Abstract
Background: Enhancer RNA (eRNA) is a non-coding RNA transcribed by enhancers and has a crucial role in controlling gene transcription. However, the potential functions of eRNA in prostate cancer (PCa) are still not fully understood.
Methods: Clinical information and expression data of eRNAs and their target genes for PCa were downloaded from TCGA. Each eRNA in PCa samples was divided into high- and low-expression groups. The Kaplan–Meier method was used to identify the eRNAs significantly associated with the overall survival of PCa. Their correlations with age, prostate-specific antigen (PSA), and tumor stage were analyzed using Spearman coefficients. Finally, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEEG) analyzes were used to predict the possible biological functions and pathways associated with eRNAs.
Results: We identified 12 eRNAs associated with PCa survival, of which PARGP1 was the most clinically significant. It was associated with biochemical recurrence in PCa patients and was also a valuable complement to distinguish between PSA 0–4 ng/ml and 4–10 ng/ml. GO, and KEEG analysis showed that herpes simplex virus-1 infection was the most biologically significant enriched process.
Conclusions: PARGP1 is associated with biochemical recurrence in prostate cancer and contributes to assessing the outcome.
Introduction
Prostate cancer (PCa) is one of the most common malignant tumors worldwide and ranks first among malignant tumors in men in the United States (Miller et al. Citation2019). In China, the morbidity of PCa ranks seventh among male cancers (Liu et al. Citation2019). According to the Global Burden of Disease 2017, there were 144,887 cases of PCa and 51,718 deaths in China (Pang et al. Citation2016). When PCa grows rapidly or metastasizes, the disease worsens, and the late metastasis and mortality rates are higher (Sebesta and Anderson Citation2017; Abdollah et al. Citation2018). Prostate-specific antigen (PSA) is a traditional screening parameter. However, PSA has low specificity, especially when the total PSA value is 4–10 ng/ml; therefore, there is an urgent need to identify new reliable diagnostic and prognostic biomarkers or therapeutic targets for PCa (Nguyen-Nielsen and Borre Citation2016; Eapen et al. Citation2019; Eskra et al. Citation2019).
Enhancer RNAs (eRNAs) are transcribed and produced by enhancers, 500–5 kb in length (Zhang et al. Citation2019). These regulatory molecules stimulate the activity of critical epigenetic regulatory enzymes, promoting gene expression. Numerous studies showed that eRNAs could be used as new targets for tumor therapy (Puc et al. Citation2015; Zhao et al. Citation2016; Bose and Berger Citation2017).
There is a paucity of research on eRNA in PCa. Therefore, in this study, we screened eRNAs and the target genes related to outcomes PCa using The Cancer Genome Atlas (TCGA) and performed clinical correlation analysis and functional enrichment to reveal their potential biological functions and signal pathways related to PCa. Our ultimate aim is to improve outcome evaluation and personalized treatment.
Materials and methods
Data download and collation
Fragments Per Kilobase Million (FPKM) files containing eRNA sequencing data, clinical data, and survival data for PCa and 32 other tumors from TCGA were downloaded from the UCSC Cancer Browser (https://xena.ucsc.edu/); the PreSTIGE algorithm was used to predict the target genes of the eRNAs, and downloaded the RNA-Seq of pca from the TCGA, screening the common genes to both for further analysis (Corradin et al. Citation2014; Vučićević et al. Citation2015). If several probes detected the same eRNA, the average value was taken as the eRNA expression value. The eRNA expression matrix was combined with patient survival data using the limma software package.
Screening survival-related eRNA
The eRNAs were divided into high and low expression groups based on the median value of expression. Kaplan–Meier survival curves were used to compare the high- and low-expression groups’ overall survival rates. The log-rank test was used to determine whether there were differences between groups for overall survival. Statistical significant differences were assumed when the p-value was < 0.05.
The Spearman method was used to identify eRNA and target gene pairs with correlation coefficients r > 0.04 and p < 0.001 in Pca (Gu et al. Citation2019). ERNAs associated with overall survival (p < 0.05) and their target gene levels (p < 0.001) were considered key eRNAs.
Clinical correlation analysis
The R software limma package was used to analyze the correlations between survival-related eRNA and target genes and biochemical recurrence status, age, Gleason score, pathological T stage, lymphatic metastasis status, and distant metastasis of PCa patients. The ggpubr package was used to draw images.
Functional annotation and enrichment analysis
To further understand the biological information of eRNA, we analyzed co-expression with other genes in PCa. The screening condition was that the correlation coefficient would be more than 0.4 and p < 0.001. The ‘ClusterProfiler’ package in R software was used for Gene Ontology (GO) functional enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis.
Statistical analysis
The eRNAs were divided into high and low expression groups based on the median value of expression. The overall survival for the study population was generated using the Kaplan–Meier method, and differences in survival were validated using the log-rank test. The Spearman method was used to identify the correlation coefficient. All statistical analyzes were performed in R, version 4.0.0 (http://www.r-project.org/) and the accompanying software package.
Results
ERNAs related to PCa outcome
Based on RNA sequencing data from PCa patients downloaded from TCGA, there were 1580 eRNAs in PCa, 37 of which were significantly associated with overall survival (Kaplan–Meier log-rank test, p < 0.05; Table ). The PreSTIGE algorithm was used to predict the target genes for these 37 eRNAs. When the levels of these 37 eRNAs were compared with the levels of their predicted target gene mRNAs, only 12 were found to be significantly correlated (Spearman’s rank correlation coefficient r > 0.4, p < 0.001, Table ).
Table 1. List of overall survival associated eRNAs derived from enhancers.
ERNA PARGP1 is the key eRNA in PCa
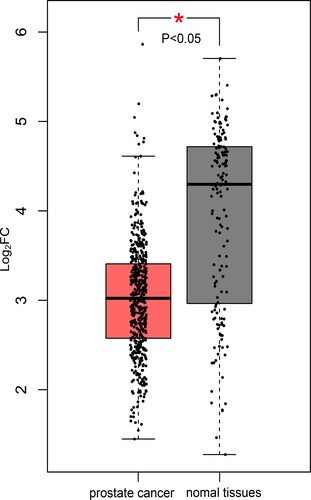
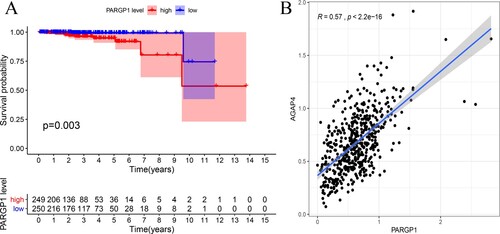
PARGP1 was found to be a critical eRNA in PCa. In PCa patients, overall survival was better in the PARGP1 low-expression group than in the high-expression group (Kaplan–Meier log-rank test, p = 0.003; Figure (a)). Levels of PARGP1 and the target gene AGAP4 mRNA were positively correlated (Figure (b), r = 0.571, p < 0.001). AGAP4 protein expression was significantly different between PCa patients and normal subjects (Figure ).
Figure 1. Impact of eRNA PARGP1 on prostate adenocarcinoma (PRAD). (a) Kaplan–Meier survival curve for patients with PARGP1-high and PARGP1-low expression. (b) Scatterplot showing the association between PARGP1 and AGAP4 levels.
The effect of PARGP1 on overall survival in the other 32 tumors was limited to prostate adenocarcinoma (PRAD), head and neck squamous cell carcinoma (HNSC), brain lower grade glioma (LGG), and pheochromocytoma and paraganglioma (PCPG) with high specificity (Table , Figure ).
Figure 3. Impact of eRNA PARGP1 on other cancers. (a) Kaplan–Meier overall survival curve for patients with PARGP1-high and PARGP1-low expression on head and neck squamous cell carcinoma (HNSC). (b) Kaplan–Meier overall survival curve for patients with PARGP1-high and PARGP1-low expression in brain lower grade glioma (LGG). (c) Kaplan–Meier overall survival curve for patients with PARGP1-high and PARGP1-low expression on pheochromocytoma and paraganglioma (PCPG).
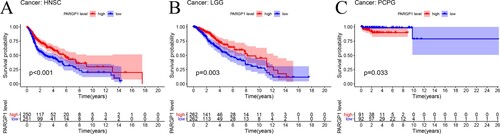
Table 2. Kaplan–Meier survival analysis and gene expression correlations for PARGP1 and AGAP4 across 33 cancer types from the Cancer Genome Atlas (TCGA) project.
PARGP1 is related to the state of biochemical recurrence of PCa
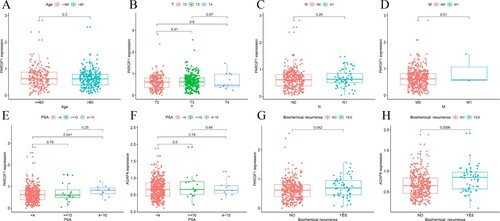
There was no significant difference in PARGP1 expression among PCa patients of different ages, pathological T-stages, N-stages, or distant metastases (Figure (a–d), p > 0.05). However, there was a significant difference in PARGP1 expression between patients with PSA less than 4 ng/ml and those with 4–10 ng/ml (Figure (e), p < 0.05). The expression of PARGP1 was significantly higher in patients with biochemical recurrence than in those without (Figure (g), p < 0.001), and the expression level of the target gene AGAP4 showed consistent results (Figure (h), p < 0.001).
Figure 4. Gene expression related to prostate cancer. (a–d) Expression levels of PARGP1 were not related to age in patients with prostate cancer or to the TNM stage of the tumor. (e–f) The difference between the expression level of PARGP1 and the value of PSA. There was a significant difference in PARGP1 expression level for PSA between 0–4 ng/ml and 4–10 ng/ml. (g–h) The expression levels of PARGP1 and AGAP4 were related to biochemical recurrence of prostate cancer (p < 0.05).
Function and pathway enrichment analysis
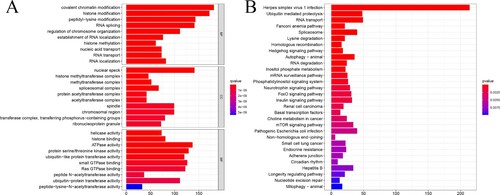
The GO and KEGG pathways were analyzed using the ‘clusterProfiler’ package in R software to explore further the molecular mechanisms of the 892 genes co-expressed with PARGP1 on the occurrence and progression of PCa. The functions of these genes were focused on biological processes, molecular functions, and cellular composition (Figure (a)). The main pathway for KEGG enrichment was infection with herpes simplex virus 1 (HSV-1) (Figure (b)).
Discussion
There is a growing body of evidence suggesting that enhancers have inherent promoter capabilities and can transcribe non-coding RNA (Kim et al. Citation2010). Enhancer transcription and its product, enhanced eRNA, have been widely studied in mouse and human cells, which possess several common characteristics. Mammalian eRNA is usually low in abundance, unspliced, and retained in the nucleus, and they are usually transcribed in both directions (Andersson et al. Citation2014; Mikhaylichenko et al. Citation2018). In human cancers, the importance of eRNAs in the activation of oncogenes or oncogenic signaling pathways has received increasing attention. For example, the activation of ESR1 can increase eRNA transcription in breast cancer (Li et al. Citation2016). Together, these findings suggest the critical role of eRNA in tumorigenesis and their potential effects in eRNA-targeted therapy (Léveillé et al. Citation2015).
Statistics show that the incidence of PCa is increasing in younger ages, with the incidence of PCa in people aged 15–40 years steadily increasing at a rate of 2% per year; men under 40 years of age are more likely to develop metastatic disease and have a higher mortality rate than older men (Bleyer et al. Citation2020; Sartor Citation2020). PSA has dramatically improved the detection of PCa as a traditional screening indicator; however, when PSA values are between 4–10 ng/ml, we need to do more specific tests to rule out other factors, including inflammation or catheterization.
In the present study, there were significant differences in expression levels of PARGP1 in patients with PSA values 0–4 ng/ml and 4–10 ng/ml, suggesting that PARGP1 can be used as a potential diagnostic marker to differentiate benign from malignant prostate lesions when the PSA is less than 10 ng/ml. Expression levels of PARGP1 in patients with biochemical recurrence were significantly higher than those without, and the target gene AGAP4 showed the same results. Eexpression of PARGP1 and AGAP4 may be associated with a significantly increased risk of biochemical recurrence. Based on these findings, we screened the functionally unannotated eRNA PARGP1 and found that it may be one of the essential eRNAs in PCa. Based on GO enrichment analysis, the predominant KEGG pathway was HSV-1 infection. HSV-1 is a neurotropic herpesvirus; the γ34.5 gene is a critical determinant of the neurovirulence of this virus (Taneja et al. Citation2001). Recent studies showed that UV-inactivated HSV-1 is involved in killing PCa cells by activating natural killer cells via Toll-like receptor 2. HSV-1 may in the future be a novel option for the treatment of PCa, especially neuroaggressive PCa (Kim et al. Citation2012; Vogel et al. Citation2014; Samudio et al. Citation2019).
Our study also has some limitations. Our analysis based on TCGA yielded PARGP1, the most critical eRNA in PCa, and AGAP4, the predicted target gene, which only provides a direction for subsequent PCa research and has not been validated in large samples. Nevertheless, based on the analysis of these large data sets, we found that PARGP1 was also associated with patient survival in PRAD, HNSC, LGG, and PCPG with high specificity, limiting the scope of the study and reducing unnecessary duplication of studies. PARGP1 may also be used as a potential diagnostic marker for the diagnosis of PCa and the detection of biochemical recurrence. Novel therapeutic approaches targeting PARGP1 expression may help induce protective immunity to treat these cancers effectively.
Conclusions
We analyzed the effect of PARGP1 on PCa outcome and found that it can be used as an ancillary means to distinguish between PSA 0–4 ng/ml and 4–10 ng/ml in patients with PCa. It is also related to the biochemical recurrence in patients with PCa. Detection of the PARGP1 expression level is critical for predicting patients’ outcome, diagnosis, and management with PCa.
Authors’ contributions
Project design and implementation were conceived by Weiguo Chen, and Feng Zhou, and Xi Zhang. Data collection, collation and analysis was performed by Qi Zhou, and Zhiyu Zhang, and Lu Ma. Manuscript drafting and editing were performed by Xiaojie Ang, and Zekun Xu. All authors read and approved the final manuscript.
Data sharing statement
Available in ‘figshare’ at https://doi.org/10.6084/m9.figshare.13498374.v3.
Ethical statement
The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Abdollah F, Dalela D, Sood A, Keeley J, Alanee S, Briganti A, Montorsi F, Peabody JO, Menon M. 2018. Impact of adjuvant radiotherapy in node-positive prostate cancer patients: the importance of patient selection. Eur Urol. 74(3):253–256.
- Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T, Ntini E. 2014. An atlas of active enhancers across human cell types and tissues. Nature. 507(7493):455–461.
- Bleyer A, Spreafico F, Barr R. 2020. Prostate cancer in young men: an emerging young adult and older adolescent challenge. Cancer. 126(1):46–57.
- Bose DA, Berger SL. 2017. eRNA binding produces tailored CBP activity profiles to regulate gene expression. RNA Biol. 14(12):1655–1659.
- Corradin O, Saiakhova A, Akhtar-Zaidi B, Myeroff L, Willis J, Cowper-Sal R, Lupien M, Markowitz S, Scacheri PC. 2014. Combinatorial effects of multiple enhancer variants in linkage disequilibrium dictate levels of gene expression to confer susceptibility to common traits. Genome Res. 24(1):1–13.
- Eapen RS, Nzenza TC, Murphy DG, Hofman MS, Cooperberg M, Lawrentschuk N. 2019. PSMA PET applications in the prostate cancer journey: from diagnosis to theranostics. World J Urol. 37(7):1255–1261.
- Eskra JN, Rabizadeh D, Pavlovich CP, Catalona WJ, Luo J. 2019. Approaches to urinary detection of prostate cancer. Prostate Cancer Prostatic Dis. 22(3):362–381.
- Gu X, Wang L, Boldrup L, Coates PJ, Fahraeus R, Sgaramella N, Wilms T, Nylander K. 2019. AP001056.1, a prognosis-related enhancer RNA in squamous cell carcinoma of the head and neck. Cancers (Basel). 11(3):347.
- Kim M, Osborne NR, Zeng W, Donaghy H, McKinnon K, Jackson DC, Cunningham AL. 2012. Herpes simplex virus antigens directly activate NK cells via TLR2, thus facilitating their presentation to CD4T lymphocytes. J Immunol. 188(9):4158–4170.
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, Markenscoff-Papadimitriou E. 2010. Widespread transcription at neuronal activity-regulated enhancers. Nature. 465(7295):182–187.
- Léveillé N, Melo CA, Agami R. 2015. Enhancer-associated RNAs as therapeutic targets. Expert Opin Biol Ther. 15(5):723–734.
- Li W, Notani D, Rosenfeld MG. 2016. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat Rev Genet. 17(4):207–223.
- Liu X, Yu C, Bi Y, Zhang ZJ. 2019. Trends and age-period-cohort effect on incidence and mortality of prostate cancer from 1990 to 2017 in China. Public Health. 172:70–80.
- Mikhaylichenko O, Bondarenko V, Harnett D, Schor IE, Males M, Viales RR, Furlong EE. 2018. The degree of enhancer or promoter activity is reflected by the levels and directionality of eRNA transcription. Genes Dev. 32(1):42–57.
- Miller KD, Nogueira L, Mariotto AB, Rowland JH, Yabroff KR, Alfano CM, Jemal A, Kramer JL, Siegel RL. 2019. Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin. 69(5):363–385.
- Nguyen-Nielsen M, Borre M. 2016. Diagnostic and therapeutic strategies for prostate cancer. Semin Nucl Med. 46(6):484–490.
- Pang C, Guan Y, Li H, Chen W, Zhu G. 2016. Urologic cancer in China. Jpn J Clin Oncol. 46(6):497–501.
- Puc J, Kozbial P, Li W, Tan Y, Liu Z, Suter T, Ohgi KA, Zhang J, Aggarwal AK, Rosenfeld MG. 2015. Ligand-dependent enhancer activation regulated by topoisomerase-I activity. Cell. 160(3):367–380.
- Samudio I, Hofs E, Cho B, Li M, Bolduc K, Bu L, Liu G, Lam V, Rennie P, Jia W, Elisia I. 2019. UV Light-inactivated HSV-1 stimulates natural killer cell-induced killing of prostate cancer cells. J Immunother. 42(5):162–174.
- Sartor O. 2020. Why is prostate cancer incidence rising in young men? Cancer. 126(1):17–18.
- Sebesta EM, Anderson CB. 2017. The surgical management of prostate cancer. Semin Oncol. 44(5):347–357.
- Taneja S, MacGregor J, Markus S, Ha S, Mohr I. 2001. Enhanced antitumor efficacy of a herpes simplex virus mutant isolated by genetic selection in cancer cells. Proc Natl Acad Sci USA. 98(15):8804–8808.
- Vogel K, Thomann S, Vogel B, Schuster P, Schmidt B. 2014. Both plasmacytoid dendritic cells and monocytes stimulate natural killer cells early during human herpes simplex virus type 1 infections. Immunology. 143(4):588–600.
- Vučićević D, Corradin O, Ntini E, Scacheri PC, Ørom UA. 2015. Long ncRNA expression associates with tissue-specific enhancers. Cell Cycle. 14(2):253–260.
- Zhang Z, Lee JH, Ruan H, Ye Y, Krakowiak J, Hu Q, Xiang Y, Gong J, Zhou B, Wang L, Lin C. 2019. Transcriptional landscape and clinical utility of enhancer RNAs for eRNA-targeted therapy in cancer. Nat Commun. 10(1):4562.
- Zhao Y, Wang L, Ren S, Wang L, Blackburn PR, McNulty MS, Gao X, Qiao M, Vessella RL, Kohli M, Zhang J. 2016. Activation of P-TEFb by androgen receptor-regulated enhancer RNAs in castration-resistant prostate cancer. Cell Rep. 15(3):599–610.