Abstract
Colon cancer (CRC) is one of the leading causes of tumor lethality in the world. CRC is mostly diagnosed at advanced stages of disease, and chemotherapy is one of the major options in treating the advanced CRC patients. However, the development of multi-drug resistance has become the obstacle to an effective control of CRC clinically. Thus, it is essentially significant for scientists and medical doctors to explore new strategies for CRC therapy. CircRNA_100920 expression was upregulated in CRC tissues and cells. Knockdown of circRNA_100920 inhibited cell proliferation in CRC cells. Clinically, circRNA_100920 was associated with survival rate, metastasis, TNM stage and tumor size. Notably, circRNA_100920 could become a potential biomarker by the receiver operating characteristic curve analysis. In addition, circRNA_100920 enhanced oncolytic adenovirus therapy in colon cancer by suppressing IFN-γ expression. CircRNA_100920, as a biomarker, may participate in the diagnosis, treatment, and prognosis of CRC.
Introduction
Colon cancer (CRC) is one of the leading causes of tumor lethality in the world (Kuipers et al. Citation2015; Siegel et al. Citation2017; Citation2019). CRC is mostly diagnosed at advanced stages of disease, and chemotherapy is one of the major options in treating the advanced CRC patients (Arraras Urdaniz et al. Citation2006; Roque et al. Citation2009; Rougier et al. Citation1997). However, the development of multi-drug resistance has become the obstacle to an effective control of CRC clinically. Thus, it is essentially significant for scientists and medical doctors to explore new strategies for CRC therapy.
In exploring for new strategies for CRC therapy, oncolytic adenovirus has shown potential and effective anti-tumor effect. For example, CRC therapy with oncolytic adenovirus is enhanced by the addition of herpes simplex virus-thymidine kinase (Wildner et al. Citation1999). A novel E1B55kDa-deleted oncolytic adenovirus, carrying microRNA-143, exerts specific antitumor efficacy on colorectal cancer cells (Luo et al. Citation2016). CD133-targeted oncolytic adenovirus demonstrates anti-tumor effect in CRC (Sato-Dahlman et al. Citation2017). Thus, it is of great to explore how to enhance therapy effect of oncolytic adenovirus.
Circular RNAs (circRNAs) are defined as a group of ncRNAs with covalently connected 3′ and 5′ ends (Jeck and Sharpless Citation2014). Due to the functional role of circRNAs in various diseases, including cancer, it has recently received widespread attention. For example, hsa_circRNA_002178 silencing retards breast cancer progression via microRNA8-mediated inhibition of COL1A1 (Liu et al. Citation2020). Hsa_circ_0004277 contributes to malignant phenotype of colorectal cancer by sponging miR-2 to upregulate the expression of PTMA (Yang et al. Citation2020). Autophagy-associated circCDYL augments autophagy and promotes breast cancer progression (Liang et al. Citation2020). Interestingly, the stable closed-loop structure of circRNAs can become a remarkable biomarker (Chen et al. Citation2017; Meng et al. Citation2017; Tian et al. Citation2018). Moreover, more and more studies revealed that circRNAs play a significant role in multi-drug resistance (Liu et al. Citation2018; Zhou et al. Citation2019). However, the relationship between circRNAs and oncolytic adenovirus still remains unclear.
In this study, we intended to identify the potential association of circRNA_100920 with the treatment of oncolytic adenovirus. The results of our study proved that circRNA_100920 enhanced oncolytic adenovirus therapy in CRC by suppressing IFN-γ expression.
Materials and methods
Cells
LoVo, HCT8, SW480, SW620, HCT116 and HT-29 cells, and non-cancerous NCM460 colon cells were collected from Cell Bank of Type Culture Collection (Shanghai City, China). All cell lines were cultured in Dulbecco’s Modified Eagle Medium (DMEM) medium (Gibco) supplemented with 10% fetal bovine serum (FBS) (Gibco) and 50 U/ mL penicillin and streptomycin sulfate at 37°C in humid air with 5% CO2.
Tissue specimens
The study was approved by the Clinical Research Ethics Committee of Affiliated Hospital of Jiangsu University, and all patients had written informed consent. 80 pairs of cancerous and adjacent normal tissues were obtained from CRC patients without receiving preoperative chemotherapy.
Bioinformatics analysis
The microarray data of circRNA profiles in CRC tissues and normal tissues were from NCBI GEO Datasets (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE142837). After applying log 2 transformation, GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/?acc=GSE142837) was performed to analyze normalized microarray data.
RNA extraction and quantitative real-time PCR (RT-qPCR)
Total RNAs were extracted from tissues and cells by using TRIzol reagent (Invitrogen), according to the manufacturer’s instructions. The expressions of circRNA_100920 were detected, using the One Step SYBR PrimeScript PLUS RT-RNA PCR Kit (TaKaRa, Dalian, China), according to the manufacturer’s instructions. The 2−ΔΔCt method was performed to detect gene expression. The primer sequences of circRNA_100920 were 5′-CTGCTTGATTTGCAGCAGCC-3′ (forward primer) and 5′-CTGCTTGCAGCTGTAGAATC-3′ (reverse primer). The primer sequences of IFN-γ were 5′-AGGCTTTATCTCAGGGGCCA-3′ (forward primer) and 5′-AGCACTGGCTCAGATTGCAG-3′ (reverse primer). The primer sequences of GADPH were 5′-GTCAACGGATTTGGTCT-3′ (forward primer) and 5′-AGTCTTCTGGGTGGCAGTGAT-3′.
Cell viability assay
The therapeutic effect of oncolytic adenovirus in CRC cells was detected by the CCK-8 assay kit (Dojindo, Kumamoto, Japan). Briefly, the HCT8/si-circRNA_100920, HCT116/si-circRNA, HCT8/si-NC, or HCT116/si-NC were seeded in 96-well plates after 0, 24, 48, 72, or 96 h, respectively. The cell vitality was examined by CCK-8 solution.
ELISA
IFN-γ ELISA kits were purchased from Sigma-Aldrich (St. Louis, MO, USA), and experiments were carried out according to the manufacturer's protocol.
Statistical analysis
All data were presented as mean ± SD (standard deviation) of at least three biological replicates, and statistically analyzed using Student’s t test. Spearman and Pearson correlation analyses were used to analyze correlation significance. The value of p < 0.05 was considered statistically significant. Statistical analysis was exercised by using SPSS software version 20.0 (SPSS, Inc., Chicago, USA) or Graphpad Prim 7 (GraphPad, La Jolla, CA, USA).
Results
CircRNA_100920 was increased in CRC by using the bioinformatics analysis
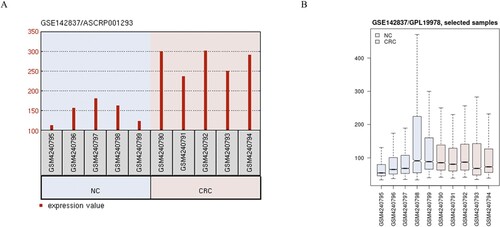
Initially, the differential expression of circRNAs was identified by analyzing microarray data in the GEO dataset (GSE142837), and we found that circRNA_100920 is highly expressed on CRC (Figure (A)). In addition, Box plots indicated normalized intensities from the cancerous and adjacent normal tissues (Figure (B)). Hence, we have chosen CircRNA_100920 for further study.
Clinical significance of circRNA_100920 in indicating CRC progression
To further examine the expression of circRNA_100920 in CRC samples, RT-qPCR were used. The results showed circRNA_100920 was also overexpressed in 80 pairs of cancerous tissues compared with adjacent normal tissues (Figure (A)). In addition, the patients were divided into high circRNA_100920 expression group (40) and low circRNA_100920 expression group (40) according to the median value (6.48), and the potential relationship between circRNA_100920 expression and clinicopathologic characteristics in 80 patients with CRC was also researched. As shown in Table , there was no correlation between circRNA_100920 expression with age and gender and high expression of circRNA_100920 that was associated with metastasis, TNM stage, and tumor size. Moreover, CRC patients with high expression of circRNA_100920 had shorter overall survival time than those with low expression of circRNA_100920 (Figure (B)), and the receiver operating characteristic (ROC) curve analysis revealed circRNA_100920 could become a potential biomarker (Figure (C)).
Figure 2. Clinical significance of circRNA_100920 in indicating CRC progression. A. CircRNA_100920 was overexpressed in 80 pairs of cancerous tissues compared with adjacent normal tissues. B. This panel showed the Kaplan–Meier survival curve relative to circRNA_100920 expression levels. C. The receiver operating characteristic (ROC) curve analysis revealed circRNA_100920 could become a potential biomarker. *p<0.05.
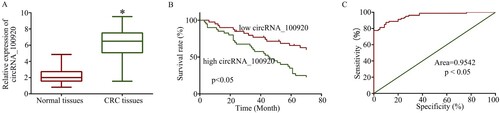
Table 1. Relationship between clinical features and circRNA_100920 expression in CRC patients.
Knockdown of circRNA_100920 inhibited the cell proliferation in CRC cells
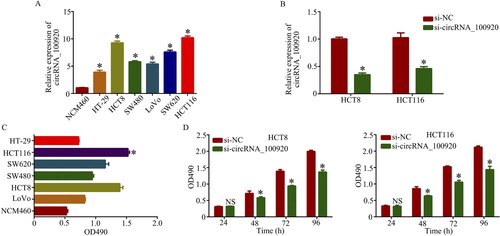
We have found overexpression of circRNA_100920 in CRC patients, and the function of circRNA_100920 should be further examined. Thus, the expression of circRNA_100920 in CRC cell was detected by RT-qPCR. The result showed the expression of circRNA_100920 in CRC cell was higher than that of NCM460 cell, and the expression of circRNA_100920 in HCT116 and HCT8 cell was highest (Figure (A)). HCT116 and HCT8 cell then were transfected with si-circRNA_100920 or si-NC to knock down circRNA_100920 expression. The results showed the circRNA_100920 expression was decreased in HCT116 and HCT8 cell after transfection with si-circRNA_100920 (Figure (B)). Moreover, byCCK8 the assay showed that the HCT116 cell proliferation in CRC cells was fastest (Figure (C)), and the cell proliferation with knockdown of circRNA_100920 was inhibited (Figure (D)). Thus, the results confirmed that knockdown of circRNA_100920 could suppress the proliferation of CRC cell, suggesting circRNA_100920 may act as an oncogene in CRC.
Figure 3. Knockdown of circRNA_100920 inhibited the cell proliferation in CRC cells. A. Relative expression of circRNA_100920 in CRC cell lines. B. Relative expression of circRNA_100920 in HCT8 and HCT116 cells transfected with si-circRNA_100980 and si-NC. C. Cell proliferation in CRC cell lines. D. Cell proliferation in HCT8 and HCT116 cells transfected with si-circRNA_100980 and si-NC. *p<0.05.
CircRNA_100920 enhanced oncolytic adenovirus therapy in CRC cells
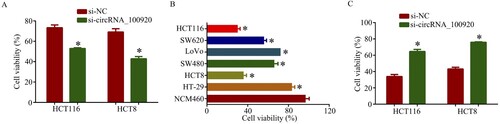
By CCK8 the assay indicated the cell viability of si-circRNA_100920 group after the treatment of cisplatin was lower than that of the si-NC group, suggesting CRC cells with high circRNA_100920 expression were less sensitive to cisplatin (Figure (A)). In addition, we found the cell viability of HCT116 after the treatment of oncolytic adenovirus H101 was lower than that of other CRC cells, suggesting H101 showed better therapeutic effect in CRC cells with high expression of circRNA_100920 (Figure (B)). Moreover, the cell viability of si-circRNA_100920 group after treatment of H101 was higher than that of the si-NC group (Figure (C)). Thus, circRNA_100920 enhanced oncolytic adenovirus therapy in CRC cells.
Figure 4. CircRNA_100920 enhanced oncolytic adenovirus therapy in CRC cells. A. Cell viability of si-circRNA_100920 group after the treatment of cisplatin (3 µg/ml) was lower than that of the si-NC group. B. Cell viability of HCT116 after the treatment of oncolytic adenovirus H101 (5 MOI) was lower than that of other CRC cells. C. Cell viability of si-circRNA_100920 group after the treatment of H101 (5 MOI) was higher than that of the si-NC group. *p<0.05.
CircRNA_100920 enhanced oncolytic adenovirus therapy in CRC cells by suppressing IFN-γ expression
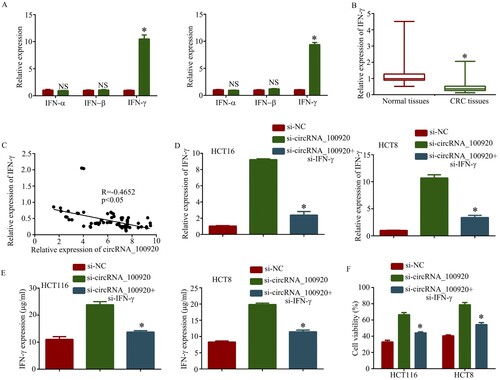
Earlier studies had indicated that interferon could prevent the function of virus (Weidner et al. Citation2010; Wheelock Citation1965). Moreover, to explore how circRNA_100920 promoted oncolytic adenovirus therapy, we detected different types of interferons. The results showed the IFN-γ expression in si-circRNA_100920 group was higher than that in the si-NC group and there was no significant difference in the IFN-α and IFN-β expression of the si-circRNA_100920 group compared with the si-NC group (Figure (A)). To further study circRNA_100920 was associated with IFN-γ, we detected IFN-γ expression in 80 pairs of cancerous tissues and adjacent normal tissues. The results showed IFN-γ expression was decreased in cancerous tissues compared with adjacent normal tissues (Figure (B)) and Spearman’s correlation analysis verified the negative correlation between circRNA_100920 expression and IFN-γ expression (Figure (C)). Subsequently, HCT116 and HCT8 cells were transfected the si-circRNA_100920 and si-IFN-γ, and RT-qPCR (Figure (D)) and ELISA (Figure (E)) assay demonstrated IFN-γ expression of transfected si-circRNA_100920 and the si-IFN-γ group was decreased compared to the transfected si-circRNA_100920 group. Moreover, knockdown of IFN-γ restored the therapeutic effect of H101 (Figure (F)). Hence, circRNA_100920 enhanced oncolytic adenovirus therapy in CRC cells by suppressing IFN-γ expression.
Figure 5. CircRNA_100920 enhanced oncolytic adenovirus therapy in CRC cells by suppressing IFN-γ expression. A. Relative expression of IFN in HCT8 and HCT116 cells transfected with si-circRNA_100980 and si-NC. B. Relative expression of IFN-γ expression in 80 pairs of cancerous tissues and adjacent normal tissues. C. Spearman’s correlation analysis verified the negative correlation between circRNA_100920 expression and IFN-γ expression. D. Relative expression of IFN-γ in HCT8 and HCT116 cells transfected with si-circRNA_100980 and si- IFN-γ was detected by RT-qPCR assay. E. Relative expression of IFN-γ in HCT8 and HCT116 cells transfected with si-circRNA_100980 and si- IFN-γ was detected by Elisa assay. F. Knockdown of IFN-γ restored therapeutic effect of H101(5 MOI). *p<0.05.
Discussion
Recently, mounting research has indicated that circRNAs are frequently dysregulated in various cancers. For example, CircMRPS35 suppresses gastric cancer progression via recruiting KAT7 to govern histone modification (Jie et al. Citation2020). Hsa_circ_0078607 suppresses ovarian cancer progression by regulating miR-518a-5p/Fas signaling pathway (Zhang et al. Citation2020). CircC3P1 suppresses hepatocellular carcinoma growth and metastasis through miR-4641/PCK1 pathway (Zhong et al. Citation2018). Notably, circRNAs also play a significant role in CRC. For example, hsa_circ_0053277 promotes the development of CRC by upregulating matrix metallopeptidase 14 via miR-2467-3p sequestration (Xiao and Liu Citation2020). Hypoxia-induced circCCDC66 promotes the tumorigenesis of CRC via the miR3140/autophagy pathway (Feng et al. Citation2020). Circ_0007142 regulates cell proliferation, apoptosis, migration, and invasion via miR-455-5p/SGK1 axis in CRC (Wen et al. Citation2021). In this study, circRNA_100920 expression was found overexpression in CRC tissues and cells and knockdown of circRNA_100920 inhibited cell proliferation in CRC cells. Clinically, circRNA_100920 was associated with survival rate, metastasis, TNM stage, and tumor size. Notably, circRNA_100920 could become a potential biomarker by the receiver operating characteristic (ROC) curve analysis. All the data indicated that circRNA_100920 regulated the development of CRC.
The development of multi-drug resistance has become the obstacle to an effective control of CRC clinically. For example, circDDX17 reduces 5-fluorouracil resistance and hinders tumorigenesis in CRC by regulating miR-31-5p/KANK1 axis (Ren et al. Citation2020). Circ-PRKDC contributes to 5-fluorouracil resistance of CRC by regulating miR-375/FOXM1 axis and wnt/β-catenin pathway (Chen et al. Citation2020). Hsa_circ_0079662 induces the resistance mechanism of the chemotherapy drug oxaliplatin through the TNF-α pathway in CRC (Lai et al. Citation2020). In exploring new strategies for CRC therapy, oncolytic adenovirus showed potential and effective anti-tumor effect in CRC. For example, E1B55kDa-deleted oncolytic adenovirus carrying microRNA-143 exerts specific antitumor efficacy on CRC cells (Luo et al. Citation2016). An oncolytic adenovirus, encoding decorin and GM-CSF, inhibits tumor growth in a colorectal tumor model by targeting pro-tumorigenic signals and via immune-activation (Liu et al. Citation2017). CRC therapy with oncolytic adenovirus is enhanced by the addition of herpes simplex virus-thymidine kinase (Wildner et al. Citation1999). In this study, we found CRC cells with high circRNA_100920 expression were less sensitive to cisplatin and circRNA_100920 enhanced oncolytic adenovirus therapy in CRC cells by suppressing IFN-γ expression.
In sum, we verified that circRNA_100920 was highly expressed in CRC tissues and CRC cells. Knocking down circRNA_100920 inhibited the proliferation of CRC cells. circRNA_100920 enhanced oncolytic adenovirus therapy in CRC cells by suppressing IFN-γ expression. Moreover, circRNA_100920, as a biomarker, may participate in the diagnosis, treatment, and prognosis of CRC.
Authors’ contributions
Shaofeng Yuan and Shengchun Dang made substantial contributions to the conception and design of the study. Xiangdong Kong, Wei Yu, Jinhong Geng, and Linwen Zeng performed the data analysis and interpretation. Shaofeng Yuan drafted the manuscript. Shengchun Dang performed the critical revision of the manuscript. All co-authors have read the manuscript and approved the final version for submission and publication to this journal.
Ethics approval and consent to participate
The study was approved by the Clinical Research Ethics Committee of Affiliated Hospital of Jiangsu University (approval no.190625). All patients provided written informed consent prior to enrollment in the study.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Arraras Urdaniz JI, Garcia RV, Aguillo MM, Burgaleta AM, de la Vega FA, Pascual ES. 2006. Quality of life assessment through the EORTC questionnaires of colorectal cancer patients in advanced disease stages. Clin Transl Oncol. 8:664–671. Epub 2006/09/29.
- Chen H, Pei L, Xie P, Guo G. 2020. Circ-PRKDC contributes to 5-fluorouracil resistance of colorectal cancer cells by regulating miR-375/FOXM1 axis and Wnt/beta-catenin pathway. Onco Targets Ther. 13:5939–5953. Epub 2020/07/02.
- Chen J, Li Y, Zheng Q, Bao C, He J, Chen B, Lyu D, Zheng B, Xu Y, Long Z, et al. 2017. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 388:208–219. Epub 2016/12/18.
- Feng J, Li Z, Li L, Xie H, Lu Q, He X. 2020. Hypoxiainduced circCCDC66 promotes the tumorigenesis of colorectal cancer via the miR3140/autophagy pathway. Int J Mol Med. 46:1973–1982. Epub 2020/10/31.
- Jeck WR, Sharpless NE. 2014. Detecting and characterizing circular RNAs. Nat Biotechnol. 32:453–461. Epub 2014/05/09.
- Jie M, Wu Y, Gao M, Li X, Liu C, Ouyang Q, Tang Q, Shan C, Lv Y, Zhang K, et al. 2020. CircMRPS35 suppresses gastric cancer progression via recruiting KAT7 to govern histone modification. Mol Cancer. 19:56. Epub 2020/03/14.
- Kuipers EJ, Grady WM, Lieberman D, Seufferlein T, Sung JJ, Boelens PG, van de Velde CJ, Watanabe T. 2015. Colorectal cancer. Nat Rev Dis Primers. 1:15065. Epub 2015/01/01.
- Lai M, Liu G, Li R, Bai H, Zhao J, Xiao P, Mei J. 2020. Hsa_circ_0079662 induces the resistance mechanism of the chemotherapy drug oxaliplatin through the TNF-alpha pathway in human colon cancer. J Cell Mol Med. 24:5021–5027. Epub 2020/04/04.
- Liang G, Ling Y, Mehrpour M, Saw PE, Liu Z, Tan W, Tian Z, Zhong W, Lin W, Luo Q, et al. 2020. Autophagy-associated circRNA circCDYL augments autophagy and promotes breast cancer progression. Mol Cancer. 19:65. Epub 2020/03/28.
- Liu T, Ye P, Ye Y, Lu S, Han B. 2020. Circular RNA hsa_circRNA_002178 silencing retards breast cancer progression via microRNA-328-3p-mediated inhibition of COL1A1. J Cell Mol Med. 24:2189–2201. Epub 2020/01/21.
- Liu Y, Dong Y, Zhao L, Su L, Luo J. 2018. Circular RNAMTO1 suppresses breast cancer cell viability and reverses monastrol resistance through regulating the TRAF4/Eg5 axis. Int J Oncol. 53:1752–1762. Epub 2018/07/18.
- Liu Z, Yang Y, Zhang X, Wang H, Xu W, Wang H, Xiao F, Bai Z, Yao H, Ma X, et al. 2017. An oncolytic adenovirus encoding decorin and granulocyte macrophage colony stimulating dactor inhibits tumor growth in a colorectal tumor model by targeting Pro-tumorigenic signals and via immune activation. Hum Gene Ther. 28:667–680. Epub 2017/05/23.
- Luo Q, Basnet S, Dai Z, Li S, Zhang Z, Ge H. 2016. A novel E1B55kDa-deleted oncolytic adenovirus carrying microRNA-143 exerts specific antitumor efficacy on colorectal cancer cells. Am J Transl Res. 8:3822–3830. Epub 2016/10/12.
- Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P, Wu M. 2017. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 16:94. Epub 2017/05/26.
- Ren TJ, Liu C, Hou JF, Shan FX. 2020. CircDDX17 reduces 5-fluorouracil resistance and hinders tumorigenesis in colorectal cancer by regulating miR-31-5p/KANK1 axis. Eur Rev Med Pharmacol Sci. 24:1743–1754. Epub 2020/03/07.
- Roque IFM, Sola I, Martin-Richard M, Lopez JJ, Bonfill Cosp X. 2009. Second-line chemotherapy in advanced and metastatic CRC. Cochrane Database Syst Rev. 1:CD006875. Epub 2009/04/17.
- Rougier P, Bugat R, Douillard JY, Culine S, Suc E, Brunet P, Becouarn Y, Ychou M, Marty M, Extra JM, et al. 1997. Phase II study of irinotecan in the treatment of advanced colorectal cancer in chemotherapy-naive patients and patients pretreated with fluorouracil-based chemotherapy. J Clin Oncol. 15:251–260. Epub 1997/01/01.
- Sato-Dahlman M, Miura Y, Huang JL, Hajeri P, Jacobsen K, Davydova J, Yamamoto M. 2017. CD133-targeted oncolytic adenovirus demonstrates anti-tumor effect in colorectal cancer. Oncotarget. 8:76044–76056. Epub 2017/11/05.
- Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RGS, Barzi A, Jemal A. 2017. Colorectal cancer statistics, 2017. CA Cancer J Clin. 67:177–193. Epub 2017/03/02.
- Siegel RL, Miller KD, Jemal A. 2019. Cancer statistics, 2019. CA Cancer J Clin. 69:7–34. Epub 2019/01/09.
- Tian M, Chen R, Li T, Xiao B. 2018. Reduced expression of circRNA hsa_circ_0003159 in gastric cancer and its clinical significance. J Clin Lab Anal. 32:e22281.Epub 2017/06/16
- Weidner JM, Jiang D, Pan XB, Chang J, Block TM, Guo JT. 2010. Interferon-induced cell membrane proteins, IFITM3 and tetherin, inhibit vesicular stomatitis virus infection via distinct mechanisms. J Virol. 84:12646–12657. Epub 2010/10/15.
- Wen T, Wu H, Zhang L, Li K, Xiao X, Zhang L, Zhang Y. 2021. Circular RNA circ_0007142 regulates cell proliferation, apoptosis, migration and invasion via miR-455-5p/SGK1 axis in colorectal cancer. Anticancer Drugs. 32:22–33. Epub 2020/09/06.
- Wheelock EF. 1965. Interferon-like virus-inhibitor induced in human leukocytes by phytohemagglutinin. Science. 149:310–311. Epub 1965/07/16.
- Wildner O, Blaese RM, Morris JC. 1999. Therapy of colon cancer with oncolytic adenovirus is enhanced by the addition of herpes simplex virus-thymidine kinase. Cancer Res. 59:410–413. Epub 1999/02/02.
- Xiao H, Liu M. 2020. Circular RNA hsa_circ_0053277 promotes the development of colorectal cancer by upregulating matrix metallopeptidase 14 via miR-2467-3p sequestration. J Cell Physiol. 235:2881–2890. Epub 2019/09/25.
- Yang L, Sun H, Liu X, Chen J, Tian Z, Xu J, Xiang B, Qin B. 2020. Circular RNA hsa_circ_0004277 contributes to malignant phenotype of colorectal cancer by sponging miR-512-5p to upregulate the expression of PTMA. J Cell Physiol. 1–12. Epub 2020/01/22.
- Zhang N, Jin Y, Hu Q, Cheng S, Wang C, Yang Z, Wang Y. 2020. Circular RNA hsa_circ_0078607 suppresses ovarian cancer progression by regulating miR-518a-5p/Fas signaling pathway. J Ovarian Res. 13:64. Epub 2020/06/07.
- Zhong L, Wang Y, Cheng Y, Wang W, Lu B, Zhu L, Ma Y. 2018. Circular RNA circC3P1 suppresses hepatocellular carcinoma growth and metastasis through miR-4641/PCK1 pathway. Biochem Biophys Res Commun. 499:1044–1049. Epub 2018/04/03.
- Zhou Y, Zheng X, Xu B, Chen L, Wang Q, Deng H, Jiang J. 2019. Circular RNA hsa_circ_0004015 regulates the proliferation, invasion, and TKI drug resistance of non-small cell lung cancer by miR-1183/PDPK1 signaling pathway. Biochem Biophys Res Commun. 508:527–535. Epub 2018/12/05.