ABSTRACT
The molecular identification of different taxa of genus Acacia plays a critical role to understand the evolutionary history of this important plant that has many medicinal and ecological benefits and distributed in many regions around the world (Africa, Asia, Australia and the Americas). This is the first study that used DNA barcoding (rbcL and matK) with 24 taxa of genus Acacia collected from Saudi Arabia for molecular identification, genetic resources preservation, detect genetic variations accompanied with a phylogenetic view and test the ability of DNA barcoding to segregate Senegalia and Vachellia from Acacia Mill taxa. Results showed that the GC ratio and transition/transversion bias (R) of rbcL (1.25) was higher than matK (0.95). This study resulted in the re-definition of some previously recorded species of Acacia into recently proposed genera,13 species under Vachellia and 2 species under Senegalia.The aim of the present study is to assess the ability of rbcL and matK loci to recognize Senegalia and Vachellia from other taxa of Acacia and assess the evolutionary relationship among them. This work will help to determine the position of Acacia collected from Saudi Arabia in the new generic classification and provide a clear view of the genetic relationships within the genera.
Introduction
DNA barcoding is an effective tool for accurate identification of plant species, estimation of the molecular evolution rate, comparative genetics studies and biodiversity research. (Khan et al. Citation2016; Harnelly et al. Citation2018; Gao et al. Citation2019). According to the progression of molecular biology, DNA sequencing has been utilized in numerous phylogenetic investigations (Hidayat and Pancoro Citation2008) because of the changes that happened in bases of nucleotide along the time. The identification of Acacia species according to phenotypic (morphological) characters is very difficult that made DNA barcoding is a reliable tool for confirming the species identity of this plant (Al-Juhani Citation2019).
Senegalia and Vachellia are genera with wide geological extents from South Africa north to the Mediterranean, Asia and Australia (Lewis Citation2005; Dharani Citation2006). Morphological characters considered for characterizing of Acacia s.l. (Seigler et al. Citation2006). In spite of the significant full scale and micromorphological contrasts among Senegalia and Vachellia, it stays hard to divide monophyletic ancestries inside these genera dependent on morphological attributes (Miller and Bayer Citation2003).
The previous studies revealed that Acacia Miller s.l. is polyphyletic and require to be divided into at least seven genera (Maslin et al. Citation2019). Some members of Acacia need re-identification and re-classification (Bouchenak-Khelladi et al. Citation2010; Miller and Seigler Citation2012). Considering this, there is a need to study associations and phylogenetic relationships of genus Acacia in many different regions around the world. Kyalangalilwa et al. (Citation2013) studied the phylogenetic relationships between the African Acacia while Boatwright et al. (Citation2015) studied the Madagascan Acacia. The phylogenetic relationships among the Australian Acacia were studied by Comben et al. (Citation2020). In Asia there are some studies in China (Maslin et al. Citation2019) and south west Asia (Ragupathy et al. Citation2014) but there is no studies in Saudi Arabia located in the middle of Asia.
So, the targets of this study are using DNA barcoding by rbcL and matK loci in 24 taxa of genus Acacia collected from Saudi Arabia to give molecular identification, preservation of its genetic resources, detect genetic variations accompanied with phylogenetic view between studied taxa, test the ability of DNA Barcoding to segregated Senegalia and Vachellia from Acacia Mill and determine the position of the Saudi Acacia in the new generic classification.
Materials and methods
Plant materials
In the present study, we tried to collect almost all taxa related to the genus Acacia in Saudi Arabia. The 24 taxa of Acacia were got from five localities in KSA (Table ) representing eighteen species, four subspecies and two varieties. One of this site was located at Riyadh from Agriculture Research Station; which comprised of five taxa of Acacia viz. Acacia nilotica subsp. Indica, A. nilotica subsp. tomentosa, A. pruinocarpa, A. tortilis subsp. raddiana and A. gerrardii var. najdensis. The second site was at Al-Qassem from Ministry of Environment, Water and Agriculture which the dominant Acacia species are 14 taxa (A. coriacea, A. cuthbertsonii, A. cyclops, A. ehrenbergiana, A. farnesiana, A. gerrardii var. iraquensis, A. johnwoodii, A. ligulate, A. salicina, A. sclerosperma, A. nilotica subsp. Indica, A. senegal, A. tortilis subsp. tortilis and A. victoriae. The third site was seeds bank in Al-Dammam (A. ampliceps). The fourth site was at Al-Taif (Taif road) which comprised of three taxa A. gerrardii var. gerrardii, A. asak and A. origena. The five site at Wadi Alaqiq of Al-Baha which comprised of one species; A. etbaica.
Table 1. DNA barcoding primers and their sequences.
Table 2. The studied taxa of genus Acacia and their Accession numbers in GenBank.
The collected taxa were identified and authenticated according to Migahid (Citation1996), Collenette (Citation1999) and Chaudhary and Al-Jowaid (Citation1999). Voucher specimens were kept in the public herbarium of College of Science, Biology Department at Taibah University. Voucher specimens and their numbers were provided in (Appendix).
DNA extraction and amplification process
The genomic DNA was extracted from seeds of the 24 studied taxa by using a DNAeasy Plant Mini Kit (QIAGEN, Santa Clarita) and according to the manufacturer’s protocol.
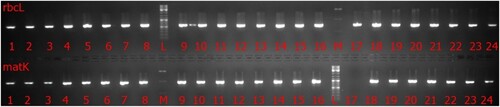
The amplification of the purified DNA was achieved using rbcL and matK primers as mentioned in (Table ). Each reaction contained 1 μl of each forward and reverse primer (Macrogen), 1 μl genomic DNA, 12 μl Thermo Scientific GeneRuler DNA Ladder Mix was used and 10 μl of distilled water, the reaction was set up in the PCR Thermal Cycler (BIO-RAD) adjusted to 35 cycles. The first step was denaturation at 94°C for 5 min, then annealing at 52°C for 30 s and finished by extension at 72°C for 2 min. A final extension was done at 72°C for 4 min. The DNA profile of taxa of genus Acacia generated by rbcl and matk was shown in Figure .
The PCR sequencing
The PCR products of the 24 taxa of Acacia were sent to Macrogen Inc. (South Korea) for purification and sequencing. In this study, all generated sequences of Acacia were deposited in GenBank (accession numbers are listed in Table ).
The sequences alignment and phylogenetic trees
The sequences of rbcL and matK of studied Acacia were subjected to BLAST of the GenBank (http://blast.ncbi.nlm.nih.gov/Blast.cgi) for identification of specimens (max identity 100%). Alignments of sequences were achieved by MUSCLE (Multiple Sequence Comparison by Log-Expectation) algorithm (Edgar Citation2004). The sequence length, (%) Variable sites, overall mean distance and transition/transversion bias (R) and nucleotide exchange rates were calculated using Maximum Likelihood Method and Tamura–Nei model. depending on MEGA6 software (Tamura and Nei Citation1993).
Routinely, gene sequences used to the topologies of unrooted phylogenetic trees without an outgroup (Sinsheimer et al. Citation2012). The maximum likelihood procedure became practical in the analysis of sequence data of reasonable size. So, MEGA6 Software was used to build the phylogenic trees by the Maximum likelihood bootstrap (MLB) analysis depending on a total of 1000 bootstrap replicates (Tamura et al. Citation2013).
Results
In this study rbcL and matK loci were used as DNA barcoding to characterize and distinguish between 24 taxa of Acacia collected from different regions in Saudi Arabia to clear the genetic diversity and phylogenetic relationship between them, that can help breeders to improve Acacia breeding programs and test the ability of these primers to distinguish between Senegalia and Vachellia from Acacia Mill and determine the position of the Saudi Acacia in the new generic classification.
The results from (Table ) showed that the sequence length of rbcL is ranged from 545 to 599 bp, while the sequence length of matK ranged from 887 to 990 bp. The GC ratio scored in locus rbcL was the highest than matK locus. The sequence length and GC ratio of rbcL or matK were almost identical in all taxa of genus Acacia (Table ). The transition/transversion bias (R) of rbcL (1.25) was higher than that of matK (0.95). The rates of transitions higher than transversions in all sequences of studied taxa (Table ).
Table 3. Statistics derived from the sequencing, alignment and BLAST processes.
Table 4 Mean nucleotide substitution rates and transition/transversion bias (R) in loci for 24 taxa of Acacia calculated by Maximum Likelihood Method.
Cladistic analysis for rbcl matrix was generated a dendrogram that was divided into two monophyletic clades, the first one comprised 13 taxa while the second one included 11 taxa. Within the first clade, Acacia gerrardii var. gerrardii, A. johnwoodii and A. ehrenbergiana formed a sister group to another one that included A. tortilis subsp. raddiana and A. etbaica, at lower level A. gerrardii var. iraquensis and A. tortilis subsp. tortilis were separated in distinct group, the two accessions of A. nilotica subsp. indica were separated at early level in another group. The remaining taxa separated as distinct lineages from A. farnesiana to A. gerrardii var. najdensis passing through A. origena and A. nilotica subsp. tomentosa.
The second clade was divided into two main sister groups, the first one included five taxa A. cyclops that separated at lower level followed by A. pruinocarpa then A. victoriae, while A. cuthbertsonii and A. coriacea occupied the highest level of separation as sister taxa. The second group could be differentiated into two sister groups; A. senegal and A. asak were nested in one group at lower position while A. ampliceps, A. ligulata, A. sclerosperma and A. salicina were occupied the higher distinct lineage.
Another dendrogram was generated by the cladistic analysis of matk matrix that divided into two main monophyletic clades, the first one comprised 12 taxa while the second one included 11 taxa. Within the first clade A. tortilis subsp. raddiana was occupied the lower position followed by A. farnesiana. A. asak and A. senegal were nested in one group at a lower position, the other taxa of the present clade were distributed into two main groups; the first one comprised A. cyclops, A. coriacea, A. cuthbertsonii and A. pruinocarpa while the second group is divided into two sister lineages one of A. ligulata and A. salicina and another of A. sclerosperma and A. ampliceps.
The second clade was differentiated into three main Lineages, the first one included three taxa A. nilotica subsp. indica, A. nilotica subsp. tomentosa as sister taxa and A. origena that was separated early. The second lineage included three taxa viz. the A. nilotica subsp. indica (second accsession), A. gerrardii var. najdensis as sister taxa and A. etbaica that was separated at a lower position. The last lineages comprised five taxa that were distributed between two sister groups; the first one of Acacia gerrardii var. gerrardii and A. gerrardii var. iraquensis while A. tortilis subsp. tortilis, A. johnwoodii and A. ehrenbergiana were nested in the second group.
Discussion
The present work represents the first study that used DNA barcoding by rbcL and matK for molecular identification, assessing the genetic relationships among 24 taxa of Acacia collected from Saudi Arabia and testing the ability of rbcL and matK loci to segregate Senegalia and Vachellia from Acacia Mill species for helping to determine the position of the Saudi Acacia in the new generic classification. Mosa et al. (Citation2019) recommended using advanced techniques for conservation of plant species and the importance to detect the genetic relationship between native plants and domesticated ones for defining their genes. Maslin et al. Citation2019 mentioned that information constructed from molecular sequence such as nuclear genes and chloroplast are helpful for understanding the phylogenetic relationships within genus Acacia.
In recent years there have been essential changes to both nomenclature and the classification of the genus previously known as Acacia Mill. Suleiman et al. (Citation2018a, Citation2018b) recommended that all Acacia species found in the Arabian Peninsula and the Middle East should be studied by DNA barcoding to discriminate closely related species in this genus.
Results from Table revealed that transition/transversion bias (R) of rbcL (1.25) was higher than that of matK (0.95). Luo et al. (Citation2016) mentioned that the transition–transversion bias differs according to the genome region. Transversions generally occurred at low rate than transitions, in which nucleotide substitution bias prefers transitions more than transversion because it is easier (Zhao and Boerwinkle Citation2002; Zhang and Gerstein Citation2003). The rates of transitions to transversions in sequences of studied taxa (Table ) indicated that the substitution mutations located in their genomes (Kimura Citation1981; Gojobori et al.Citation1982).
The statistical differences observed between rbcl and matk loci, indicated the presence of different evolutionary events between the two regions, allowing each other to complement the discrimination between Acacia species. Across the genome of Acacia rbcL and matK were efficient loci in sequence quality and in species segregation capability (Kress and Erickson Citation2008; Piredda et al. Citation2011; de Vere et al. Citation2012). Sequence data of the two loci was used to reconstruct the phylogenetic trees using the Maximum Likelihood Tree Method (Figures and ) for clarification of the phylogenetic relationships between studied taxa. These loci have great discriminatory power for developing the phylogenetic relationships between many plant species (Dong et al. Citation2014; Al-Hemaid et al. Citation2015; Bolson et al. Citation2015; Tang et al. Citation2016; Khan et al. Citation2016; Shinwari et al. Citation2018).
Figure 2 Phylogeny tree of 24 taxa of Acacia based on rbcL locus using Maximum Likelihood Tree Method.
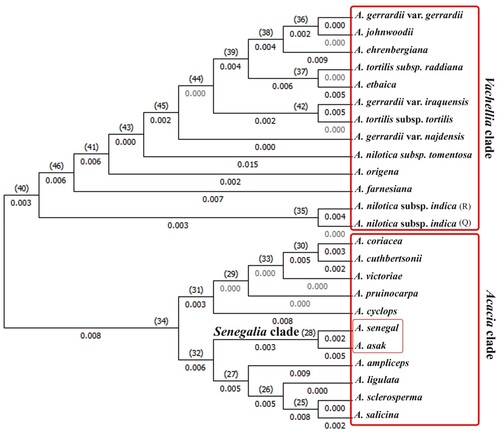
Figure 3 Phylogeny tree of 24 taxa of Acacia based on matK locus using Maximum Likelihood Tree Method.
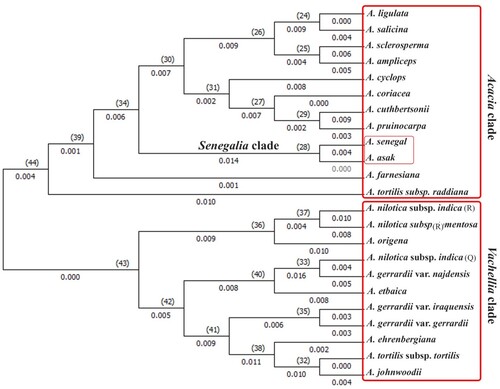
This study demonstrated the ability of both rbcl and matk to separate Senegalia and Vachellia from other studied taxa of Acacia. The rbcL phylogenetic tree (Figure ) segregated studied taxa into two main clades, the first one included Vachellia, the second clade viz. Acacia clade that included two Senegalia species. Although the majority of Acacia s.s. form a single clade, the placement of the two species of Senegalia demonstrates the polyphylly of Acacia clade (Kyalangalilwa et al. Citation2013). The matk phylogenetic tree (Figure ) clarified that Senegalia lineage was nested within Acacia s.s. clade in addition to two taxa from Vachellia clade viz. A. farnesiana and A. tortilis subsp. Raddiana. This result can be explained that the matK sequence of these taxa is not complete gene region (Elansary et al. Citation2011) and not all the portions of gene region evolve at the same rate (Schori and Showalter Citation2011). Elansary (Citation2013) revealed that A. farnesiana showed several segregation sites that make it easily to discriminate from other species accompanied within this genus. A. farnesiana shared some similarities with subg. Acacia (Miller and Bayer Citation2003) and this data support the multiple origins of this species in which Maslin et al. (Citation2003) demonstrated the A. farnesiana from African-Asian subg. Acacia formed a clade separated from American member of A. farnesiana. While the second clade included the remaining taxa of Vachellia that is distantly related from other members of Acacia s.l. (Luckow et al. Citation2003; Miller and Bayer Citation2003; Seigler et al. Citation2006; Ragupathy Citation2009; Steven and Subramanyam Citation2009). Depending upon previous results, rbcL was recommended as the first option to reveal sequence variability, because it had a high evolutionary rate. An accelerated evolution was observed within Acacia genome through the high transition/transversion bias (R = 1.25), suitable sequence length, and clear interspecific divergence.
This study resulted in the re-definition of some previously recorded species of Acacia into recently proposed genera, 13 species under Vachellia and 2 species under Senegalia and that demonstrated the ability of rbcL and matK loci to recognize Senegalia and Vachellia from other taxa of Acacia Mil.
Conclusion
This study is the first of its kind to assess DNA barcoding for characterization and re-definition of almost taxa of genus Acacia collected from Saudi Arabia. It is concluded that both two loci (rbcL and matK) have considerable interspecific divergence and can recognize and bolster the recently proposed genera of Senegalia and Vachellia from Acacia Mill. rbcL was a more applicable barcode than matK for the authentication and diversification of Acacia species and detect possible evolutionary relationship among studied taxa of genus Acacia. The new identified sequences from multiple plant taxa provide critical data for further evolutionary studies and these should be tested with other related plant taxa that found in another countries to broaden its genetic background.
Author Contributions
Amal M.E. Abdel-Hamid and Usama K. Abdel-Hameed designed the study and wrote the manuscript. All the authors performed experiments. Amal M.E. Abdel-Hamid analyzed the data and submitted the DNA sequences on GenBank database. All the authors validate and approved the manuscript for publication.
Availability of data
The data of this study are public and available in the NCBI GenBank database (https://www.ncbi.nlm.nih.gov/) and all samples were recorded by their accession numbers. The studied taxa of genus Acacia and their Accession numbers in GenBank.
Acknowledgements
The authors appreciate Agriculture Research Station (Riyadh-Saudi Arabia), Saudi Ministry of Environment, Water and Agriculture and seeds bank for supplying seeds of Acacia.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Al-Hemaid FMA, Ali MA, Lee J, Kim S, Oliur Rahman M. 2015. Molecular evolutionary relationships of Euphorbia scordifolia Jacq. within the genus inferred from analysis of internal transcribed spacer sequences. Bangladesh J Plant Taxon. 22(2):111–118.
- Al-Juhani WS. 2019. Evaluation of the capacity of the DNA barcode ITS2 for identifying and discriminating dry land plants. GenMol Res. 18(1):32–46.
- Boatwright JS, Maurin O, van der Bank M. 2015. Phylogenetic position of Madagascan species of Acacia sl and new combinations in Senegalia and Vachellia (Fabaceae, Mimosoideae, Acacieae). Bot J Lin Soc. 179(2):288–294.
- Bolson M, Smidt EdC, Brotto ML, Silva-Pereira V, Gomory D. 2015. ITS and t-rnH-psbA as efficient DNA barcodes to identify threatened commercial woody angiosperms from southern Brazilian atlantic rainforests. PLoS One. 10(12):e0143049. doi:10.1371/journal.pone.0143049
- Bouchenak-Khelladi Y, Maurin O, Hurter J, Van der Bank M. 2010. The evolutionary history and biogeography of Mimosoideae (Leguminosae): an emphasis on African acacias. Mol Phylogen Evol. 57:495–508.
- Chaudhary SA, Al-Jowaid AAA. 1999. Vegetation of the kingdom of Saudi Arabia, Riyadh (Saudi Arabia) Ministry of Agriculture and Water.
- Collenette S. 1999. Wildflowers of Saudi Arabia. Riyadh: National Commission for wildlife Conservation and development (NCWCD). pp. 799.
- Comben DF, McCulloch GA, Brown GK, Walter GH. 2020. Phylogenetic placement and the timing of diversification in Australia’s endemic Vachellia (Caesalpinioideae, Mimosoid Clade, Fabaceae) species. Austr SystBot. 33(1):103–109.
- de Vere N, Rich TCG, Ford CR, Trinder SA, Long C, Moore CW, Satterthwaite D, Davies H, Allainguillaume J, Ronca S, et al. 2012. DNA barcoding the native flowering plants and conifers of Wales. PLoS One. 7:e37945. doi:10.1371/journal.pone.0037945
- Dharani N. 2006. Field guide to Acacias of East Africa. Cape Town: Struik Publishers.
- Dong W, Cheng T, Li C, Xu C, Long P, Chen C, Zhou S. 2014. Discriminating plants using the DNA barcode rbcl b: an appraisal based on a large data set. Mol Ecol Res. 14:336–343.
- Dunning LT, Savolainen V. 2010. Broad-scale amplification of matK for DNA barcoding plants, a technical note. Bot J Linn Soc. 164(1):1–9.
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucl Acid Res. 32(5):1792–1797.
- Elansary HO. 2013. Towards a DNA barcode library for Egyptian flora, with a preliminary focus on ornamental trees and shrubs of two major gardens. DNA Barcodes. 1:46–55.
- Elansary HO, Mostafa GG, Elansary DO, Hussein A. 2011. Assessment of genetic diversity within the genus Acacia grown in Egypt and studying its relation to leaf tannin and phenolic contents. Seventh Pl. Breed. Conf. May 4-5, 2011, Alexandria, Egypt. Egypt J Plant Breed. 2(Special issue):243–250.
- Fay MF, Cameron KM, Prance GT, Lledó MD, Chase MW. 1997. Familial relationships of Rhabdodendron (Rhabdodendraceae): plastid rbcL sequences indicate a caryophyllid placement. Kew Bull. 52: 923–932.
- Fazekas AJ, Kuzmina ML, Newmaster SG, Hollingsworth PM. 2012. DNA barcoding methods for land plants. Meth Mol Biol. 858:223–252.
- Ford CS, Ayres KL, Toomey N, Haider NADIA, van Alphen Stahl J, Kelly LJ, Wikström N, Hollingsworth PM, Duff RJ, Hoot SB, et al. 2009. Selection of candidate coding DNA barcoding regions for use on land plants. BotJ Linn Soc. 159(1):1–11.
- Gao Z, Wang X, Wei X, Liu Y, Han J. 2019. DNA Mini-Barcoding: A Derived Barcoding Method for Herbal Molecular Identification. Front Plant Sci. 10:987:1–11. doi:10.3389/fpls.2019.00987
- Gojobori T, Li WH, Graur D. 1982. Patterns of nucleotide substitution in pseudogenes and functional genes. J Mol Evol. 18:360–369.
- Harnelly E, Thomy Z, Fathiya N. 2018. Phylogenetic analysis of Dipterocarpaceae in Ketambe Research Station, Gunung Leuser National Park (Sumatra, Indonesia) based on rbcL and matK genes. Biodivers J Biol Divers. 19(3):1074–1080.
- Hidayat T, Pancoro A. 2008. Molecular phylogenetic studies provide a basic knowledge of improving genetic resources. Agrobiogen. 4:35–40.
- Khan MQ, Khalil AT, Shinwari ZK. 2016. Searching for DNA Barcodes in Plants. Am-Euras J Agric Environ Sci. 15(4):504–513.
- Kimura M. 1981. Estimation of evolutionary distances between homologous nucleotide sequences. Proc Natl Acad Sci. 78:454–458.
- Kress JW, Erickson DL. 2008. DNA barcodes: genes, genomics, and bioinformatics. Proc Natl Acad Sci. 105:2761–2762.
- Kress WJ and Erickson DL 2007. A two-locus global DNA barcode for land plants: the coding rbcL gene complements the non-coding t-rnH-psbA spacer region. PLoS One. 2:e508. doi:10.1371/journal.pone.0000508
- Kyalangalilwa B, Boatwright JS, Daru BH, Maurin O, van der Bank M. 2013. Phylogenetic position and revised classification of Acacia sl (Fabaceae: Mimosoideae) in Africa, including new combinations in Vachellia and Senegalia. Bot J Linn Soc. 172(4):500–523.
- Levin RA, Wagner WL, Hoch PC, Nepokroeff M, Pires JC, Zimmer EA, Sytsma KJ. 2003. Family-level relationships of Onagraceae based on chloroplast rbcL and ndhF data. Am J Bot. 90(1):107–15.
- Lewis GP. 2005. Acacieae. In: Lewis GP, Schrire BD, Mackinder B, Lock JM, editor. Legumes of the world. Kew: Royal Botanical Gardens; p. 187–191.
- Luckow M, Miller JT, Murphy DJ, Livshultz T. 2003. A phylogenetic analysis of the Mimosoideae (Leguminosae) based on chloroplast DNA sequence data. In: Klitgaard BB, Bruneau A, editor. Advances in Legume Systematics, Part 10, Higher Level Systematics, Royal Botanic Gardens, Kew, United Kingdom.
- Luo G, Li X, Han Z, Zhang Z, Yang Q, Guo H, Fang J. 2016. Transition and transversion mutations are biased towards GC in transposons of Chilo suppressalis (Lepidoptera: Pyralidae). Genes. 7:1–12.
- Maslin BR, Ho BC, Sun H, Bai L. 2019. Revision of Senegalia in China, and notes on introduced species of Acacia, Acaciella, Senegalia and Vachellia (Leguminosae: Mimosoideae). Plant Divers. 41(6):353–480.
- Maslin BR, Miller J, Seigler DS. 2003. Overview of the generic status of Acacia (Leguminosae: Mimosoideae). Austral Syst Bot. 16:1–18. doi:10.1071/SB02008.
- Migahid AM. 1996. Flora of Saudi Arabia. University Libraries. King Saudi University Press,Saudi Arabia.
- Miller JT, Bayer RJ. 2003. Molecular phylogenetics of Acacia subgenera Acacia and Aculeiferum (Fabaceae: Mimosoideae), based on the chloroplast matK coding sequence and flanking trnK intron spacer regions. Austr Syst Bot. 16:27–33.
- Miller JT, Seigler DS. 2012. Evolutionary and taxonomic relationships of Acacia s.l. (Leguminosae: Mimosoideae). Austr Syst Bot. 25:217–224.
- Mosa KA, Gairola S, Jamdade R, El-Keblawy A, Al Shaer KI, Al Harthi EK, Shabana HA, Mahmoud T. 2019. The Promise of Molecular and Genomic Techniques for Biodiversity Research and DNA Barcoding of the Arabian Peninsula Flora. Front Plant Sci. 9:1929. doi:10.3389/fpls.2018.01929.
- Piredda RSM, Attimonelli M, Bellarosa R, Schirone B, Simeone MC. 2011. Prospects of barcoding the Italian wild dendroflora: oaks reveal severe limitations to tracking species identity. Mol Ecol Resour. 11:72–83.
- Ragupathy SGNS. 2009. Ethnobotany genomics–use of DNA barcoding to explore cryptic diversity in economically important plants. Ind J Sci Technol. 2:(5):1–8.
- Ragupathy S, Seigler DS, Ebinger JE, Maslin BR. 2014. New combinations in Vachellia and Senegalia (Leguminosae: Mimosoideae) for south and west Asia. Phytotaxa. 162(3):174–180.
- Schori M, Showalter AM. 2011. DNA barcoding as a means for identifying medicinal plants of Pakistan. Pak J Bot. 43:1–4.
- Seigler DS, Ebinger JE, Miller JT. 2006. The genus Senegalia (Fabaceae: Mimosoideae) from the new world. Phytologia. 88:34–94.
- Shinwari ZK, Jan SA, Khalil AT, Khan A, Ali M, Qaiser M, Zahra NB. 2018. Identification and phylogenetic analysis of selected medicinal plant species from Pakistan: DNA barcoding approach. Pak J Bot. 50(2):553–560.
- Sinsheimer JS, Little RJ, Lake JA. 2012. Rooting gene trees without outgroups: EP rooting. Genome Biol Evol. 4(8):821–831. doi:10.1093/gbe/evs047.
- Steven G, Subramanyam R. 2009. Testing plant barcoding in a sister species complex of pantropical Acacia (Mimosoideae, Fabaceae). Mol Ecol Res. 9:172–180.
- Suleiman MK, Dixon K, Commander L, Nevill P, Bhat NR, Jacob Sheena, Islam MA, Thomas R. 2018a. Seed germinability and longevity influences regeneration of Acacia gerrardii. Plant Ecol. 219(5):591–609.
- Suleiman MK, Quoreshi AM, Bhat NR, Manuvel AJ. 2018b. Species identification of Vachellia pachyceras from Kuwait and its relatives Vachellia gerrardii and Vachellia tortilis based on multilocus plastid gene sequences. Edinburgh J Bot. 75(1):73–90.
- Tamura K, Nei M. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 10:512–526.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Tang Y, Wu Y, Huang R, Chao N, Liu Y, Xu P, Li K, Cai D, Luo Y. 2016. Molecular identification of Uncaria (Gouteng) through DNA barcoding. Chin Med. 11(1):1–14.
- Wight R, Arnott GAW. 1834. Prodromus. Florae peninsulae Indiae Orientalis. London: Parbury, AllenandCo.. Vol. 272, pp. 1–18. doi:10.1071/SB02008.
- Zhang Z, Gerstein M. 2003. Patterns of nucleotide substitution, insertion and deletion in the human genome inferred from pseudogenes. Nucl Acid Res. 31:5338–5348.
- Zhao Z, Boerwinkle E. 2002. Neighboring-nucleotide effects on single nucleotide polymorphisms: A study of 2.6 million polymorphisms across the human genome. Gen Res. 12:1679–1686.