ki C-S, Lee S-Y, Kim S-J. 2008. Combined use of myeloid-related protein 8/14 and procalcitonin as diagnostic markers for acute allograft rejection in kidney transplantation recipients. Transpl Immunol. 18:338–343. doi: https://doi.org/10.1016/j.trim.2007.10.004
- Kalil AC, Metersky ML, Klompas M, Muscedere J, Sweeney DA, Palmer LB, Napolitano LM, O’Grady NP, Bartlett JG, Carratalà J. 2016. Management of adults with hospital-acquired and ventilator-associated pneumonia: 2016 clinical practice guidelines by the infectious diseases society of America and the American thoracic society. Clin Infect Dis. 63:e61–e111. doi: https://doi.org/10.1093/cid/ciw353
- Kaur H. 2018. Recent developments in cell-SELEX technology for aptamer selection. Biochim Biophys Acta. 1862:2323–2329. doi: https://doi.org/10.1016/j.bbagen.2018.07.029
- Keramat F, Basir HRG, Abdoli E, Aghdam AS, Poorolajal J. 2018. Association of serum procalcitonin and C-reactive protein levels with CURB-65 criteria among patients with community-acquired pneumonia. Int J Gen Med. 11:217. doi: https://doi.org/10.2147/IJGM.S165190
- Kermany DS, Goldbaum M, Cai W, Valentim CC, Liang H, Baxter SL, McKeown A, Yang G, Wu X, Yan F. 2018. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 172:1122–1131. doi: https://doi.org/10.1016/j.cell.2018.02.010
- Khor CC, Chapman SJ, Vannberg FO, Dunne A, Murphy C, Ling EY, Frodsham AJ, Walley AJ, Kyrieleis O, Khan A. 2007. A Mal functional variant is associated with protection against invasive pneumococcal disease, bacteremia, malaria and tuberculosis. Nat Genet. 39:523. doi: https://doi.org/10.1038/ng1976
- Kim M, Lee SM, Song J-W, Do K-H, Lee HJ, Lim S, Choe J, Park KJ, Park HJ, Kim HJ. 2017. Added value of prone CT in the assessment of honeycombing and classification of usual interstitial pneumonia pattern. Eur J Radiol. 91:66–70. doi: https://doi.org/10.1016/j.ejrad.2017.03.018
- Kinghorn AB, Fraser LA, Liang S, Shiu SC-C, Tanner JA. 2017. Aptamer bioinformatics. Int J Mol Sci. 18:2516. doi: https://doi.org/10.3390/ijms18122516
- Kitsios GD, Fitch A, Manatakis DV, Rapport SF, Li K, Qin S, Huwe J, Zhang Y, Evankovich J, Bain W. 2018. Respiratory microbiome profiling for etiologic diagnosis of pneumonia in mechanically ventilated patients. Front Microbiol. 9:1413. doi: https://doi.org/10.3389/fmicb.2018.01413
- Ku C-L, Von Bernuth H, Picard C, Zhang S-Y, Chang H-H, Yang K, Chrabieh M, Issekutz AC, Cunningham CK, Gallin J. 2007. Selective predisposition to bacterial infections in IRAK-4–deficient children: IRAK-4–dependent TLRs are otherwise redundant in protective immunity. J Exp Med. 204:2407–2422. doi: https://doi.org/10.1084/jem.20070628
- Kumar A, Lodha R. 2018. Biomarkers for diagnosing ventilator associated pneumonia: is that the way forward? Indian J Pediatr. 85:411–412. doi: https://doi.org/10.1007/s12098-018-2672-6
- Lee K-Y. 2017. Pneumonia, acute respiratory distress syndrome, and early immune-modulator therapy. Int J Mol Sci. 18:388. doi: https://doi.org/10.3390/ijms18020388
- Lee EY, Wong GC, Ferguson AL. 2018. Machine learning-enabled discovery and design of membrane-active peptides. Bioorg Med Chem. 26:2708–2718. doi: https://doi.org/10.1016/j.bmc.2017.07.012
- Leijte GP, Rimmelé T, Kox M, Bruse N, Monard C, Gossez M, Monneret G, Pickkers P, Venet F. 2020. Monocytic HLA-DR expression kinetics in septic shock patients with different pathogens, sites of infection and adverse outcomes. Crit Care. 24:1–9. doi: https://doi.org/10.1186/s13054-020-2830-x
- Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, Ren R, Leung KS, Lau EH, Wong JY. 2020. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N Engl J Med. 33(4):997–1001.
- Lin C-J, Chang Y-C, Tsou M-T, Chan H-L, Chen Y-J, Hwang L-C. 2020. Factors associated with hospitalization for community-acquired pneumonia in home health care patients in Taiwan. Aging Clin Exp Res. 32:149–155. doi: https://doi.org/10.1007/s40520-019-01169-8
- Ling W. 2020. C-reactive protein levels in the early stage of COVID-19. Med Mal Infect.
- Lippi G, Plebani M. 2020. Procalcitonin in patients with severe coronavirus disease 2019 (COVID-19): A meta-analysis. Clin Chim Acta.
- Liu H, Grantham ML, Pekosz A. 2018. Mutations in the Influenza A virus M1 protein enhance virus budding to complement lethal mutations in the M2 cytoplasmic tail. J Virol. 92:e00858–17.
- Liu L, Oza S, Hogan D, Perin J, Rudan I, Lawn JE, Cousens S, Mathers C, Black RE. 2015. Global, regional, and national causes of child mortality in 2000–13, with projections to inform post-2015 priorities: an updated systematic analysis. The Lancet. 385:430–440. doi: https://doi.org/10.1016/S0140-6736(14)61698-6
- Marik PE, Kaplan D. 2003. Aspiration pneumonia and dysphagia in the elderly. Chest. 124:328–336. doi: https://doi.org/10.1378/chest.124.1.328
- McLaughlin JM, Jiang Q, Isturiz RE, Sings HL, Swerdlow DL, Gessner BD, Carrico RM, Peyrani P, Wiemken TL, Mattingly WA. 2018. Effectiveness of 13-valent pneumococcal conjugate vaccine against hospitalization for community-acquired pneumonia in older US adults: a test-negative design. Clin Infect Dis. 67:1498–1506.
- Mendelson F, Griesel R, Tiffin N, Rangaka M, Boulle A, Mendelson M, Maartens G. 2018. C-reactive protein and procalcitonin to discriminate between tuberculosis, Pneumocystis jirovecii pneumonia, and bacterial pneumonia in HIV-infected inpatients meeting WHO criteria for seriously ill: a prospective cohort study. BMC Infect Dis. 18:1–11. doi: https://doi.org/10.1186/s12879-018-3303-6
- Messiaen PE, Cuyx S, Dejagere T, van der Hilst JC. 2017. The role of CD 4 cell count as discriminatory measure to guide chemoprophylaxis against Pneumocystis jirovecii pneumonia in human immunodeficiency virus-negative immunocompromised patients: a systematic review. Transpl Infect Dis. 19:e12651. doi: https://doi.org/10.1111/tid.12651
- Molefe PF, Masamba P, Oyinloye BE, Mbatha LS, Meyer M, Kappo AP. 2018. Molecular application of aptamers in the diagnosis and treatment of cancer and communicable diseases. Pharmaceuticals. 11:93. doi: https://doi.org/10.3390/ph11040093
- Momosaki R, Yasunaga H, Matsui H, Horiguchi H, Fushimi K, Abo M. 2016. Predictive factors for oral intake after aspiration pneumonia in older adults. Geriatr Gerontol Int. 16:556–560. doi: https://doi.org/10.1111/ggi.12506
- Moreno I, Cicinelli E, Garcia-Grau I, Gonzalez-Monfort M, Bau D, Vilella F, De Ziegler D, Resta L, Valbuena D, Simon C. 2018. The diagnosis of chronic endometritis in infertile asymptomatic women: a comparative study of histology, microbial cultures, hysteroscopy, and molecular microbiology. Am J Obstet Gynecol. 218(602):e1–602.
- Morisset J, Johannson KA, Jones KD, Wolters PJ, Collard HR, Walsh SL, Ley B. 2018. Identification of diagnostic criteria for chronic hypersensitivity pneumonitis. An international modified Delphi survey. Am J Respir Crit Care Med. 197:1036–1044. doi: https://doi.org/10.1164/rccm.201710-1986OC
- Murdoch DR, O’Brien KL, Driscoll AJ, Karron RA, Bhat N, Group PMW, Team PC. 2012. Laboratory methods for determining pneumonia etiology in children. Clin Infect Dis. 54:S146–S152. doi: https://doi.org/10.1093/cid/cir1073
- Nascimento-Carvalho AC, Vilas-Boas AL, Fontoura MSH, Xu M, Vuorinen T, Söderlund-Venermo M, Ruuskanen O, Nascimento-Carvalho CM, Group PES. 2018. Serologically diagnosed acute human bocavirus 1 infection in childhood community-acquired pneumonia. Pediatr Pulmonol. 53:88–94. doi: https://doi.org/10.1002/ppul.23891
- Nerenberg KA, Zarnke KB, Leung AA, Dasgupta K, Butalia S, McBrien K, Harris KC, Nakhla M, Cloutier L, Gelfer M. 2018. Hypertension Canada’s 2018 guidelines for diagnosis, risk assessment, prevention, and treatment of hypertension in adults and children. Can J Cardiol. 34:506–525. doi: https://doi.org/10.1016/j.cjca.2018.02.022
- Ngari CG, Malonza DM, Muthuri GG. 2014. A model for childhood pneumonia dynamics.
- Nguyen T, Tran T, Roberts C, Fox G, Graham S, Marais B. 2017. Risk factors for child pneumonia-focus on the Western Pacific Region. Paediatr Respir Rev. 21:95–101.
- Para RA, Fomda BA, Jan RA, Shah S, Koul PA. 2018. Microbial etiology in hospitalized North Indian adults with community-acquired pneumonia. Lung India. 35:108.
- Park KS. 2018. Nucleic acid aptamer-based methods for diagnosis of infections. Biosens Bioelectron. 102:179–188. doi: https://doi.org/10.1016/j.bios.2017.11.028
- Parker AR, Bradley C, Harding S, Sánchez-Ramón S, Jolles S, Kiani-Alikhan S. 2018. Measurement and interpretation of Salmonella typhi Vi IgG antibodies for the assessment of adaptive immunity. J Immunol Methods. 459:1–10. doi: https://doi.org/10.1016/j.jim.2018.05.013
- Pulido M, Moreno-Martínez P, González-Galán V, Cuenca FF, Pascual Á, Garnacho-Montero J, Antonelli M, Dimopoulos G, Lepe J, McConnell M. 2018. Application of BioFire FilmArray blood culture identification panel for rapid identification of the causative agents of ventilator-associated pneumonia. Clin Microbiol Infect. 24:1213. doi: https://doi.org/10.1016/j.cmi.2018.06.001
- Rajpurkar P, Irvin J, Zhu K, Yang B, Mehta H, Duan T, Ding D, Bagul A, Langlotz C, Shpanskaya K. 2017. Chexnet: radiologist-level pneumonia detection on chest x-rays with deep learning. arXiv preprint arXiv:1711.05225.
- Ramirez JA, Wiemken TL, Peyrani P, Arnold FW, Kelley R, Mattingly WA, Nakamatsu R, Pena S, Guinn BE, Furmanek SP. 2017. Adults hospitalized with pneumonia in the United States: incidence, epidemiology, and mortality. Clin Infect Dis. 65:1806–1812. doi: https://doi.org/10.1093/cid/cix647
- Rawson TM, Charani E, Moore LS, Gilchrist M, Georgiou P, Hope W, Holmes AH. 2018. Exploring the use of C-reactive protein to estimate the pharmacodynamics of vancomycin. Ther Drug Monit. 40:315. doi: https://doi.org/10.1097/FTD.0000000000000507
- Reglinski M, Ercoli G, Plumptre C, Kay E, Petersen FC, Paton JC, Wren BW, Brown JS. 2018. A recombinant conjugated pneumococcal vaccine that protects against murine infections with a similar efficacy to Prevnar-13. NPJ Vaccines. 3:1–11. doi: https://doi.org/10.1038/s41541-018-0090-4
- Rudan I, Lawn J, Cousens S, Rowe AK, Boschi-Pinto C, Tomašković L, Mendoza W, Lanata CF, Roca-Feltrer A, Carneiro I. 2005. Gaps in policy-relevant information on burden of disease in children: a systematic review. The Lancet. 365:2031–2040. doi: https://doi.org/10.1016/S0140-6736(05)66697-4
- Sabri MZ, Abdul Hamid AA, Sayed Hitam SM, Rahim A, Zulkhairi M. 2019a. In silico screening of aptamers configuration against hepatitis B surface antigen. Adv Bioinform. 2019. doi: https://doi.org/10.1155/2019/6912914
- Sabri MZ, Hamid A, Azzar A, Sayed Hitam SM, Rahim A, Zulkhairi M. 2019b. In silico screening of aptamers configuration against hepatitis B surface antigen. Adv Bioinform. 2019. doi: https://doi.org/10.1155/2019/6912914
- Sager R, Kutz A, Mueller B, Schuetz P. 2017. Procalcitonin-guided diagnosis and antibiotic stewardship revisited. BMC Med. 15:15. doi: https://doi.org/10.1186/s12916-017-0795-7
- Salisbury ML, Gu T, Murray S, Gross BH, Chughtai A, Sayyouh M, Kazerooni EA, Myers JL, Lagstein A, Konopka KE. 2019. Hypersensitivity pneumonitis: radiologic phenotypes are associated with distinct survival time and pulmonary function trajectory. Chest. 155:699–711. doi: https://doi.org/10.1016/j.chest.2018.08.1076
- Schiffner-Rohe J, Witt A, Hemmerling J, von Eiff C, Leverkus F-W. 2016. Efficacy of PPV23 in preventing pneumococcal pneumonia in adults at increased risk – a systematic review and meta-analysis. PloS one. 11:e0146338. doi: https://doi.org/10.1371/journal.pone.0146338
- Schürch A, Arredondo-Alonso S, Willems R, Goering RV. 2018. Whole genome sequencing options for bacterial strain typing and epidemiologic analysis based on single nucleotide polymorphism versus gene-by-gene-based approaches. Clin Microbiol Infect. 24:350–354. doi: https://doi.org/10.1016/j.cmi.2017.12.016
- Schwartz S, Kontoyiannis DP, Harrison T, Ruhnke M. 2018. Advances in the diagnosis and treatment of fungal infections of the CNS. Lancet Neurol. 17:362–372. doi: https://doi.org/10.1016/S1474-4422(18)30030-9
- Shehabi Y, Seppelt I. 2008. Pro/Con debate: is procalcitonin useful for guiding antibiotic decision making in critically ill patients? Crit Care. 12:211. doi: https://doi.org/10.1186/cc6860
- Shen K, Yang Y, Wang T, Zhao D, Jiang Y, Jin R, Zheng Y, Xu B, Xie Z, Lin L. 2020a. Diagnosis, treatment, and prevention of 2019 novel coronavirus infection in children: experts’ consensus statement. World J Pediatr. 1–9.
- Shen M, Zhou Y, Ye J, Al-Maskri AAA, Kang Y, Zeng S, Cai S. 2020b. Recent advances and perspectives of nucleic acid detection for coronavirus. J Pharm Anal.
- Shields RK, Nguyen MH, Chen L, Press EG, Kreiswirth BN, Clancy CJ. 2018. Pneumonia and renal replacement therapy are risk factors for ceftazidime-avibactam treatment failures and resistance among patients with carbapenem-resistant Enterobacteriaceae infections. Antimicrob Agents Chemother. 62:e02497–17.
- Slagman A, Searle J, Müller C, Möckel M. 2015. Temporal release pattern of copeptin and troponin T in patients with suspected acute coronary syndrome and spontaneous acute myocardial infarction. Clin Chem. 61:1273–1282. doi: https://doi.org/10.1373/clinchem.2015.240580
- Sommerfeld CG, Weiner DJ, Nowalk A, Larkin A. 2018. Hypersensitivity pneumonitis and acute respiratory distress syndrome from e-cigarette use. Pediatrics. 141:e20163927. doi: https://doi.org/10.1542/peds.2016-3927
- Son YG, Shin J, Ryu HG. 2017. Pneumonitis and pneumonia after aspiration. J Dental Anesthes Pain Med. 17:1–12. doi: https://doi.org/10.17245/jdapm.2017.17.1.1
- Stockmann C, Ampofo K, Killpack J, Williams DJ, Edwards KM, Grijalva CG, Arnold SR, McCullers JA, Anderson EJ, Wunderink RG. 2018. Procalcitonin accurately identifies hospitalized children with low risk of bacterial community-acquired pneumonia. J Pediatric Infect Dis Soc. 7:46–53. doi: https://doi.org/10.1093/jpids/piw091
- Suzuki M, Dhoubhadel BG, Ishifuji T, Yasunami M, Yaegashi M, Asoh N, Ishida M, Hamaguchi S, Aoshima M, Ariyoshi K. 2017. Serotype-specific effectiveness of 23-valent pneumococcal polysaccharide vaccine against pneumococcal pneumonia in adults aged 65 years or older: a multicentre, prospective, test-negative design study. Lancet Infect Dis. 17:313–321. doi: https://doi.org/10.1016/S1473-3099(17)30049-X
- Tahar R, Sayang C, Foumane VN, Soula G, Moyou-Somo R, Delmont J, Basco LK. 2013. Field evaluation of rapid diagnostic tests for malaria in Yaounde, Cameroon. Acta Trop. 125:214–219. doi: https://doi.org/10.1016/j.actatropica.2012.10.002
- Tejera A, Santolaria F, Diez M-L, Alemán-Valls M-R, González-Reimers E, Martínez-Riera A, Milena-Abril A. 2007. Prognosis of community acquired pneumonia (CAP): value of triggering receptor expressed on myeloid cells-1 (TREM-1) and other mediators of the inflammatory response. Cytokine. 38:117–123. doi: https://doi.org/10.1016/j.cyto.2007.05.002
- Tekerek NU, Akyildiz BN, Ercal BD, Muhtaroglu S. 2018. New biomarkers to diagnose ventilator associated pneumonia: pentraxin 3 and surfactant protein D. Indian J Pediatr. 85:426–432. doi: https://doi.org/10.1007/s12098-018-2607-2
- Thornton CR. 2008. Development of an immunochromatographic lateral-flow device for rapid serodiagnosis of invasive aspergillosis. Clin Vaccine Immunol 15:1095–1105. doi: https://doi.org/10.1128/CVI.00068-08
- Tincho M, Gabere M, Pretorius A. 2016. In silico identification and molecular validation of putative antimicrobial peptides for HIV therapy. J AIDS Clin Res. 7. doi: https://doi.org/10.4172/2155-6113.1000606
- Tjalvin G, Svanes Ø, Bertelsen RJ, Hollund BE, Aasen TB, Svanes C, Kirkeleit J. 2018. Hypersensitivity pneumonitis in fish processing workers diagnosed by inhalation challenge. ERJ Open Res. 4:00071–02018. doi: https://doi.org/10.1183/23120541.00071-2018
- Tong S, Amand C, Kieffer A, Kyaw MH. 2018. Trends in healthcare utilization and costs associated with pneumonia in the United States during 2008–2014. BMC Health Serv Res. 18:1–8.
- Torres MD, Sothiselvam S, Lu TK, de la Fuente-Nunez C. 2019. Peptide design principles for antimicrobial applications. J Mol Biol. 431:3547–3567. doi: https://doi.org/10.1016/j.jmb.2018.12.015
- Torres A, Zhong N, Pachl J, Timsit J-F, Kollef M, Chen Z, Song J, Taylor D, Laud PJ, Stone GG. 2018. Ceftazidime-avibactam versus meropenem in nosocomial pneumonia, including ventilator-associated pneumonia (REPROVE): a randomised, double-blind, phase 3 non-inferiority trial. Lancet Infect Dis. 18:285–295. doi: https://doi.org/10.1016/S1473-3099(17)30747-8
- Tucker AT, Leonard SP, DuBois CD, Knauf GA, Cunningham AL, Wilke CO, Trent MS, Davies BW. 2018. Discovery of next-generation antimicrobials through bacterial self-screening of surface-displayed peptide libraries. Cell. 172:618–628. doi: https://doi.org/10.1016/j.cell.2017.12.009
- Veltri D, Kamath U, Shehu A. 2018. Deep learning improves antimicrobial peptide recognition. Bioinformatics. 34:2740–2747. doi: https://doi.org/10.1093/bioinformatics/bty179
- Viriyakosol S, Kapoor M, Okamoto S, Covel J, Soltow QA, Trzoss M, Shaw KJ, Fierer J. 2019. APX001 and other Gwt1 inhibitor prodrugs are effective in experimental Coccidioides immitis pneumonia. Antimicrob Agents Chemother. 63:e01715–18.
- Von Bernuth H, Picard C, Jin Z, Pankla R, Xiao H, Ku C-L, Chrabieh M, Mustapha IB, Ghandil P, Camcioglu Y. 2008. Pyogenic bacterial infections in humans with MyD88 deficiency. Science. 321:691–696. doi: https://doi.org/10.1126/science.1158298
- Wagner K, Springer B, Imkamp F, Opota O, Greub G, Keller PM. 2018. Detection of respiratory bacterial pathogens causing atypical pneumonia by multiplex Lightmix® RT-PCR. Int J Med Microbiol. 308:317–323. doi: https://doi.org/10.1016/j.ijmm.2018.01.010
- Wattanathum A, Manocha S, Groshaus H, Russell JA, Walley KR. 2005. Interleukin-10 haplotype associated with increased mortality in critically ill patients with sepsis from pneumonia but not in patients with extrapulmonary sepsis. Chest. 128:1690–1698. doi: https://doi.org/10.1378/chest.128.3.1690
- Wazny K, Zipursky A, Black R, Curtis V, Duggan C, Guerrant R, Levine M, Petri Jr WA, Santosham M, Scharf R. 2013. Setting research priorities to reduce mortality and morbidity of childhood diarrhoeal disease in the next 15 years. PLoS Med. 10:e1001446. doi: https://doi.org/10.1371/journal.pmed.1001446
- Williams M, Tincho M, Gabere M, Uys A, Meyer M, Pretorius A. 2016. Molecular validation of putative antimicrobial peptides for improved human immunodeficiency virus diagnostics via HIV protein p24. J AIDS Clin Res. 7:571.
- Wunderink RG, Self WH, Anderson EJ, Balk R, Fakhran S, Courtney DM, Qi C, Williams DJ, Zhu Y, Whitney CG. 2018. Pneumococcal community-acquired pneumonia detected by serotype-specific urinary antigen detection assays. Clin Infect Dis. 66:1504–1510. doi: https://doi.org/10.1093/cid/cix1066
- Yang Y, Hao Q, Flaherty JH, Cao L, Zhou J, Su L, Shen Y, Dong B. 2018. Comparison of procalcitonin, a potentially new inflammatory biomarker of frailty, to interleukin-6 and C-reactive protein among older Chinese hospitalized patients. Aging Clin Exp Res. 30:1459–1464. doi: https://doi.org/10.1007/s40520-018-0964-3
- Yoshida M, Hinkley T, Tsuda S, Abul-Haija YM, McBurney RT, Kulikov V, Mathieson JS, Reyes SG, Castro MD, Cronin L. 2018. Using evolutionary algorithms and machine learning to explore sequence space for the discovery of antimicrobial peptides. Chem. 4:533–543. doi: https://doi.org/10.1016/j.chempr.2018.01.005
- Zar HJ, Andronikou S, Nicol MP. 2017. Advances in the diagnosis of pneumonia in children. Br Med J. 358:j2739. doi: https://doi.org/10.1136/bmj.j2739
- Zeng H, Xu C, Fan J, Tang Y, Deng Q, Zhang W, Long X. 2020. Antibodies in infants born to mothers with COVID-19 pneumonia. Jama.
- Zhu H, Wang L, Fang C, Peng S, Zhang L, Chang G, Xia S, Zhou W. 2020. Clinical analysis of 10 neonates born to mothers with 2019-nCoV pneumonia. Transl Pediatr. 9:51. doi: https://doi.org/10.21037/tp.2020.02.06
- Zilberberg MD, Nathanson BH, Sulham K, Fan W, Shorr AF. 2017. Carbapenem resistance, inappropriate empiric treatment and outcomes among patients hospitalized with Enterobacteriaceae urinary tract infection, pneumonia and sepsis. BMC Infect Dis. 17:279. doi: https://doi.org/10.1186/s12879-017-2383-z
