Abstract
Osteoarthritis (OA) is the most common form of arthritis and a leading cause of chronic pain and disability in the elderly. However, the detailed molecular mechanisms of OA remain unclear. Therefore, uncovering the molecular mechanisms underlying OA progression is crucial for the development of OA therapy. Of note, for the last 6 years (especially in 2020 and 2021), increasing numbers of studies have suggested that aberrantly expressed circular RNAs (circRNAs) are involved in the pathogenesis of OA. Until now, circRNAs have been attracting high attention as microRNA (miRNA) sponges that sequester miRNAs from their endogenous targets, meanwhile alternative functions of circRNAs have been also suggested. Moreover, it has been reported that circulating or tissue-specific circRNAs can be used for diagnosing different types of human diseases, including cancers, neurological diseases, and inflammatory diseases. Given the putative involvement of circRNAs in the pathogenesis of OA, it looks promising that some of the dysregulated circRNAs may have the potential to serve as therapeutic targets and/or diagnostic biomarkers for OA. This review summarizes the current progress on dysregulated circRNAs and their pathological implications in knee OA, which may provide a direction for revealing their therapeutic potential in OA.
Introduction
Since the discovery of microRNA (miRNA), overwhelming evidence has been suggesting that alterations in miRNA expression levels are associated with many human diseases (Paul et al. Citation2018). MiRNAs are small non-coding RNAs (ncRNAs) of ∼22 nucleotides, which interact with complementary sequences within the target mRNAs to negatively regulate gene expression (O’Brien et al. Citation2018). NcRNAs are generally defined as functional RNA molecules which are not translated (Zhang et al. Citation2019). Until recently, different kinds of ncRNAs, including long ncRNAs (lncRNAs, >200 nucleotides) and circular RNAs (circRNAs) have been discovered by the latest technologies such as high-throughput RNA sequencing (RNA-seq) and bioinformatics analysis (Zhang et al. Citation2019).
CircRNAs are a type of ncRNAs characterized by covalently closed structures without 5′ caps and 3′ tails (Szabo and Salzman Citation2016). Unlike linear ncRNAs, such as miRNAs and lncRNAs, circRNAs are produced from precursor mRNA through a non-canonical splicing process called back-splicing (Szabo and Salzman Citation2016). Currently, three models of circRNA biogenesis have been suggested: spliceosome-mediated biogenesis, intron-involved biogenesis, and RNA-binding protein (RBP)-mediated biogenesis (Liu D et al. Citation2021; Zhang W et al. Citation2021). CircRNAs are generally categorized into three types based on the genomic sequence origin: exonic circRNAs, intronic circRNAs, and exon-intron circRNAs (Chen et al. Citation2021). Exonic circRNAs are the most abundant circRNAs (more than 80% of the known circRNAs) and mainly located in the cytoplasm, whereas intronic circRNAs and exon-intron circRNAs mainly reside in the nucleus (Liu D et al. Citation2021; Mao G et al. Citation2021; Zhang W et al. Citation2021). Compared with their corresponding linear transcripts, circRNAs are more highly expressed and stable because the absence of free ends makes circRNAs resistant to exonuclease activity (Holdt et al. Citation2018). In addition, circRNAs are broadly expressed in eukaryotic cells with conserved and tissue-specific patterns (Salzman et al. Citation2012). Interestingly, tissue-specific regulation of circRNAs has been reported in various diseases, including cancers (Vo et al. Citation2019), neurological diseases (Floris et al. Citation2017), and inflammatory diseases (Chen et al. Citation2019).
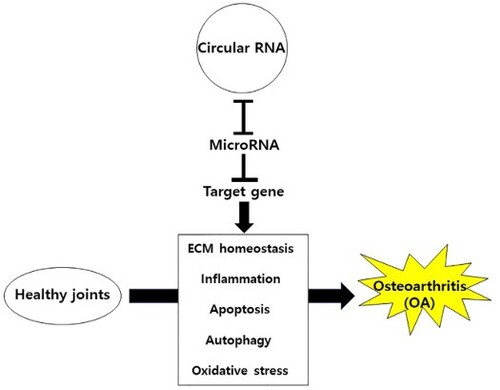
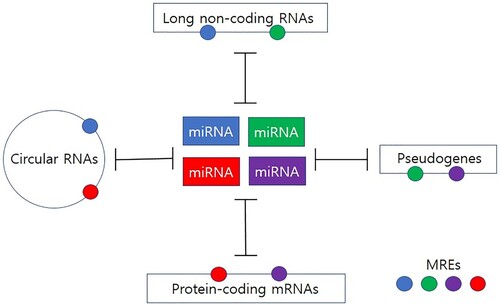
Recent studies have shown that, besides mRNA (Tay et al. Citation2011), other endogenous RNAs including pseudogene (Poliseno et al. Citation2010), lncRNA (Cesana et al. Citation2011), and circRNA (Memczak et al. Citation2013) can compete for shared miRNAs (Figure ): These competitive endogenous RNAs (ceRNAs) act as miRNA sponges through competitive binding of common miRNA response element (MRE), thereby cross-regulating each other (Su et al. Citation2013). Indeed, most circRNAs are considered to function as miRNA sponges. Meanwhile, other functional roles of circRNAs such as controlling protein localization, regulating alternative splicing, and interacting with proteins to act as scaffolds have been also proposed (Chen et al. Citation2021; Mao X et al. Citation2021), suggesting that circRNAs may play essential roles in diverse biological processes.
Figure 1. Network of competitive endogenous RNAs (ceRNAs). MicroRNAs (miRNAs) negatively regulate gene expression through binding to specific miRNA response elements (MREs) within the target transcripts. Different classes of ceRNAs (i.e. mRNAs, pseudogenes, long non-coding RNAs, and circular RNAs) with common MREs can compete for binding to a shared pool of miRNAs, thereby reducing the availability of miRNAs. MREs are represented by small circles with the same colors to their targeting miRNAs.
Osteoarthritis (OA) is the most common arthritis and a degenerative joint disease characterized by the degradation of articular cartilage (Neogi Citation2013). Articular cartilage is a specialized connective tissue protecting the ends of the bones to help joints to move smoothly (Sophia Fox et al. Citation2009). It consists of articular chondrocytes and the extracellular matrix (ECM) (Sophia Fox et al. Citation2009). Chondrocytes are the only active cells in articular cartilage essential for the maintenance of the ECM (Sophia Fox et al. Citation2009). Articular cartilage ECM is mainly composed of proteins (e.g. type II Collagen) and glycosaminoglycans (e.g. aggrecan). Articular cartilage preservation is crucial for joint health because it has a limited capacity for intrinsic repair (Sophia Fox et al. Citation2009). OA occurs when the protective cartilage is gradually damaged and commonly affects joints in knees, hips, and spine (Neogi Citation2013). Until now, the detailed mechanisms of OA are largely unknown. Therefore, uncovering important new contributors to OA progression is crucial for the development of OA therapy.
Because it is generally considered that OA cannot be reversed, the main goal of OA treatment is to reduce symptomatic synovial joint pain and improve function (Sofat and Kuttapitiya Citation2014). Although non-surgical options including pharmacological treatments, exercise, self-management, and alternative medicine are usually recommended and used for OA management (Khan et al. Citation2020), it is often criticized that the effectiveness of these treatments is largely dependent on subjective factors such as the subjective judgment of patients. Given biomarkers are objectively measurable indicators to assess biological/clinical status, identification and utilization of specific biomarkers for OA are critical for evaluating the effectiveness of treatments for OA.
Recently, advances in RNA-seq technologies and bioinformatics can facilitate discovering a large set of circRNAs presumably involved in the pathogenesis of OA (Zhou Z et al. Citation2018; Li H et al. Citation2019; Wang et al. Citation2019b; Xiao et al. Citation2019), suggesting that some of the dysregulated (i.e. aberrantly expressed) circRNAs may be useful therapeutic targets and/or biomarkers for OA. This review summarized current understanding of the connection between dysregulated circRNAs and knee OA, providing hints that may help determine their therapeutic potential in OA.
Clinical significance
Currently, no disease-modifying treatment for OA is available. Thus, it is important to identify factors and elucidate their roles in OA pathogenesis.
Emerging evidence suggests that dysregulated circRNAs are involved in the progression of OA by reducing the availability of miRNAs.
Overexpression or knockdown of some dysregulated circRNAs in animal models of OA suggests that some dysregulated circRNAs have therapeutic potential for the treatment of OA.
Identification of differentially expressed circRNAs in body fluids of OA patients suggests that circRNAs have the potential to be used as diagnostic biomarkers for OA.
Literature search
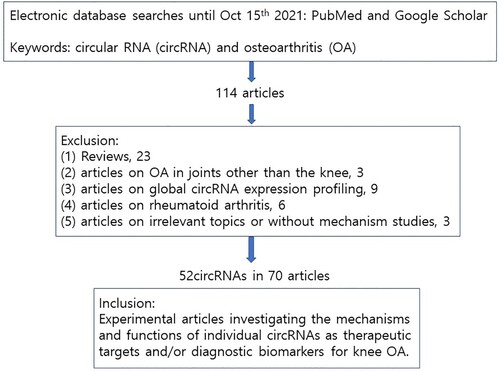
PubMed and Google Scholar were searched for articles published until October 15th 2021, using the keywords including ‘circular RNA’ or ‘circRNA’ and ‘osteoarthritis’. In the sections of Dysregulated circRNAs and their roles in OA and Dysregulated circRNAs as potential diagnostic biomarkers for OA, this review included all articles that investigated the mechanisms and functions of individual circRNAs which have the potential to serve as therapeutic targets and/or diagnostic biomarkers for knee OA. As summarized in Figure , the following articles were excluded in the sections: (1) review articles, (2) articles on OA in joints other than the knee, (3) articles on global circRNA expression profiling, (4) articles on rheumatoid arthritis, and (5) articles on irrelevant topics or without mechanism studies.
Involvement of circRNAs on the mechanisms involved in the pathogenesis of OA
Although the underlying mechanisms of OA are not yet fully understood, it has been considered that inflammation (Chen et al. Citation2017), ECM homeostasis (Maldonado and Nam Citation2013), oxidative stress (Marchev et al. Citation2017), apoptosis (Hwang and Kim Citation2015), and autophagy (Song et al. Citation2017) are major mechanisms involved in OA pathogenesis (Yu and Sun Citation2018).
A common feature of OA progression is inflammation-induced changes in ECM homeostasis (Maldonado and Nam Citation2013; Chen et al. Citation2017). It has been shown that levels of pro-inflammatory cytokines (e.g. interleukin-1β (IL-1β) and tumor necrosis factor-α (TNF-α)) and damage associated molecular patterns (DAMPs) were upregulated in the development of OA (Chen et al. Citation2017). These inflammatory molecules activated inflammatory pathways, thereby resulting in ECM degradation via upregulation of ECM degradation enzymes such as matrix metalloproteinases (MMPs) and a disintegrin and metalloproteinase with thrombospondin motifs (ADAMTS) (Maldonado and Nam Citation2013; Chen et al. Citation2017). Besides, it is also known that chronic inflammation along with aging can increase oxidative stress, thereby accumulating reactive oxygen species (ROS) in articular cartilage (Marchev et al. Citation2017). Since chronic oxidative stress in OA joints can induce cartilage abnormalities and/or degradation (Marchev et al. Citation2017), the maintenance of redox homeostasis in articular cartilage is important to prevent the initiation and progression of OA.
Another common feature of OA progression is chondrocyte death (Yu and Sun Citation2018). Maintenance of the chondrocyte survival is critical for the functional integrity of the ECM: Apoptosis-induced chondrocyte death plays an important role in the functional loss of the ECM (Hwang and Kim Citation2015). Moreover, it has been suggested that autophagy, a cellular recycling process, is also involved in OA pathogenesis (Song et al. Citation2017). There may exist a complex crosstalk between autophagy and apoptosis: Autophagy seems to be enhanced or reduced in OA cartilage in a context-dependent manner (Hwang and Kim Citation2015; Song et al. Citation2017). Nevertheless, recent studies suggest that autophagy may play a protective role in the progression of OA (Cheng et al. Citation2017; Zhou J et al. Citation2019; Luo et al. Citation2019).
Consistently, recent studies of upregulated or downregulated circRNAs in OA cartilage suggest that dysregulated circRNAs are also mainly associated with articular cartilage ECM homeostasis, inflammation, proliferation/apoptosis, autophagy, and oxidative stress as described in the following section and summarized in Table and Figure .
Figure 3. Mechanisms of circRNAs in the pathogenesis of OA. Upregulated (black) or downregulated (red) circRNAs participate in the progression of OA mainly via interaction with target miRNAs to regulate downstream gene expression. Dysregulated circRNAs are involved in the progression of OA through mechanisms including inflammation, extracellular matrix (ECM) homeostasis, oxidative stress, apoptosis/proliferation, and autophagy. CircRNAs whose function has been tested in vivo are marked with an asterisk.
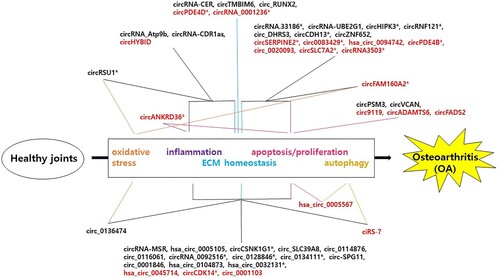
Table 1. Dysregulated circular RNAs (circRNAs) and their roles in osteoarthritis.
Dysregulated circRNAs and their roles in OA
Upregulated circRNAs in OA
CircRNA-CER (chondrocyte extracellular matrix related)
Liu et al. reported that 71 circRNAs were differentially expressed (16 were upregulated and 55 were downregulated) in cartilages isolated from the knee joints of OA patients by microarray analysis (Liu et al. Citation2016). Among these circRNAs, upregulation of circRNA-CER (also known as circRNA-100876) was further confirmed by reverse transcription-quantitative polymerase chain reaction (RT-qPCR). The authors also showed that the expression levels of circRNA-CER and MMP-13, a well-known ECM-degrading enzyme (Li H et al. Citation2017) were increased in chondrocytes induced by IL-1 and TNF-α. By network analysis of circRNAs-miRNAs-mRNAs, they identified 5 binding sites for miRNAs (miR-636, miR-665, miR-217, miR-646, and miR-136) in circRNA-CER. Then, they demonstrated that miR-136 can also target the 3′-UTR of MMP-13 by luciferase reporter assay. Knockdown of circRNA-CER by small interfering RNA (siRNA) suppressed MMP-13 expression and increased ECM formation, which were reversed by inhibiting miR-136. Altogether, their results suggested that circRNA-CER participated in chondrocyte ECM degradation process by blocking miR-136 which inhibits MMP-13 expression.
CircRNAs-MSR (mechanical stress related)
Liu et al. showed that 104 circRNAs were differentially expressed (44 were upregulated and 60 were downregulated) in OA cartilages, then confirmed the upregulation of circRNAs-MSR (also known as circRNA_100226) by RT-qRCR (Liu et al. Citation2017). The authors also found that mechanical stress increased the expression of circRNA-MSR in chondrocytes. Using bioinformatics analysis, the authors identified miR-875 as a common miRNA that can target both circRNA-MSR and TNF-α although it was not experimentally verified. Moreover, knockdown of circRNA-MSR by siRNA suppressed TNF-α expression and increased ECM formation (Liu et al. Citation2017). Taken together, their results suggested that circRNA-MSR could function as a negative regulator of miR-875 which inhibits TNF-α expression, resulting in ECM degradation.
In a separate study, Jia and Wei also reported that circRNA-MSR inhibited cell proliferation but enhanced apoptosis, inflammation, and ECM degradation in lipopolysaccharide (LPS)-induced chondrocytes through miR-643/Mitogen-activated protein kinase kinase 6 (MAP2K6) axis (Jia and Wei Citation2021). The regulatory network of circRNA-MSR/miR-643/MAP2K6 was validated by RNA pull down and luciferase activity assays and further confirmed by a series of loss and gain-of-function experiments. Altogether, these results suggest that circRNA-MSR may function as a sponge of both miR-875 and miR-643 to promote the progression of OA.
Has_circ_0005105
Wu et al. reported that has_circ_0005105 was upregulated by IL-1β in human chondrocytes (Wu Y et al. Citation2017). Then, the authors demonstrated that upregulation of has_circ_0005105 and downregulation of miR-26a were negatively correlated. Using bioinformatics analysis, they found that has_circ_0005105 has miR-26a binding site. Moreover, dual luciferase assay and western blot analysis revealed that hsa_circ_0005105 can inhibit miR-26a, thereby upregulating the expression of nicotinamide phosphoribosyl transferase (NAMPT) (Wu Y et al. Citation2017), an enzyme that triggers MMP-13 synthesis and release (Gosset et al. Citation2008). The authors also demonstrated that has_circ_0005105 inhibited the expression of type II Collagen and aggrecan but promoted the expression of MMP-13 and inflammatory factors (Prostaglandin E2, IL-6, IL-8), whereas miR-26a showed the opposite effect. Moreover, concomitant knockdown of NAMPT lessened the effect of has_circ_0005105 overexpression (Wu Y et al. Citation2017). Their results suggested that has_circ_0005105 promoted ECM degradation via miR-26a-NAMPT signaling.
Recently, using similar approaches as Wu J et al. (Citation2017), two independent studies demonstrated that hsa_circ_0005105 can interact with other miRNAs: Shi et al. showed that hsa_circ_0005105 (also known as circRNA SEC24 homolog A, COPII coat complex component (circSEC24A)) reduced proliferation and enhanced apoptosis and inflammation through miR-142-5p/SRY-related HMG-box transcription factor 5 (SOX5) axis in IL-1β treated chondrocytes (Shi et al. Citation2021). Zhang et al. also showed that hsa_circ_0005105 (circSEC24A) inhibited proliferation and enhanced apoptosis and ECM degradation through miR-26b-5p/DNA methyltransferase 3 alpha (DNMT3A) axis in IL-1β-induced chondrocytes (Zhang Z et al. Citation2021). Collectively, these results suggest that hsa_circ_0005105 may contribute to the progression of OA through functioning as a sponge of miR-26a, miR-142-5p, and miR-26b-5p.
CircRNA_Atp9b
Zhou et al. reported that circRNA_Atp9b (also known as circ_15898) expression was upregulated in IL-1β treated mouse chondrocytes (Zhou ZB et al. Citation2018). Knockdown of circRNA_Atp9b increased the expression of type II Collagen while reducing the generation of MMP-13 and inflammatory factors (IL-6, COX-2), suggesting that circRNA_Atp9b may promote ECM degradation and production of inflammatory factors (Zhou ZB et al. Citation2018). Using bioinformatics analysis and dual-luciferase assay, they demonstrated that circRNA_Atp9b targeted miR-138-5p although they did not further investigate the potential target of miR-138-5p. Their results suggested that circR-NA_Atp9b controlled the progression of OA by modulating ECM synthesis and chondrocyte inflammation, which may be mediated by miR-138-5p.
CircRNA.33186
Zhou et al. reported that circRNA.33186 was highly upregulated in both chondrocytes induced by IL-1β and cartilage tissues induced by destabilized medial meniscus (DMM), suggesting that circRNA.33186 may be associated with the development of OA (Zhou ZB et al. Citation2019). Using bioinformatics analysis and luciferase activity assay, the authors showed that circRNA.33186 can inhibit miR-127-5p and increase MMP-13 expression. Consistently, knockdown of circRNA.33186 inhibited apoptosis and MMP-13 expression, but enhanced proliferation and type II Collagen expression in IL-1β treated chondrocytes, suggesting that circRNA.33186 regulated chondrocyte apoptosis/proliferation and ECM homeostasis (Zhou ZB et al. Citation2019).
Importantly, they also showed that in vivo inhibition of circRNA.33186 alleviated DMM-induced OA by intra-articular (IA) lentivirus injection with circRNA.33186 siRNA (Zhou ZB et al. Citation2019). Taken together, their findings suggested that circRNA.33186 contributed to the development of OA by blocking miR-127-5p which inhibits MMP-13.
Circ_0136474
Using RNA-seq and microarray data collected from public databases, Li et al. showed that circ_0136474 and miR-127-5p were negatively correlated pairs: Circ_0136474 showed a strong increase while miR-127-5p was downregulated in OA cartilage tissues (Li Z et al. Citation2019). The authors predicted the targeted relationship between miR-127-5p and MMP-13 by bioinformatics analysis and then verified the relationship by dual-luciferase reporter assay. Overexpression of circ_0136474 promoted apoptosis and suppressed proliferation by inhibiting miR-127-5p and enhancing MMP-13, whereas knockdown of circ_0136474 or inhibition of MMP-13 showed the opposite effects via downregulation of inflammatory mediators (IL-1β, TNF-α, IL-17) and upregulation of type II Collagen (Li Z et al. Citation2019). Their results suggested that circ_0136474 promoted cell apoptosis and suppressed cell proliferation in OA by blocking miR-127-5p which inhibits MMP-13 expression.
In a separate study, Zhu et al. also reported that circ_0136474 promoted IL-1β-induced proliferation inhibition, apoptosis, and oxidative stress through miR-766-3p/DNMT3A axis (Zhu H et al. Citation2021). The regulatory network of circ_0136474/miR-766-3p/DNMT3A was validated by luciferase reporter and RNA immunoprecipitation assays and further confirmed by a series of loss and gain-of-function experiments focusing on IL-1β-induced chondrocyte oxidative injury (Zhu H et al. Citation2021). Altogether, these results suggest that circ_0136474 may function as a sponge of both miR-127-5p and miR-766-3p to promote the progression of OA.
CircRNA-UBE2G1
Chen et al. reported that the expression levels of circRNA-UBE2G1 and Hypoxia-inducible factor-1 alpha (HIF-1α) were increased but miR-373 expression was decreased in OA tissues (Chen G et al. Citation2020). Moreover, LPS-induced apoptosis was reduced by circRNA-UBE2G1 inhibition, while miR-373 inhibition or HIF-1α overexpression reversed the effect of circRNA-UBE2G1 inhibition (Chen G et al. Citation2020). They also demonstrated that circRNA-UBE2G1 can sequester miR-373 which inhibits HIF-1α, suggesting that circRNA-UBE2G1 promoted the progression of LPS-induced OA through controlling the miR-373/HIF-1α axis.
In a separate study, Fu et al. also reported that circRNA-UBE2G1 (also known as circ_0008956) can promote IL-1β-induced OA progression through miR-149-5p/NAMPT axis (Fu S et al. Citation2021). Using bioinformatics analysis, the authors predicted that miR-149-5p could bind to both circRNA-UBE2G1 and NAMPT and verified the interactions by dual-luciferase assay. To test the functional roles of circRNA-UBE2G1 and miR-149-5p, they investigated the effects of circRNA-UBE2G1 knockdown or miR-149-5p overexpression on cell viability, apoptosis, and ECM degradation. They showed that knockdown of circRNA-UBE2G1 or overexpression of miR-149-5p reduced IL-1β-induced chondrocyte damage, and these effects were reversed by concomitant knockdown of miR-149-5p or overexpression of NAMPT respectively (Fu S et al. Citation2021). Altogether, these results suggest that circRNA-UBE2G1 may contribute to the progression of OA through functioning as a sponge of both miR-373 and miR-149-5p.
CircRNA-CDR1as (the antisense cerebellar degenerative-related protein-1)
Zhang et al. found that circRNA-CDR1as was highly upregulated and miR-641 was downregulated in OA chondrocytes (Zhang W et al. Citation2020). Knockdown of circRNA-CDR1as using siRNA increased type II Collagen but reduced MMP-13 and IL-6 levels, while co-transfection of the miR-641 inhibitor partially reversed the effects. Bioinformatics analysis and experimental verification such as RNA pull down and luciferase activity assays showed that both circRNA-CDR1as and Fibroblast growth factor-2 (FGF-2) interact with miR-641 (Zhang W et al. Citation2020). Indeed, FGF-2 was upregulated in OA chondrocytes and knockdown of FGF-2 using siRNA increased type II Collagen but reduced the expression of MMP-13 and IL-6. Moreover, knockdown of circRNA-CDR1as using siRNA decreased the levels of FGF-2, p-Mitogen-activated protein kinase (MEK)1/2, and p-Extracellular signal-regulated kinase (ERK)1/2 (Zhang W et al. Citation2020). Therefore, their results suggested that circRNA-CDR1as acted as a sponge of miR-641 to promote ECM degradation and inflammation through FGF-2 mediated MEK/ERK pathway.
CircPSM3
Previously, it was reported that circPSMC3 inhibited the proliferation and metastasis of gastric cancer by directly targeting miR-296-5p (Rong et al. Citation2019). Thus Ni et al. investigated the roles of circPSM3 and miR-296-5p in the development of OA chondrocytes. They found that circPSM3 expression was upregulated while miRNA-296-5p was downregulated in both OA cartilage tissues and OA chondrocytes (Ni et al. Citation2020). They also showed that knockdown of circPSM3 or overexpression of miRNA-296-5p promoted chondrocyte proliferation and differentiation. The dual-luciferase results showed that circPSM3 targets miRNA-296-5p. Consistently, miRNA-296-5p inhibitors reversed the effects caused by knockdown of circPSM3, while overexpression of miR-296-5p promoted the effects caused by knockdown of circPSM3 (Ni et al. Citation2020). Thus, their results suggested that circPSM3 inhibited chondrocyte proliferation and differentiation by targeting miR-296-5p.
CircVCAN
Ma et al. reported that circVCAN was overexpressed in both OA tissues and OA chondrocyte (Ma et al. Citation2020). However, knockdown of circVCAN using siRNA promoted apoptosis and inhibited cell proliferation, while overexpression of circVCAN inhibited apoptosis and promoted cell proliferation in OA chondrocytes, suggesting that circVCAN might be a part of compensatory responses against apoptosis in OA. Knockdown of circVCAN inhibited the mRNA and protein levels of p50, p52, and p65, which are related to the nuclear factor-κB (NF-κB) pathway, while overexpression of circVCAN exerted opposite effects (Ma et al. Citation2020), suggesting that circVCAN may be involved in the activation of NF-κB pathway in OA chondrocytes. Moreover, PDTC (NF-κB inhibitor) significantly reversed the effect of overexpression of circVCAN. Altogether, their results suggested that circVCAN affected the proliferation and apoptosis of OA chondrocytes through NF-κB pathway.
CircHIPK3
Wu et al. reported that circHIPK3 was upregulated while miR-124 level was downregulated in both OA cartilage tissues and OA chondrocytes (Wu et al. Citation2020). However, knockdown of circHIPK3 or overexpression of miR-124 induced OA chondrocyte apoptosis, suggesting that circHIPK3 might be a part of compensatory responses against apoptosis in OA. The dual-luciferase reporter assays showed that circHIPK3 interacts with miR-124 and SOX8 is a downstream target molecule of miR-124 in OA chondrocytes (Wu et al. Citation2020). In addition, miR-124 inhibitors reversed the effect of circHIPM3 knockdown and SOX8 knockdown reversed the effect of miR-124 inhibitors on OA cell apoptosis, suggesting that circHIPK3 inhibited OA chondrocyte apoptosis via miR-124/SOX8 axis.
Recently, Li et al. also investigated the effects of extracellular vesicles (EVs) derived from mesenchymal stem cells (MSCs) overexpressing circHIPK3 (MSCs-circHIPK3-EVs) in the pathogenesis of OA (Li S et al. Citation2021) because circHIPK3 was known to be more abundant in EVs (Zheng Q et al. Citation2016). The authors demonstrated that MSCs-circHIPK3-EVs reduced IL-1β-induced proliferation inhibition, apoptosis, and ECM degradation via regulating miR-124-3p/Myosin heavy chain 9 (MYH9) axis: The regulatory network of circHIPK3/miR-124-3p/MYH9 was validated by luciferase activity assays and further confirmed by a series of loss and gain-of-function experiments (Li S et al. Citation2021). More importantly, IA-injection of MSCs-EVs, MSCs-circHIPK3-EVs, or circHIPK3 showed similar results as in vitro and reduced the degenerative changes in the cartilage tissues in collagenase-induced OA mice, suggesting that circHIPK3 is involved in MSCs-EVs-mediated protective effects against the progression of OA in vivo (Li S et al. Citation2021).
CircTMBIM6
Bai et al. reported that the expression levels of circTMBIM6 and MMP-13 were upregulated while miR-27a expression was downregulated in both cartilage tissues of OA patients and the chondrocyte model induced by IL-1β and TNF-α (Bai et al. Citation2020). They also showed that overexpression of circTMBIM6 inhibited miR-27a expression and upregulated MMP-13 expression, while knockdown of circTMBIM6 showed the opposite effect. Consistently, overexpression of miR-27a inhibited MMP-13 expression while inhibition of miR-27a induced upregulation of MMP-13. Thus, their results suggested that circTMBIM6 promoted OA-induced ECM degradation via miR-27a/MMP-13 axis.
CircRNF121
Based on bioinformatics analyses using data collected from public databases, Wang et al. hypothesized that Lymphoid enhancer-binding factor 1 (LEF1), might regulate the progression of OA (Wang T et al. Citation2020). Then, they showed that the expression of LEF1 was increased in both OA cartilage tissues and IL-1β treated chondrocytes. In addition, the authors revealed that circRNF121 (also known as has_circ_0023404) and the LEF1 expressions are positively correlated in the OA cartilage tissues (Wang T et al. Citation2020). Overexpression of circRNF121 inhibited chondrocyte proliferation and induced chondrocyte apoptosis and ECM degradation, while knockdown of circRNF121 exerted opposite effects. Using bioinformatics analysis and dual-luciferase assay, miR-665 was identified as a target of circRNF121 and Myeloid differentiation factor 88 (MYD88) (Wang T et al. Citation2020). Overexpression or knockdown of MYD88 exerted similar effects of circRNF121 overexpression or knockdown, while overexpression of miR-665 reversed those effects caused by overexpression of circRNF121 or MYD88. The authors also showed that circRNF121/miR-665/MYD88 axis regulated NF-кB signaling pathway.
Importantly, in vivo injection of lentivirus expressing MYD88 or circRNF121 showed that circRNF121 and MYD88 induced the destruction of cartilage tissues in the DMM model, while miR-665 agomir injection showed the opposite effect (Wang T et al. Citation2020). Collectively, their results suggested that LEF1 regulated the progression of OA by controlling the circRNF121/miR-665/MYD88 axis through NF-B pathway.
CircRSU1
Yang et al. reported the upregulation of circRSU1 (also known as hsa_circ_0006577) in H2O2-stimulated human articular chondrocytes using RNA-seq (Yang et al. Citation2021). Knockdown of circRSU1 reduced H2O2-stimulated induction of COX2/iNOS and mRNA levels of MMPs and pro-inflammatory factors whereas overexpression of circRSU1 showed the opposite effect, suggesting that circRSU1 regulated ROS generation and oxidative stress-induced inflammation and ECM degradation. Using target prediction databases and dual-luciferase reporter assay, the authors showed the targeted relationship between miR-93-5p and circRSU1 or mitogen-activated protein kinase kinase kinase 8 (MAP3K8) (Yang et al. Citation2021). Moreover, they further demonstrated that the circRSU1/miR-93-5p/MAP3K8 axis could modulate MEK/ERK and NF-κB pathways.
Importantly, in vivo overexpression of circRSU1 using adeno-associated virus (AAV) promoted ROS generation and ECM degradation in DMM-operated mice (Yang et al. Citation2021), suggesting that circRSU1 plays an important role in regulating the progression of OA in vivo.
CircCSNK1G1
Xiao et al. showed that the expression of circCSNK1G1 was increased in both the cartilage tissues of OA patients and OA rats. First, the authors tested the in vivo significance of circCSNK1G1 using IA lentivirus injection with circCSNK1G1 siRNA: In vivo knockdown of circCSNK1G1 improved the damage of cartilage tissues and decreased chondrocyte apoptosis and the expression of inflammatory factors in DMM-induced OA rats (Xiao J et al. Citation2021). Bioinformatic prediction and experimental verification demonstrated that circCSNK1G1 interacts with miR-4428 which targets Fucosyltransferase 2 (FUT2), an enzyme mediates the transfer of fucose on the glycans. Consistent with the in vivo data, knockdown of circCSNK1G1 or FUT2 inhibited the expression of MMP-13 and increased cell viability in IL-1β treated chondrocytes. In addition, miR-4428 mimics showed anti-apoptotic effects and knockdown of miR-4428 reduced the effects caused by knockdown of circCSNK1G1 in IL-1β treated chondrocytes. Their results suggested that circCSNK1G1 promoted the ECM degradation and apoptosis in the progression of OA through miR-4428/FUT2 axis.
Circ_DHRS3
Consistent with a previous circRNA expression profile (Xiao et al. Citation2019), Jiang et al. showed that the expression of circ_DHRS3 (also known as circ_0010024 from Dehydrogenase/Reductase 3 (DHRS3) gene) was increased in both OA cartilage tissues and IL-1β treated chondrocytes. Then, the authors showed that knockdown of circ_DHRS3 restored the expression of a proliferation marker (Ki67) and inhibited the expression of MMP-13 and apoptotic markers (Bcl-2, Bax, and Cleaved PARP) in IL-1β-treated chondrocytes. They predicted the targeted relationships between miR-183-5p and circ_DHRS3 or Gremlin 1 (GREM1), DAN Family BMP Antagonist, and then verified the relationships by dual-luciferase reporter assay (Jiang R et al. Citation2021). Consistently, miR-183-5p mimics showed similar effects to knockdown of circ_DHRS3 and miR-183-5p inhibitor reversed the effect caused by knockdown of circ_DHRS3 in IL-1β-treated chondrocytes. Also, concomitant overexpression of GREM1 reversed the effect of miR-183-5p mimics in IL-1β-treated chondrocytes. Their results suggested that Circ_DHRS3 may facilitate OA progression by regulating proliferation, apoptosis and ECM degradation through miR-183-5p/GREM1 axis.
Circ_SLC39A8
Yu et al. reported that the expression of circ_SLC39A8 (also known as hsa_circ_0002782) was increased in OA cartilage tissues. In IL-1β-treated chondrocytes, knockdown of circ_SLC39A8 increased cell proliferation and prevent apoptosis, inflammation, and ECM loss (Yu J et al. Citation2021). Bioinformatic prediction and experimental validation demonstrated that circ_SLC39A8 acted as a sponge of miR-591 to target interleukin-1-receptor-associated kinase 3 (IRAK3). Consistently, knockdown of miR-591 reversed the effects caused by knockdown of circ_SLC39A8 and overexpression of IRAK3 reduced the effects caused by overexpression of miR-591. Their results suggested that circ_SLC39A8 may be involved in the progression of OA through miR-591/IRAK3 axis.
Circ_0114876
Wang et al. reported that circ_0114876 was upregulated in both OA tissues and IL-1β-induced chondrocytes. Using Pearson’s correlation analysis, bioinformatic prediction, and experimental validation such as dual-luciferase reporter assay and RNA immunoprecipitation assay, the authors showed that circ_0114876 interacts with miR-671 as a sponge to target TNF receptor-associated factor 2 (TRAF2). For functional investigation, they performed a series of loss and gain-of-function experiments: knockdown of circ_0114876 enhanced cell viability and the expression of ECM anabolic factors (type II Collagen, aggrecan) and inhibited apoptosis and the expression of pro-inflammatory factors (IL-6, IL-8) and MMP-13 in IL-1β-induced chondrocytes (Wang Q et al. Citation2021). Moreover, concomitant knockdown of miR-671 reversed the effects caused by knockdown of circ_0114876, and concomitant overexpression of TRAF2 reduced the effects caused by overexpression of miR-671. These results suggested that circ_0114876 may contribute to OA progression via regulating miR-671/TRAF2 axis.
Circ_0116061
Based on previous results (Wu et al. Citation2008; Wu J et al. Citation2017; Li Z et al. Citation2019), Zheng et al. further investigated the expression of circ_0116061, miR-200b-3p, and Smad ubiquitin regulatory factor 2 (SMURF2), an E3 ubiquitin ligase involved in the regulation of transforming growth factor-β (TGF-β) signaling (Bai and Ying Citation2020), by RT-qPCR: The expression of circ_0116061 and SMURF2 was increased and the expression of miR-200b-3p was decreased in OA cartilage tissues (Zheng et al. Citation2021). Then, the authors validated these correlated expression patterns by dual-luciferase assays, suggesting that circ_0116061 acted as a sponge of miR-200b-3p which targets SMURF2. Functionally, knockdown of circ_0116061 or SMURF2 increased proliferation and inhibited apoptosis and inflammation in OA chondrocytes. Moreover, miR-200b-3p inhibitor or overexpression of SMURF2 reversed the effects caused by knockdown of circ_0116061. Their results suggested that circ_0116061 may play a role in the progression of OA through miR-200b-3p/SMURF2 axis.
In a separate study, Guo et al. also showed that circ_0116061 (also known as exosomal circRNA bromodomain and WD repeat domain containing 1, circ-BRWD1) promoted IL-1β-induced cell death, inflammation, and ECM degradation via regulating miR-1277/TNF receptor-associated factor 6 (TRAF6) axis (Guo et al. Citation2021). For functional investigation of circ_0116061/miR-1277/TRAF6 network, the authors performed loss and gain-of-function approaches similar to those of Zheng et al. using IL-1β treated chondrocytes. Moreover, they showed that exosome treatment increased the level of circ_0116061, but GW4869 (a blocker of exosome generation and release) treatment decreased the level of circ_0116061 in IL-1β treated chondrocytes, suggesting that the transmission of circ_0116061 is mediated by exosomes (Guo et al. Citation2021). Collectively, these results suggested that circ_0116061 may promote the progression of OA by acting as a sponge of both miR-200b-3p and miR-1277.
CircRNA_0092516
Huang et al. showed that the expression of circRNA_0092516 was increased in both OA cartilage tissues and IL-1β treated chondrocytes. Moreover, the authors revealed the negative correlation between circRNA_0092516 and miR-337-3p expression levels and the positive correlation between circRNA_0092516 and the phosphatase and tensin homolog (PTEN) expression levels by the Pearson correlation analysis (Huang et al. Citation2021). Consistently, knockdown of circRNA_0092516 decreased the protein levels of PTEN and p-NF-κB p65 while co-transfection of miR-337-3p inhibitor reversed the effect. Using bioinformatics analysis and dual-luciferase assay, miR-337-3p was confirmed to interact with circRNA_0092516. Moreover, Knockdown of circRNA_0092516 increased chondrocyte proliferation and inhibited apoptosis, inflammation, and ECM degradation while miR-337-3p inhibitor reversed the effects caused by knockdown of circRNA_0092516.
Importantly, in vivo injection of lentivirus expressing circRNA_0092516 siRNA showed that knockdown of circRNA_0092516 improved the damage of cartilage tissues in the medial meniscectomy tear (MMT)-induced OA model, while co-injection of miR-337-3p inhibitor reversed the effect (Huang et al. Citation2021). Altogether, these results suggested that circRNA_0092516 facilitated the progression of OA through miR-337-3p/PTEN axis.
CircCDH13
Zhou et al. identified circCDH13 (also known as hsa_circ_0040646) as a highly upregulated circRNA among 769 differentially expressed circRNAs by microarray analysis and RT-qPCR using samples of OA and normal cartilage tissues. Knockdown of circCDH13 decreased IL-1β-induced apoptosis and the expression of ECM-degrading enzymes and increased the expression of ECM anabolic factors while overexpression of circCDH13 showed opposite effects (Zhou Z et al. Citation2021). Bioinformatic prediction and experimental verification such as RNA pull down and luciferase assays showed that both circCDH13 and PTEN interact with miR-296-3p. Consistently, PTEN overexpression showed similar effects to circCDH13 overexpression. Moreover, miR-296-3p mimics reversed the effects caused by overexpression of circCDH13 or PTEN.
More importantly, in vivo knockdown of circCDH13 using AAV-short hairpin (sh) RNA decreased chondrocyte apoptosis and improved the damage of cartilage tissues in DMM-induced OA mice (Zhou Z et al. Citation2021). These results suggested that circCDH13 promoted the progression of OA through miR-296-3p/PTEN axis.
Circ_0128846
Since it was previously suggested that circ_0128846 was upregulated in OA tissues (Li Z et al. Citation2019), Liu et al. investigated the role and mechanism of circ_0128846 in OA chondrocytes. Functionally, knockdown of circ_0128846 enhanced cell viability and decreased the protein expression levels of apoptotic markers, inflammation factors, and ECM degradation enzyme (Liu C et al. Citation2021). Bioinformatic prediction and experimental verification suggested that circ_0128846 can function as a sponge of miR-127-5p to target NAMPT. Consistently, concomitant knockdown of miR-127-5p reversed the effects caused by knockdown of circ_0128846, and concomitant overexpression of NAMPT reversed the effects caused by overexpression of miR-127-5p, suggesting that circ_0128846 may contribute to OA progression via miR-127-5p/NAMPT axis.
In a separate study, Li et al. also reported that circ_0128846 inhibited cell viability but enhanced apoptosis, inflammation, and ECM degradation through miR-140-3p/Janus kinase 2 (JAK2) axis (Li H et al. Citation2021). The regulatory network of circ_0128846/miR-140-3p/JAK2 was validated by dual-luciferase and RNA immunoprecipitation assays and further confirmed by a series of loss and gain-of-function experiments. More importantly, knockdown of circ_0128846 in OA rats reduced the progression of OA and showed similar results as in vitro (Li H et al. Citation2021). Collectively, these results suggest that circ_0128846 may function as a sponge of both miR-127-5p and miR-140-3p to promote the progression of OA.
Circ_0134111
Based on the previous results (Wang et al. Citation2019b), Wu et al. reconfirmed that the expression of circ_0134111 was increased in both OA cartilage tissues and IL-1β-induced chondrocytes by RT-qPCR. Then, the authors showed that knockdown of circ_0134111 restored IL-1β-induced proliferation inhibition, apoptosis, inflammation, and ECM degradation (Wu R et al. Citation2021). Bioinformatic prediction and experimental verification demonstrated that circ_0134111 interacts with miR-515-5p which targets suppressor of cytokine signaling 1 (SOCS1). For the functional investigation, they further showed that miR-515-5p inhibitor reversed the effects caused by knockdown of circ_0134111 while miR-515-5p mimics showed similar effects to knockdown of circ_0134111 in IL-1β-treated chondrocytes. Consistently, concomitant overexpression of SOCS1 reversed the effects caused by miR-515-5p mimics in IL-1β-treated chondrocytes (Wu R et al. Citation2021). These results suggested that circ_0134111 may be involved in the progression of OA through miR-515-5p/SOCS1 axis.
Moreover, by performing a series of loss and gain-of-function approaches similar to those of Wu R et al. (Citation2021), Zhang et al. showed that circ_0134111 participated in chondrocyte apoptosis and ECM degradation through miR-224-5p/C–C motif chemokine ligand 2 (CCL2) axis (Zhang L et al. Citation2021). Interestingly, it was also reported that miR-224-5p regulates CCL1 expression in human chondrocytes: Liu and Zhang found another regulatory network of circ_0134111/miR-224-5p/CCL1 (Liu and Zhang Citation2021). Importantly, Liu and Zhang showed that knockdown of circ_0134111 reduced the degenerative changes in the cartilage tissues and decreased the expression levels of CCL1 and pro-inflammatory cytokines in DMM-induced OA rats (Liu and Zhang Citation2021). Altogether, these results suggest that circ_0134111 may facilitate the progression of OA through targeting both miR-515-5p and miR-224-5p.
Circ-SPG11
Liu et al. showed that the expression of circRNA spastic paraplegia 11 (circ-SPG11) was increased in both OA tissues and IL-1β treated chondrocytes (Liu Y et al. Citation2021). Functionally, knockdown of circ-SPG11 enhanced cell viability and inhibited apoptosis, inflammation, and ECM degradation in IL-1β treated chondrocytes. Bioinformatic prediction and experimental verification such as dual-luciferase and RNA immunoprecipitation assays demonstrated that circ-SPG11 interacts with miR-337-3p which targets ADAMTS5. To further investigate the functional relationship of circ-SPG11/miR-337-3p/ADAMTS5 network, the authors performed the following loss and gain-of-function experiments: (1) Overexpression of miR-337-3p showed similar effects to knockdown of circ-SPG11. (2) Concomitant knockdown of miR-337-3p or overexpression of ADAMTS5 reversed the effects caused by knockdown of circ-SPG11 or overexpression of miR-337-3p respectively. (3) IL-1β treatment induced the upregulation of ADAMTS5 mRNA and protein levels, which was reduced by concomitant knockdown of circ-SPG11. (4) IL-1β treatment and IL-1β+si-circ-SPG11 + anti-miR-337-3p showed similar effect on the expression levels of ADAMTS5 mRNA and protein. Collectively, these results suggested that circ-SPG11 may promote the progression of OA through regulating miR-337-3p/ADAMTS5 axis.
CircZNF652
Based on the previous study (Liu et al. Citation2020) and their own preliminary microarray analysis, Yuan et al. investigated the role of circZNF652 (also known as hsa_circ_0003258) in the development of OA (Yuan et al. Citation2021). The authors showed that the expression of circZNF652 and PTEN was increased in both synovial fluid samples from OA patients and LPS-treated chondrocytes: They also analyzed the correlation between circZNF652 and PTEN by Pearson’s correlation coefficient. Functionally, overexpression of circZNF652 increased the expression of PTEN, MMP13, and NF-KB while knockdown of circZNF652 showed opposite effects. In addition, overexpression of circZNF652 or PTEN increased the apoptosis in LPS-treated chondrocytes while knockdown of circZNF652 showed the opposite effect. Consistently, overexpression of PTEN reversed the anti-apoptotic effect caused by knockdown of circZNF652 in LPS-treated chondrocytes (Yuan et al. Citation2021). These results suggested that circZNF652 promoted LPS-stimulated apoptosis in chondrocytes through PTEN upregulation although the mechanical relationship between circZNF652 upregulation and PTEN upregulation is obscure.
Circ_0001846
Based on the previous result that the expression of circ_0001846 was increased in OA patients (Yu et al. Citation2018), Zhu et al. investigated the role of circ_0001846 in IL-1β-induced chondrocyte model of OA (Zhu C et al. Citation2021). First, the authors reconfirmed that the expression of circ_0001846 was increased in both OA tissues and IL-1β-treated human chondrocytes. Interestingly, they found that IL-1β treatment increased the expression of circ_0001846 in exosome. Moreover, exosome treatment increased circ_0001846 level in chondrocytes, but GW4869 treatment decreased circ_0001846 level in IL-1β-treated chondrocytes, suggesting that exosomes may mediate the transmission of circ_0001846 (Zhu C et al. Citation2021). Then, they demonstrated that circ_0001846 interacts with miR-149-5p which targets Wingless-type MMTV integration site family (Wnt), member 5B (WNT5B), a conserved secretory protein of Wnt family using bioinformatics analysis and experimental validation (Zhu C et al. Citation2021). Functionally, knockdown of circ_0001846 or miR-149-5p mimics reduced IL-1β-induced chondrocyte damages, such as cell viability, invasion, migration, apoptosis, inflammation, and ECM loss. Moreover, concomitant transfection with miR-149-5p inhibitors or overexpression of WNT5B reversed the effects caused by knockdown of circ_0001846 or miR-149-5p mimics respectively. Consistently, knockdown of circ_0001846 reduced WNT5B upregulation in IL-1β-treated chondrocytes while miR-149-5p inhibitors reversed the effect (Zhu C et al. Citation2021). Altogether, their results suggested that exosomal circ_0001846 may contribute to IL-1β-induced OA progression through miR-149-5p/WNT5B axis.
Downregulated circRNAs in OA
Has_circ_0045714
Li et al. reported that TNF-α inhibited hsa_circ_0045714 expression and upregulated miR-193b in human chondrocytes (Li BF et al. Citation2017). The authors also showed that overexpression of hsa_circ_0045714 promoted cell proliferation and the expression of ECM anabolic factors (type II Collagen, aggrecan) whereas overexpression of miR-193b exerted opposite effects. Using luciferase reporter assay, they confirmed that insulin-like growth factor 1 receptor (IGF1R) is a target of miR-193b (Li BF et al. Citation2017). Moreover, the authors demonstrated that overexpression of miR-193b or knockdown of IGF1R inhibited the effect of has_circ_0045714 overexpression, while IGF1R overexpression reversed the effect of miR-193b overexpression. These results suggested that has_circ_0045714 controlled chondrocyte proliferation/apoptosis and ECM synthesis via miR-193b-IGF1R signaling (Li BF et al. Citation2017).
Recently, Jiang et al. reported that circ_0045714 relieved TNF-α-induced reduction in viability, apoptosis, inflammation, and ECM degradation through miR-218-5p/ GTPase HRas (also known as transforming protein p21, HRAS) axis (Jiang H et al. Citation2021). For functional investigation of the regulatory network of circ_0045714/miR-218-5p/HRAS, the authors performed a series of loss and gain-of-function experiments: Overexpression of miR-218-5p reversed the effects caused by overexpression of circ_0045714, and knockdown of HRAS reduced the protective effects of miR-218-5p inhibition in TNF-α-induced chondrocytes, suggesting that miR-218-5p/HRAS axis mediated the protective effect of circ_0045714 in TNF-α-induced chondrocytes (Jiang H et al. Citation2021). Using similar approaches as Jiang H et al. (Citation2021), Ding et al. also reported that circ_0045714 enhanced cell proliferation and ECM synthesis, but reduced apoptosis and inflammation in IL-1β-stimulated human articular chondrocytes by blocking miR-331-3p which inhibits Phosphoinositide 3-kinase (PI3K) regulatory subunit 3 (PIK3R3), an inhibitor of PI3K (Ding et al. Citation2021).
Moreover, Fang et al. reported the downregulation of hsa_circ_0045714 (also known as circUNK) in the cartilage tissues of rabbit knee OA model (Fang et al. Citation2021). Interestingly, the authors demonstrated that aerobic exercise and glucosamine synergistically improved the cartilage damage and increased the expression of hsa_circ_0045714 in knee OA tissues. Consistently, they showed that overexpression of hsa_circ_0045714 reduced apoptosis and enhanced cell viability and ECM synthesis, while knockdown of hsa_circ_0045714 showed the opposite effects in iodoacetic acid-induced OA chondrocytes (Fang et al. Citation2021). Collectively, all these results suggested that hsa_circ_0045714 may contribute to reducing the progression of OA.
CiRS-7
Based on previous results that ciRS-7/miR-7 axis is involved in cancer signaling pathways (Hansen et al. Citation2013), Zhou et al. further investigated the roles of the ciRS-7/miR-7 axis in OA. They found that ciRS-7 was downregulated while miR-7 was upregulated in blood samples from OA patients and IL-1β-induced human chondrocytes (Zhou X et al. Citation2019). They also showed that IL-1β could induce apoptosis and inflammation in chondrocytes. Knockdown of ciRS-7 using siRNA or overexpression of miR-7 using miR-7 mimic promoted apoptosis and inflammation in OA chondrocytes (Zhou X et al. Citation2019).
In a separate study (Zhou et al. Citation2020), the authors also showed that overexpression of ciRS-7 restored IL-1β-induced upregulation of ECM-degrading enzymes (MMP-3, MMP-13, and ADAMTS5) and downregulation of autophagy-related proteins (LC3 and Beclin1), suggesting that ciRS-7 also regulated ECM degradation and autophagy deficiency in the OA cell model. Using RNA-seq analysis and various combinations of loss and gain-of-function experiments, they also showed that miR-7 mimics increased the expression of IL-17A, thereby increasing the levels of p-PI3 K, p-Akt, and p-mTOR although the exact mechanical relationship between miR-7 upregulation and IL-17A upregulation is still ambiguous (Zhou et al. Citation2020). Moreover, in vivo IA injection of lentivirus expressing miR-7-siRNA improved the damage of cartilage tissues, decreased the expression of MMP-13, and increased the expression of LC3 and Beclin1 in DMM-induced OA rats (Zhou et al. Citation2020). These results suggested that ciRS-7/miR-7 axis also participated in reducing ECM degradation and autophagy deficiency through regulating IL-17A-mediated PI3 K/AKT/mTOR pathway in the progression of OA.
Altogether, their results suggested that ciRs-7 played a protective role via antagonizing miR-7 in the progression of OA. Interestingly, ciRS-7 is also known as Cdr1as. Thus, downregulation of ciRS-7 in blood samples from OA patients (Zhou X et al. Citation2019) and upregulation of circRNA-CDR1as in OA chondrocytes (Zhang W et al. Citation2020) seem inconsistent. Thus, further investigation will be required to clarify this discrepancy.
CircSERPINE2
By RNA-seq and RT-qPCR, Shen et al. identified 14 downregulated circRNAs in OA cartilage tissues (Shen et al. Citation2019). Then, they further confirmed the reduced expression of circSERPINE2 (also known as hsa_circ_0008365) in IL-1β or TNF-α treated primary human chondrocytes (Shen et al. Citation2019). Knockdown of circSERPINE2 induced rapid apoptosis of chondrocytes and promoted ECM degradation, while overexpression of circSERPINE2 suppressed apoptosis. In addition, the authors demonstrated that circSERPINE2 interacts with miR-1271, and E26 transformation-specific-related gene (ERG) is a target of miR-1271 using bioinformatics analyses and validation experiments such as RNA pull down and luciferase activity assays (Shen et al. Citation2019).
Importantly, IA injection of adeno-associated virus (AAV)-circSERPINE2 reduced the de-generative changes in the cartilage matrix and increased the ECM composition in anterior cruciate ligament transection (ACLT)-induced OA rabbits (Shen et al. Citation2019). These results suggested that circSERPINE2 inhibited apoptosis and protected ECM components from degradation through miR-1271-ERG pathway.
Moreover, using similar approaches as Shen et al., Zhang et al. also reported that circSERPINE2 reduced ECM degradation and apoptosis through regulating miR-495/TGF-β receptor 2 (TGFBR2) axis in IL-1β-induced chondrocytes (Zhang Q et al. Citation2020). Taken together, all these results suggested that circSERPINE2 can play a protective role against OA progression through sequestration of both miR-1271 and miR-495.
Circ9119
Chen et al. found that circ9119 was downregulated in OA cartilage by microarray analysis using samples from healthy controls and OA patients, which was reconfirmed by RT-qPCR using samples from different OA patients and IL-1β-induced chondrocytes (Chen C et al. Citation2020). Moreover, overexpression of circ9119 restored the proliferative rate and apoptosis of IL-1β-induced chondrocytes based on MTT assay and flow cytometry. The authors predicted the targeted relationships between miR-26a and circ9119 or PTEN by bioinformatics analysis, and then verified the relationships by dual-luciferase reporter assay (Chen C et al. Citation2020). Consistently, overexpression of circ9119 restored the change in the expression of miR-26a and PTEN in IL-1β-induced chondrocytes. Altogether, their findings suggested that the miR-26a/PTEN axis mediated the protective effect of circ9119 in IL-1β-treated chondrocytes.
CircCDK14
Shen et al. found that circCDK14 (also known as hsa_circ_0001722) was downregulated in OA tissues in their previous study (Shen et al. Citation2019), thus they reconfirmed the result by comparing the expression levels of circCDK14 in weight-bearing and the non-weight bearing areas of human cartilage samples (Shen et al. Citation2020). Overexpression and knockdown studies suggested that circCDK14 could play a protective role against OA through controlling inflammation, ECM metabolism, and cell apoptosis/proliferation in human chondrocytes. The authors also demonstrated that miR-125a-5p/Mothers against decapentaplegic homolog 2 (Smad2) axis mediated the protective effect of circCDK14 in human chondrocytes through a series of experiments including RNA immunoprecipitation, luciferase assay, fluorescence in situ hybridization, western blot, and RT-qPCR (Shen et al. Citation2020).
Importantly, the authors further investigated the effects of circCDK14 in vivo using ACLT-induced OA rabbits: IA injection of AAV-circCDK14 decreased the percentage of TUNEL-positive cells, reduced the expression of MMP-3, MMP-13, and increased the expression of Smad2, SOX9, and type II Collagen in chondrocytes, while miR-125a-5p overexpression showed the opposite effect (Shen et al. Citation2020), suggesting the possibility that circCDK14 can prevent the progression of OA in vivo.
Hsa_circ_0005567
Previously, hsa_circ_0005567 was identified as one of downregulated circRNAs in OA knee condyle by illumina sequencing platform (Xiao et al. Citation2019). Meanwhile, Zhang et al. showed that overexpression of hsa_circ_0005567 reduced IL-1β-induced chondrocyte apoptosis while knockdown of hsa_circ_0005567 showed the opposite effect using flow cytometry and western blot analysis of Caspase-3, Bax, and anti-apoptotic Bcl-2 (Zhang J et al. Citation2020). Moreover, the authors showed that overexpression of hsa_circ_0005567 induced autophagy activation while 3-methyladenine, an inhibitor of autophagy, reversed the effects of hsa_circ_0005567 overexpression in IL-1β-induced chondrocytes using GFP-LC3 immunofluorescence and western blot analysis of autophagy markers (LC3 and Beclin-1) (Zhang J et al. Citation2020). Bioinformatics analysis predicted that hsa_circ_0005567 contained binding sites for miR-495 and Autophagy-related 14 (ATG14), an autophagy marker, could be a target gene of miR-495, which was confirmed by RNA pull down and luciferase reporter assays (Zhang J et al. Citation2020). Consistently, hsa_circ_0005567 overexpression-induced pro-autophagic and anti-apoptotic effects were reversed by overexpression of miR-495 or knockdown of ATG14 in IL-1β-treated chondrocytes. Taken together, their results suggested that hsa_circ_0005567 may contribute to reducing the progression of OA by regulating autophagy activation and apoptosis through miR-495/ATG14 axis.
CircPDE4D
Wu et al. reported that circPDE4D was downregulated in both OA cartilage tissues and TNF-α treated chondrocytes. The authors predicted the targeted relationships between miR-103a-3p and circPDE4D or FGF18, and then verified the relationships by dual-luciferase reporter assay (Wu Y et al. Citation2021). Functionally, knockdown of circPDE4D promoted ECM degradation but showed no effects on chondrocyte proliferation and apoptosis. In addition, knockdown of miR-103a-3p or overexpression of FGF18 reversed the effects caused by knockdown of circPDE4D.
More importantly, in vivo IA injection of AAV-circPDE4D improved the damage of cartilage tissues and performance for pain and endurance in DMM-induced OA mice (Wu Y et al. Citation2021). Their findings suggested that circPDE4D can play a protective role against ECM degradation through miR-103a-3p/FGF18 axis.
CircANKRD36
Zhou et al. revealed decreased expression of circANKRD36 in the cartilage tissues of OA patients and IL-1β treated human chondrocytes (Zhou JL et al. Citation2021). Then, the authors showed that overexpression of circANKRD36 promoted cell proliferation and inhibited the expression of apoptotic markers and pro-inflammatory factors whereas knockdown of circANKRD36 exerted opposite effects in IL-1β treated human chondrocytes. Using bioinformatics analysis and dual-luciferase assay, they demonstrated that circANKRD36 interacts with miR-599 which targets Casz1, a conserved zinc-finger transcription factor. Consistently, miR-599 inhibitor reduced the effects caused by knockdown of circANKRD36, and Casz1 overexpression reduced miR-599-induced apoptosis and inflammation.
Importantly, IA injection of AAV-circANKRD36 reduced the degenerative changes in the cartilage tissues of ACLT-induced OA mice (Zhou JL et al. Citation2021). Altogether, their findings suggested that the miR-599/Casz1 axis mediated the protective effect of circANKRD36 against OA.
Circ0083429
Based on the previous results (Shen et al. Citation2019), Yao et al. found circ0083429 as one of the obviously downregulated circRNAs in OA cartilage tissues and reconfirmed the downregulation of circ0083429 by RT-qPCR. Knockdown of circ0083429 increased the expression of ECM degradation enzymes and reduced the expression of ECM anabolic factors and cell proliferation, while overexpression of circ0083429 showed opposite effects in human chondrocytes (Yao et al. Citation2021). Using bioinformatics analysis and luciferase reporter and RNA pulldown assays, the authors demonstrated that miR-346 can bind to both circ0083429 and Smad3. Consistently, overexpression of miR-346 showed similar effects caused by knockdown of circ0083429. Moreover, the expression levels of Smad3 mRNA and protein were decreased by knockdown of circ0083429, and concomitant overexpression of Smad3 reversed the changes in ECM-related protein expression caused by knockdown of circ0083429.
Importantly, injection of AAV-circ0083429 into the joint cavities of the knee improved the damage of cartilage tissues, reduced the expression of ECM degradation enzymes, and increased the expression of ECM anabolic factors in the ACLT-Induced OA mouse Model (Yao et al. Citation2021). Altogether, their findings suggested that circ0083429 can prevent the progression of OA by regulating ECM homeostasis through miR-346/Smad3 axis.
CircADAMTS6
Using microarray analysis, Fu et al. identified 585 differentially expressed circRNAs (205 were upregulated and 380 were downregulated) in IL-1β-treated chondrocytes (Fu Q et al. Citation2021). Then, circADAMTS6 (also known as hsa_circ_0008667) was selected as a highly downregulated circRNA. The authors demonstrated that circADAMTS6 can interact with miR-431-5p by bioinformatics analysis and luciferase assay although they did not further investigate the potential downstream target of miR-431-5p. Functionally, overexpression of circADAMTS6 or miR-431-5p inhibitor reduced apoptosis in IL-1β-stimulated chondrocytes while knockdown of circADAMTS6 or miR-431-5p mimics showed the opposite effect, suggesting that circADAMTS6 may have an anti-apoptotic role in IL-1β-induced OA chondrocytes via targeting miR-431-5p.
Hsa_circ_0094742
Based on the previous result that the expression of hsa_circ_0094742 was decreased in OA patients (Wang et al. Citation2019b), Sun et al. further investigated the role of hsa_circ_0094742 using immortalized cartilage cells (CHON-001 cells). The authors showed that overexpression of hsa_circ_0094742 inhibited apoptosis and promoted proliferation and ECM synthesis in IL-1β treated chondrocytes. Then, the authors predicted the targeted relationships between miR-127-5p and hsa_circ_0094742 or Latexin (LXN), a carboxypeptidase A inhibitor, and verified the relationships by dual-luciferase reporter assay (Sun et al. Citation2021). Consistently, miR-127-5p agomir reversed the effects caused by overexpression of hsa_circ_0094742 in IL-1β treated chondrocytes. These results suggested that miR-127-5p/LXN axis may mediate the protective effect of hsa_circ_0094742 in IL-1β-induced OA chondrocytes.
CircPDE4B
Shen et al. reported that the expression of circPDE4B was decreased in chondrocytes of OA patients. The authors also showed that IL-1β or TNF-α decreased the expression of circPDE4B in both human and mouse chondrocytes, and knockdown of Fused in sarcoma (FUS), an RNA-binding protein involved in circRNA production (Errichelli et al. Citation2017) downregulated circPDE4B, but not mPDE4B (Shen et al. Citation2021). Functionally, knockdown of circPDE4B increased the expression of ECM degradation enzymes and decreased chondrocyte viability and the expression of ECM anabolic factors, compared with negative control siRNA.
Interestingly, AGO2 RNA immunoprecipitation assay revealed that circPDE4B does not bind to AGO2, while it was shown that circPDE4B can interact with both the RIC8 guanine nucleotide exchange factor A (RIC8A) and midline 1 (MID1, an E3 ligase) using RNA pull down, RNA immunoprecipitation, co-immunoprecipitation assays (Shen et al. Citation2021). Knockdown of circPDE4B increased the half-life of RIC8A protein and decreased the polyubiquitination of RIC8A, meanwhile knockdown of MID1 decreased RIC8A ubiquitylation, suggesting that circPDE4B may promote MID1-mediated degradation of RIC8A by acting as a scaffold to form a ternary complex: Consistently, RIC8A knockdown showed opposite effects of circPDE4B knockdown. In addition, the phosphorylation level of p38 was increased by circPDE4B knockdown but decreased by RIC8A knockdown. Moreover, RIC8A knockdown reversed circPDE4B knockdown-induced activation of p38 pathway, suggesting that circPDE4B may function through the RIC8A-p38 axis.
Importantly, AAV-mediated in vivo overexpression of circPDE4B in DMM-induced OA mice improved the damage of cartilage tissues and behavioral performance for pain (Shen et al. Citation2021). These results suggested that circPDE4B can function as a scaffold to promote MID1-mediated degradation of RIC8A, thereby inhibiting RIC8A-involved activation of p38 pathway to prevent the progression of OA.
Circ_0020093
Previously, circ_0020093 was identified as one of the significantly downregulated circRNAs in OA tissues (Shen et al. Citation2019). Thus, Feng et al. investigated the functional significance of circ_0020093 in OA using IL-1β-induced human chondrocytes. The regulatory network of circ_0020093/miR-23b/Sprouty 1 (SPRY1) was predicted by bioinformatics analysis and verified by luciferase reporter and RNA pull down assays (Feng et al. Citation2021). The authors also showed that overexpression of circ_0020093 or miR-23b inhibitor reduced IL-1β-induced apoptosis and ECM loss, which was reversed by concomitant overexpression of miR-23b or knockdown of SPRY1 respectively (Feng et al. Citation2021). These results suggested that circ_0020093 may contribute to preventing OA progression through miR-23b/SPRY1 axis.
Moreover, using similar approaches as Feng et al., Wang et al. also reported that circ_0020093 (also known as circATRNL1 derived from attractin like 1 (ATRNL1) gene) relieved IL-1β-induced apoptosis and ECM degradation through regulating Krüppel-like factor 5 (KLF5) via sequestering miR-153-3p (Wang KF et al. Citation2021). Collectively, these studies suggested that circ_0020093 may prevent the progression of OA through functioning as a sponge of both miR-23b and miR-153-3p.
Circ_0001103
Based on the previous result that the expression of circ_0001103 was decreased in OA tissues (Shen et al. Citation2019), Zhang et al. investigated the functional significance of circ_0001103 in IL-1β-induced chondrocyte model of OA. The authors reconfirmed that the expression of circ_0001103 was decreased in both OA tissues and IL-1β treated human chondrocytes by RT-qPCR. Then, they showed that overexpression of circ_0001103 reduced IL-1β-induced chondrocyte damages, such as proliferation inhibition, apoptosis, inflammation, and ECM loss. Using bioinformatics analysis and experimental validation (i.e. dual-luciferase reporter assay and RNA immunoprecipitation assay), they demonstrated that circ_0001103 interacts with miR-375 which targets Sirtuin 1 (SIRT1), a nicotinamide adenine dinucleotide (NAD)-dependent deacetylase (Zhang M et al. Citation2021). Consistently, miR-375 inhibitor showed similar effects to overexpression of circ_0001103. Moreover, concomitant overexpression of miR-375 or knockdown of SIRT1 reversed the effects caused by overexpression of circ_0001103 or miR-375 inhibitor respectively. Their results suggested that circ_0001103 may play a protective role against IL-1β-induced OA progression through miR-375/SIRT1 axis.
CircSLC7A2
CircSLC7A2 (also known as hsa_circ_0005805) was previously reported as a highly downregulated circRNA (Shen et al. Citation2019), thus Ni et al. investigated its functional significance using chondrocyte and in vivo mouse models of OA. Interestingly, the authors showed that knockdown of circSLC7A2 induced chondrocyte apoptosis and ECM degradation, while knockdown of linear SLC7A2 caused the opposite effects compared with IL-1β treatment as a positive control, suggesting that circSLC7A2 and linear SLC7A2 may play opposite roles in the progression of OA (Ni et al. Citation2021). In addition, they showed that the circularization of circSLC7A2 is mediated by FUS. The regulatory network of circSLC7A2/miR-4498/Tissue inhibitor of metalloproteinase 3 (TIMP3) was predicted by bioinformatics analysis and verified by luciferase reporter and RNA pull down assays. Consistently, miR-4498 inhibitor reversed OA-like phenotypes caused by circSLC7A2 knockdown, and overexpression of TIMP3 reversed the effects of miR-4498 mimics similar to circSLC7A2 knockdown.
Importantly, the authors further investigated the effects of circSLC7A2 in vivo using ACLT-induced OA mice: IA injection of AAV-circSLC7A2 improved the damage of cartilage tissues and behavioral performances in the hotplate and treadmill tests (Ni et al. Citation2021). Altogether, these results suggested that circSLC7A2 can prevent the progression of OA through miR-4498/TIMP3 axis.
CircRNA3503
Based on the idea that melatonin mediates the beneficial effects of sleep on tissue healing (Elkhenany et al. Citation2018), Tao et al. found circRNA3503 as an upregulated circRNA in chondrocytes following melatonin treatment (Tao et al. Citation2021). The authors also showed that the expression of circRNA3503 was decreased by TNF-α or IL-1β treatment. In addition, melatonin treatment or overexpression of circRNA3503 reversed IL-1β-induced apoptosis and ECM loss, meanwhile knockdown of circRNA3503 inhibited the effects of melatonin on IL-1β-induced apoptosis and ECM loss. Using bioinformatic analysis, they predicted two regulatory networks of circRNA3503: circRNA3503/hsa-miR-181c-3p/Peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α) and circRNA3503/hsa-let-7b-3p/SOX9. Then, the authors confirmed the functional relationship of each network by a series of loss and gain-of-function experiments using miRNA mimics, miRNA inhibitors, and circRNA3503 overexpression.
Importantly, the authors made small EVs (sEVs) from synovium mesenchymal stem cells (SMSC) overexpressing circRNA3503 (circRNA3503-OE-sEVs) and showed that circRNA3503-OE-sEVs inhibited chondrocyte apoptosis and enhanced ECM homeostasis and chondrocyte proliferation using in vitro co-culture and in vivo surgical models of OA (Tao et al. Citation2021): The effect of circRNA3503-OE-sEVs on chondrocyte proliferation is attributed to Wnt5a/b derived from SMSC-sEVs, but not circRNA3503 because overexpression of circRNA3503 did not show any effect on chondrocyte proliferation but SMSC-sEVs showed the ability to enhance chondrocyte proliferation through Wnt5a/b pathway. Furthermore, based on the gelation characteristics at body temperature of the polymer aqueous solutions, the authors demonstrated the effectiveness of poly(D,L-lactide)-poly(ethylene glycol)-poly(D,L-lactide) copolymer (PLEL) for sustained delivery of circRNA3503 in vitro and in vivo (Tao et al. Citation2021). These results suggested that circRNA3503 can play a protective role against apoptosis and ECM degradation through functioning as a miRNA sponge for hsa-miR-181c-3p and hsa-let-7b-3p.
CircHYBID
Liao et al. identified 450 differentially expressed circRNAs (200 were upregulated and 250 were downregulated) in damaged cartilage samples by RNA sequencing and expression profile analysis (Liao et al. Citation2021). Then, circHYBID (also known as has_circ_00003893) was selected as a downregulated circRNA in damaged cartilage samples because hyaluronan (HA) metabolism is involved in maintaining tissue integrity (Avenoso et al. Citation2018) and the host gene of circHYBID encodes HA-binding protein involved in HA depolymerization (HYBID) (also known as cell migration-inducing hyaluronidase 1, CEMIP). Interestingly, IL-1β treatment induced the upregulation of HYBID expression, but the downregulation of circHYBID, HA synthase 2 (HAS2), and HA accumulation. Functionally, circHYBID overexpression decreased HYBID, IL-6, and TNF-α expression but increased HAS2 expression and HA accumulation in IL-1β-treated chondrocytes (Liao et al. Citation2021). In addition, the authors predicted the targeted relationships between circHYBID and miR-29b-3p and between miR-29b-3p and TGF-β1, and then verified the relationships by dual-luciferase reporter assay (Liao et al. Citation2021). These results suggested that circHYBID may contribute to reducing the progression of OA by regulating HA metabolism in chondrocytes through miR-29b-3p/TGF-β1 axis.
CircRNA_0001236
To investigated the role of mesenchymal stem cell (MSC)-derived exosomal circRNAs (MSC-Exos) in the pathogenesis of OA, Mao et al. detected differentially expressed exosomal circRNAs in chondrogenesis-induced human bone marrow-derived MSCs (hMSCs) by using circRNA microarrays (Mao G et al. Citation2021). The authors identified exosomal circRNA_0001236 as one of the highly upregulated MSC-Exos after chondrogenic differentiation and found that the expression level of circRNA_0001236 was decreased in OA cartilage tissues. Then, using bioinformatics analysis and luciferase reporter assay, they demonstrated that circRNA_0001236 interacts with miR-3677-3p which targets SOX9 (Mao G et al. Citation2021). Functionally, overexpression of circ_0001236 increased the expression of chondrogenic markers, Collagen type II alpha 1 chain (Col2a1) and SOX9, but decreased the expression of MMP-13 while miR-3677-3p mimics reversed the effects caused by circ_0001236 overexpression.
Importantly, IA-injection of MSC-circ_0001236-Exos reduced the degenerative changes in the cartilage tissues and the progression of OA while miR-3677-3p mimics reversed the effects in DMM-induced OA mice (Mao G et al. Citation2021). Altogether, their results suggested that exosomal circRNA_0001236 may have a protective role in the progression of OA by regulating ECM homeostasis through the miR-3677-3p/SOX9 axis.
CircFADS2
Based on their own preliminary sequencing data, Zhang et al. investigated the role of circ FADS2 and miR-195-5p in OA. The authors showed that the expression level of circFADS2 was decreased but the expression level of miR-195-5p was increased in both synovial fluid samples from OA patients and LPS-treated chondrocytes (Zhang H et al. Citation2021). Consistently, Pearson’s correlation coefficient showed that the expression of circFADS2 and miR-195-5p were negatively correlated. Interestingly, using methylation-specific PCR, the authors demonstrated that overexpression of circFADS2 decreased miR-195-5p expression through methylation (Zhang H et al. Citation2021). Functionally, overexpression of circFADS2 reduced the apoptosis but overexpression of miR-195-5p increased the apoptosis in LPS-treated chondrocytes. Moreover, concomitant overexpression of miR-195-5p inhibited the anti-apoptotic effect caused by overexpression of circFADS2 (Zhang H et al. Citation2021). These results suggested that circFADS2 inhibited LPS-stimulated apoptosis in chondrocytes by downregulating miR-195-5p through methylation although the mechanism by which circFADS2 mediates methylation of the miR-195-5p gene is ambiguous.
CircFAM160A2
Since it was reported that SIRT3 prevented chondrocyte degeneration through regulating mitochondrial homeostasis against oxidative stress (Wang et al. Citation2018), Bao et al. investigated the upstream regulatory mechanism of SIRT3 in the progression of OA. Using RNA-seq data analysis and dual luciferase assay, the authors demonstrated that circFAM160A2 (also known as hsa_circ_0020990) interacts with miR-505-3p which targets SIRT3 (Bao et al. Citation2021). The expression level of circFAM160A2 was significantly decreased in OA tissues. Functionally, circFAM160A2 overexpression and miR-505-3p overexpression showed the opposite effects in chondrocytes: miR-505-3p overexpression caused ECM degradation, apoptosis, and mitochondrial dysfunction while concomitant overexpression of SIRT3 or miR-505-3p reduced the effects caused by miR-505-3p overexpression or circFAM160A2 overexpression respectively. Consistently, overexpression of circFAM160A2 increased the expression level of SIRT3 protein (Bao et al. Citation2021).
More importantly, in vivo injection of AAV-circFAM160A2 showed similar results as in vitro. Moreover, Safranin staining showed that AAV-circFAM160A2 reduced the degenerative changes in the cartilage tissues of ACLT-induced OA mice (Bao et al. Citation2021). Altogether, their results suggested that circFAM160A2 may have a protective role against OA progression by regulating ECM homeostasis, apoptosis, and mitochondrial homeostasis through miR-505-3p/SIRT3 axis.
Dysregulated circRNAs as potential diagnostic biomarkers for OA
Currently, early diagnosis of OA is difficult because the diagnosis of OA largely depends on symptoms, physical findings, and imaging examinations. As described in the above section, dysregulated circRNAs has been shown as novel contributors in OA pathogenesis. Meanwhile, given circRNAs are considered as promising biomarkers for various human diseases such as cardiometabolic diseases and cancers (Zhang et al. Citation2018), there is increasing attention on clinical application of circRNAs as early diagnostic biomarkers for OA as follows.
Hsa_circ_0104873, Hsa_circ_0104595, and Hsa_circ_0101251
By circRNA microarray analysis, Yu et al. identified 29 upregulated and 18 downregulated circRNAs in synovial fluid between OA patients and healthy controls (Yu et al. Citation2018). To validate the microarray data, the authors performed RT-qPCR in an independent cohort and showed that hsa_circ_0104873, hsa_circ_0104595, hsa_circ_0101748, hsa_circ_0101251, and hsa_circ_0102061 were significantly upregulated in the synovial fluid of OA patients (Yu et al. Citation2018). The authors also performed receiver operating characteristics (ROC) curves and the area under the ROC curves (AUC) to investigate the diagnostic value of the 5 candidate circRNAs for OA: ROC analysis showed that hsa_circ_0104873 (AUC:0.683), hsa_circ_0104595 (AUC:0.708), and hsa_circ_0101251 (AUC:0.754) can be used as novel biomarkers for OA with high degrees of specificity (Yu et al. Citation2018). Moreover, they confirmed that these circRNAs were positively correlated with the degree of radiographic grading and symptomatic severity of OA patients by spearman’s rank correlation analysis (Yu et al. Citation2018). These results suggested that hsa_circ_0104873, hsa_circ_0104595, and hsa_circ_0101251 may have the potential to be used as diagnostic biomarkers for OA.
Moreover, Xi et al. recently investigated the functional significance and mechanism of hsa_circ_0104873 (also known as circRNA IQ motif-containing GTPase-activating protein 1 (circ-IQGAP1), according to Yu et al. Citation2018) in IL-1β treated chondrocytes (Xi et al. Citation2021). Using bioinformatics analysis, dual-luciferase assay, and RNA immunoprecipitation assay, the authors showed that miR-671-5p interacts with both hsa_circ_0104873 and Transcription factor 4 (TCF4). Consistently, the expression of hsa_circ_0104873 and TCF4 was increased and miR-671-5p expression was decreased in both OA cartilage tissues and IL-1β treated chondrocytes. For the functional investigation, they also showed that knockdown of hsa_circ_0104873 or overexpression of miR-671-5p decreased IL-1β-induced reduction in viability, apoptosis, inflammation, and ECM degradation. In addition, concomitant knockdown of miR-671-5p or overexpression of TCF4 reversed the effects caused by knockdown of hsa_circ_0104873 or overexpression of miR-671-5p respectively (Xi et al. Citation2021). Collectively, these results suggested that hsa_circ_0104873 may be a promising biomarker and therapeutic target for OA.
Hsa_circ_0032131
Using microarray data and RT-qPCR, Wang et al. reported that 1380 circRNAs were differentially expressed (215 were upregulated and 1165 were downregulated) in OA chondrocytes (Wang et al. Citation2019b). The microarray data were further validated using RT-qRCR and a predicted circRNA-miRNA network was also constructed. Their results suggested that some of the dysregulated circRNAs including hsa_circ_0032131 might be involved in the pathogenesis of OA. In a separate study, the authors measured the blood levels of hsa_circ_0032131 in OA patients and healthy controls by RT-qPCR and determined whether hsa_circ_0032131 has potential as a diagnostic biomarker for OA: The ROC curve showed that hsa_circ_0032131 held diagnostic value for OA (AUG:0.8455) (Wang et al. Citation2019a). These results suggested that hsa_circ_0032131 may have potential as a diagnostic biomarker for OA.
Recently, Zhao et al. reported that hsa_circ_0032131 (also known as circRNA protein kinase C eta (circ-PRKCH)) can promote the progression of OA through miR-140-3p/ADAM10 axis (Zhao et al. Citation2021). They showed that the expression of hsa_circ_0032131 and ADAM10 was increased while miR-140-3p expression was decreased in OA tissues and LPS-induced chondrocytes. Then, they verified the targeted relationships between miR-140-3p and hsa_circ_0032131 or ADAM10 by dual-luciferase reporter assay. The authors also showed that knockdown of hsa_circ_0032131 or overexpression of miR-140-3p reduced LPS-induced proliferation inhibition, apoptosis, inflammation, and ECM loss in chondrocytes, while these effects were reversed by concomitant knockdown of miR-140-3p or overexpression of ADAM10 respectively.
Moreover, using similar approaches as Zhao et al., Xu and Ma also reported that hsa_circ_0032131 can promote the progression of OA in IL-1β-induced chondrocytes through miR-502-5p/Peroxiredoxin 3 (PRDX3, an antioxidative protein) axis (Xu and Ma Citation2021). Importantly, the authors demonstrated that in vivo knockdown of hsa_circ_0032131 reduced the symptoms of OA in the DMM model (Xu and Ma Citation2021). Collectively, these results suggest that hsa_circ_0032131 may have potential not only as a diagnostic biomarker but also as a therapeutic target for OA.
Hsa_circ_101178
By RT-qPCR, Chen reported that the expression level of hsa_circ_101178 was upregulated in both serum and synovial fluid of OA patients (Chen Citation2020). Moreover, both Western Ontario McMaster University Osteoarthritis Index (WOMAC) pain scores and Kellgren-Lawrence (KL) grading system were positively correlated with serum hsa_circ_101178. ROC analysis also showed that serum hsa_circ_101178 can separate the patients with OA from the healthy controls (AUG: 0.835) (Chen Citation2020). Thus, these data suggested that serum hsa_circ_101178 may be a potential diagnostic biomarker for OA.
Circ_RUNX2
Wang et al. reported that the expression level of circ_RUNX2 (also known as hsa_circ_0005526) was significantly increased in the sera of OA patients. In addition, the ROC curve showed that circ_RUNX2 held diagnostic value for OA (AUG:0.82) (Wang C et al. Citation2021). Using bioinformatics analysis and luciferase reporter assay, the authors also demonstrated that circ_RUNX2 can interact with 4 miRNAs (miR-498, miR-924, miR-361-3p, miR-665) which may be involved in the ECM-receptor interaction pathway (Wang C et al. Citation2021). These results suggested that circ_RUNX2 may have the potential to be used as a diagnostic biomarker for OA and circ_RUNX2/miRNA networks may contribute to the progression of OA.
Conclusion
OA is a leading cause of chronic pain in the elderly population (Cross et al. Citation2014). Currently, joint replacement is still the main treatment option for severe OA because disease-modifying treatment for OA is not available yet. Thus, to find novel clinical options for treating OA patients, it is important to identify new factors and elucidate their roles in the pathogenesis of OA.
The recent development of RNA-seq and bioinformatics contributes to revealing the potential involvement of circRNAs in the pathogenesis of OA. As summarized in Table and Figure , dysregulated circRNAs in OA have been shown to participate in the progression of OA through mechanisms such as inflammation, ECM homeostasis, oxidative stress, apoptosis/proliferation, and autophagy mostly by reducing the availability of their target miRNAs, meanwhile circPDE4B was shown to act as a scaffold to control the degradation of the target protein (RIC8A) (Shen et al. Citation2021). Especially, in vivo studies to overexpress (circHIPK3, circRNF121, circRSU1, circSERPINE2, circCDK14, circPDE4D, circANKRD36, circ0083429, circPDE4B, circSLC7A2, circRNA_0001236, and circFAM160A2) or knockdown (circRNA.33186, circCSNK1G1, circRNA_0092516, circCDH13, circ_0128846, circ_013 4111, and hsa_circ_0032131) specific circRNA successfully demonstrated the effect of specific circRNA on the cartilage in animal models of OA, suggesting that these circRNAs have promising therapeutic potential for the treatment of OA. Nevertheless, relevant studies are still limited. Besides the use of viruses, other in vivo methods using carriers such as cholesterol molecules (Chen Q et al. Citation2010), receptor-specific pegylated immunoliposomes (Pardridge Citation2004), and EVs (Tkach and Théry Citation2016) have already been developed to effectively deliver miRNAs. Given EVs can deliver their cargo (e.g. ncRNAs including miRNA and circRNA) to specific recipient cells (Tu et al. Citation2020), in vivo studies using EVs may be useful to evaluate the therapeutic effect of circRNAs for the treatment of OA: In fact, circ_0116061 (Guo et al. Citation2021) and circ_0001846 (Zhu C et al. Citation2021) were revealed as exosomal circRNAs which can promote OA progression in vitro. Moreover, it was shown that small EVs (sEVs) from synovium MSC overexpressing circRNA3503 (circRNA3503-OE-sEVs, Tao et al. Citation2021), MSCs-derived EVs overexpressing circHIPK3 (MSCs-circHIPK3-EVs, Li S et al. Citation2021), and exosomes secreted by circRNA_0001236-overexpressing MSCs (MSC-circRNA_0001236-Exos, Mao G et al. Citation2021) can prevent OA progression in vivo, suggesting that MSC-derived circRNAs could be a potential alternative to MSC therapy for OA, especially given the safety concerns about stem cell-based therapy.
Although circRNAs have been considered as potential biomarkers for various diseases (Zhang Z et al. Citation2018), it has not yet been clarified whether circRNAs can be used for diagnosing OA. As summarized in Figure , the following circRNAs were identified as promising diagnostic biomarkers for OA based on ROC curve analysis: hsa_circ_0104873, hsa_circ_0104595, hsa_circ_0101251 (in synovial fluid, Yu et al. Citation2018), hsa_circRNA_0032131 (in peripheral blood, Wang et al. Citation2019a), hsa_circ_101178 (in both serum and synovial fluid, Chen Citation2020), and circ_RUNX2 (in serum, Wang C et al. Citation2021). However, their putative regulatory roles of the circRNA-miRNA-mRNA axis in the progression of OA have not yet been fully explored except hsa_circ_0104873 and hsa_circRNA_0032131 (Table , Figures and ). Thus, future studies, including prospective cohort studies, may be needed to further demonstrate the reliability of utilizing these circRNAs as novel diagnostic biomarkers for OA.
Figure 4. CircRNAs as potential diagnostic biomarkers for OA. Dysregulated circRNAs in the body fluids such as blood/serum and synovial fluid from OA patients have been identified as potential biomarkers for OA based on AUC-ROC curve analysis. ADAM10: A-disintegrin and metallopeptidase domain 10; AUC: The area under the ROC curves; PRDX3: Peroxiredoxin 3; ROC: Receiver operating characteristics; TCF4: Transcription factor 4.
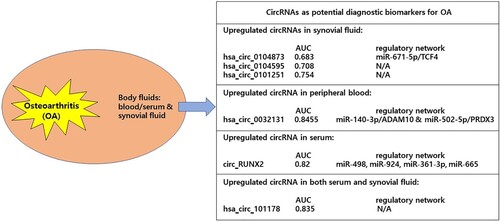
In conclusion, the current findings on dysregulated circRNAs in OA suggest that circRNAs may play important roles in the progression of OA and have the potential to be utilized as novel therapeutic targets and diagnostic biomarkers for OA. In future, more detailed research to fully clarify the roles of circRNAs in the pathogenesis of OA may contribute to the early diagnosis and treatment of OA.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
References
- Avenoso A, D’Ascola A, Scuruchi M, Mandraffino G, Calatroni A, Saitta A, Campo S, Campo GM. 2018. Hyaluronan in experimental injured/inflamed cartilage: In vivo studies. Life Sci. 193:132–140.
- Bai Y, Ying Y. 2020. The post-translational modifications of Smurf2 in TGF-beta signaling. Front Mol Biosci. 7:128.
- Bai ZM, Kang M-M, Zhou X-F, Wang D. 2020. CircTMBIM6 promotes osteoarthritis-induced chondrocyte extracellular matrix degradation via miR-27a/MMP13 axis. Eur Rev Med Pharmacol Sci. 24(15):7927–7936.
- Bao J, Lin C, Zhou X, Ma D, Ge L, Xu K, Moqbel SAA, He Y, Ma C, Ran J, Wu L. 2021. circFAM160A2 promotes mitochondrial stabilization and apoptosis reduction in osteoarthritis chondrocytes by targeting miR-505-3p and SIRT3. Oxid Med Cell Longev. 2021:5712280.
- Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. 2011. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 147(2):358–369.
- Chen C. 2020. Serum hsa_circ_101178 as a potential biomarker for early prediction of osteoarthritis. Clin Lab. 66(8).
- Chen C, Yin P, Hu S, Sun X, Li B. 2020. Circular RNA-9119 protects IL-1β-treated chondrocytes from apoptosis in an osteoarthritis cell model by intercepting the microRNA-26a/PTEN axis. Life Sci. 256:117924.
- Chen D, Shen J, Zhao W, Wang T, Han L, Hamilton JL, Im H-J. 2017. Osteoarthritis: toward a comprehensive understanding of pathological mechanism. Bone Res. 5:16044.
- Chen G, Liu T, Yu B, Wang B, Peng Q. 2020. CircRNA-UBE2G1 regulates LPS-induced osteoarthritis through miR-373/HIF-1a axis. Cell Cycle. 19(13):1696–1705.
- Chen G, Tang W, Wang S, Long C, He X, Yang D, Peng S. 2021. Promising diagnostic and therapeutic circRNAs for skeletal and chondral disorders. Int J Biol Sci. 17(5):1428–1439.
- Chen Q, Butler D, Querbes W, Pandey RK, Ge P, Maier MA, Zhang L, Rajeev KG, Nechev L, Kotelianski V, et al. 2010. Lipophilic siRNAs mediate efficient gene silencing in oligodendrocytes with direct CNS delivery. J Control Release. 144(2):227–232.
- Chen X, Yang T, Wang W, Xi W, Zhang T, Li Q, Yang A, Wang T. 2019. Circular RNAs in immune responses and immune diseases. Theranostics. 9(2):588–607.
- Cheng N-T, Meng H, Ma L-F, Zhang L, Yu H-M, Wang Z-Z, Guo A. 2017. Role of autophagy in the progression of osteoarthritis: The autophagy inhibitor, 3-methyladenine, aggravates the severity of experimental osteoarthritis. Int J Mol Med. 39:1224–1232.
- Cross M, Smith E, Hoy D, Nolte S, Ackerman I, Fransen M, Bridgett L, Williams S, Guillemin F, Hill CL, et al. 2014. The global burden of hip and knee osteoarthritis: estimates from the global burden of disease 2010 study. Ann Rheum Dis. 73(7):1323–1330.
- Ding R, Zhou J, Xu J, Lu H, Zhang T, Xiang X, Shi Z. 2021. Circ_0045714/miR-331-3p interaction affects IL-1β-evoked human articular chondrocyte injury through regulating PIK3R3 in a ceRNA regulatory cascade. J Orthop Surg Res. 16(1):595.
- Elkhenany H, AlOkda A, El-Badawy A, El-Badri N. 2018. Tissue regeneration: impact of sleep on stem cell regenerative capacity. Life Sci. 214:51–61.
- Errichelli L, Dini Modigliani S, Laneve P, Colantoni A, Legnini I, Capauto D, Rosa A, De Santis R, Scarfò R, Peruzzi G, et al. 2017. FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat Commun. 8:14741.
- Fang L, Lin L, Lv Y, Huang Z, Lin X, Wang X, Chen B. 2021. The mechanism of aerobic exercise combined with glucosamine therapy and circUNK in improving knee osteoarthritis in rabbits. Life Sci. 275:119375.
- Feng M, Jing L, Cheng J, An S, Huang J, Yan Q. 2021. Circ_0020093 ameliorates IL-1β-induced apoptosis and extracellular matrix degradation of human chondrocytes by upregulating SPRY1 via targeting miR-23b. Mol Cell Biochem. 476(10):3623–3633.
- Floris G, Zhang L, Follesa P, Sun T. 2017. Regulatory role of circular RNAs and neurological disorders. Mol Neurobio. 54(7):5156–5165.
- Fu Q, Li L, Wang B, Wu J, Li H, Han Y, Xiang D, Chen Y, Zhu J. 2021. CircADAMTS6/miR-431-5p axis regulate interleukin-1β induced chondrocyte apoptosis. J Gene Med. 23(2):e3304.
- Fu S, Fan Q, Xu J, Yu S, Sun M, Ji Y, Liu D. 2021. Circ_0008956 contributes to IL-1β-induced osteoarthritis progression via miR-149-5p/NAMPT axis. Int Immunopharmacol. 98:107857.
- Gosset M, Berenbaum F, Salvat C, Sautet A, Pigenet A, Tahiri K, Jacques C. 2008. Crucial role of visfatin/pre–B cell colony-enhancing factor in matrix degradation and prostaglandin E2 synthesis in chondrocytes: possible influence on osteoarthritis. Arthritis Rheum. 58(5):1399–1409.
- Guo Z, Wang H, Zhao F, Liu M, Wang F, Kang M, He W, Lv Z. 2021. Exosomal circ-BRWD1 contributes to osteoarthritis development through the modulation of miR-1277/TRAF6 axis. Arthritis Res Ther. 23(1):159.
- Hansen TB, Kjems J, Damgaard CK. 2013. Circular RNA and miR-7 in cancer. Cancer Res. 73(18):5609–5612.
- Holdt LM, Kohlmaier A, Teupser D. 2018. Molecular roles and function of circular RNAs in eukaryotic cells. Cell Mol Life Sci. 75(6):1071–1098.
- Huang Z, Ma W, Xiao J, Dai X, Ling W. 2021. CircRNA_0092516 regulates chondrocyte proliferation and apoptosis in osteoarthritis through the miR-337-3p/PTEN axis. J Biochem. 169(4):467–475.
- Hwang HS, Kim HA. 2015. Chondrocyte apoptosis in the pathogenesis of osteoarthritis. Int J Mol Sci. 16(11):26035–26054.
- Jia Z, Wei QJ. 2021. CircRNA-MSR regulates LPS-induced C28/I2 chondrocyte injury through miR-643/MAP2K6 signaling pathway. Cartilage. 13(2_suppl):785S–795S.
- Jiang H, Dai J, Zhang C, Sun H, Tang X. 2021. Circ_0045714 alleviates TNF-α-induced chondrocyte injury and extracellular matrix degradation through miR-218-5p/HRAS axis. J Bioenerg Biomembr. 53(1):97–107.
- Jiang R, Gao H, Cong F, Zhang W, Song T, Yu Z. 2021. Circ_DHRS3 positively regulates GREM1 expression by competitively targeting miR-183-5p to modulate IL-1β-administered chondrocyte proliferation, apoptosis and ECM degradation. Int Immunopharmacol. 91:107293.
- Khan S, Logan PC, Asokan A, Handford C, Moores T. 2020. The assessment and management of the arthritic knee: an update. Cureus. 12(11):e11582.
- Li BF, Zhang Y, Xiao J, Wang F, Li M, Guo X-z, Xie H-b, Xia H, Chen B. 2017. Hsa_circ_0045714 regulates chondrocyte proliferation, apoptosis and extracellular matrix synthesis by promoting the expression of miR-193b target gene IGF1R. Hum Cell. 30(4):311–318.
- Li H, Liu Z, Guo X, Zhang M. 2021. Circ_0128846/miR-140-3p/JAK2 network in osteoarthritis development. Immunol Invest. Sep 21:1–19. Epub ahead of print.
- Li H, Wang D, Yuan Y, Min J. 2017. New insights on the MMP-13 regulatory network in the pathogenesis of early osteoarthritis. Arthritis Res Ther. 19(1):248.
- Li H, Yang HH, Sun ZG, Tang HB, Min JK. 2019. Whole-transcriptome sequencing of knee joint cartilage from osteoarthritis patients. Bone Joint Res. 8(7):290–303.
- Li S, Liu J, Liu S, Jiao W, Wang X. 2021. Mesenchymal stem cell-derived extracellular vesicles prevent the development of osteoarthritis via the circHIPK3/miR-124-3p/MYH9 axis. J Nanobiotechnol. 19(1):194.
- Li Z, Yuan B, Pei Z, Zhang K, Ding Z, Zhu S, Wang Y, Guan Z, Cao Y. 2019. Circ_0136474 and MMP-13 suppressed cell proliferation by competitive binding to miR-127-5p in osteoarthritis. J Cell Mol Med. 23(10):6554–6564.
- Liao HX, Zhang Z-H, Chen H-L, Huang Y-M, Liu Z-L, Huang J. 2021. CircHYBID regulates hyaluronan metabolism in chondrocytes via hsa-miR-29b-3p/TGF-β1 axis. Mol Med. 27(1):56.
- Liu C, Cheng P, Liang J, Zhao X, Du W. 2021. Circular RNA circ_0128846 promotes the progression of osteoarthritis by regulating miR-127-5p/NAMPT axis. J Orthop Surg Res. 16(1):307.
- Liu D, Liang Y-H, Yang Y-T, He M, Cai Z-J, Xiao W-F, Li Y-S. 2021. Circular RNA in osteoarthritis: an updated insight into the pathophysiology and therapeutics. Am J Transl Res. 13(1):11–23.
- Liu Q, Zhang X, Hu X, Dai L, Fu X, Zhang J, Ao Y. 2016. Circular RNA related to the chondrocyte ECM regulates MMP13 expression by functioning as a MiR-136 ‘sponge’ in human cartilage degradation. Sci Rep. 6:22572.
- Liu Q, Zhang X, Hu X, Yuan L, Cheng J, Jiang Y, Ao Y. 2017. Emerging roles of circRNA related to the mechanical stress in human cartilage degradation of osteoarthritis. Mol Ther Nucleic Acids. 7:223–230.
- Liu X, Zhao P, Ge W. 2020. Knockdown of circular RNA circZNF652 remits LPS-induced inflammatory damage by regulating miR-181a. Biofactors. 46(6):1031–1040.
- Liu Y, Li Q, Gao Z, Lei F, Gao X. 2021. Circ-SPG11 knockdown hampers IL-1β-induced osteoarthritis progression via targeting miR-337-3p/ADAMTS5. J Orthop Surg Res. 16(1):392.
- Liu Y, Zhang Y. 2021. Hsa_circ_0134111 promotes osteoarthritis progression by regulating miR-224-5p/CCL1 interaction. Aging. 13(16):20383–20394.
- Luo P, Gao F, Niu D, Sun X, Song Q, Guo C, Liang Y, Sun W. 2019. The role of autophagy in chondrocyte metabolism and osteoarthritis: a comprehensive research review. Biomed Res Int. 2019:5171602.
- Ma HR, Mu WB, Zhang KY, Zhou HK, Jiang RD, Cao L. 2020. CircVCAN regulates the proliferation and apoptosis of osteoarthritis chondrocyte through NF-κB signaling pathway. Eur Rev Med Pharmacol Sci. 24(7):6517–6525.
- Maldonado M, Nam J. 2013. The role of changes in extracellular matrix of cartilage in the presence of inflammation on the pathology of osteoarthritis. Biomed Res Int. 2013:1–10.
- Mao G, Xu Y, Long D, Sun H, Li H, Xin R, Zhang Z, Li Z, Yang Z, Kang Y. 2021. Exosome-transported circRNA_0001236 enhances chondrogenesis and suppress cartilage degradation via the miR-3677-3p/Sox9 axis. Stem Cell Res Ther. 12(1):389.
- Mao X, Cao Y, Guo Z, Wang L, Xiang C. 2021. Biological roles and therapeutic potential of circular RNAs in osteoarthritis. Mol Ther Nucleic Acids. 24:856–867.
- Marchev AS, Dimitrova PA, Burns AJ, Kostov RV, Dinkova-Kostova AT, Georgiev MI. 2017. Oxidative stress and chronic inflammation in osteoarthritis: can NRF2 counteract these partners in crime? Ann N Y Acad Sci. 1401(1):114–135.
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. 2013. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 495(7441):333–338.
- Neogi T. 2013. The epidemiology and impact of pain in osteoarthritis. Osteoarthritis Cartilage. 21(9):1145–1153.
- Ni JL, Dang XQ, Shi ZB. 2020. CircPSM3 inhibits the proliferation and differentiation of OA chondrocytes by targeting miRNA-296-5p. Eur Rev Med Pharmacol Sci. 24(7):3467–3475.
- Ni W, Jiang C, Wu Y, Zhang H, Wang L, Yik JHN, Haudenschild DR, Fan S, Shen S, Hu Z. 2021. CircSLC7A2 protects against osteoarthritis through inhibition of the miR-4498/TIMP3 axis. Cell Prolif. 54(6):e13047.
- O’Brien J, Hayder H, Zayed Y, Peng C. 2018. Overview of MicroRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol. 9:402.
- Pardridge WM. 2004. Intravenous, non-viral RNAi gene therapy of brain cancer. Expert Opin Biol Ther. 4(7):1103–1113.
- Paul P, Chakraborty A, Sarkar D, Langthasa M, Rahman M, Bari M, Singha RS, Malakar AK, Chakraborty S. 2018. Interplay between miRNAs and human diseases. J Cell Physiol. 233(3):2007–2018.
- Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. 2010. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 465(7301):1033–1038.
- Rong D, Lu C, Zhang B, Fu K, Zhao S, Tang W, Cao H. 2019. CircPSMC3 suppresses the proliferation and metastasis of gastric cancer by acting as a competitive endogenous RNA through sponging miR-296-5p. Mol Cancer. 18:25.
- Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO, Preiss T. 2012. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 7(2):e30733.
- Shen P, Yang Y, Liu G, Chen W, Chen J, Wang Q, Gao H, Fan S, Shen S, Zhao X. 2020. CircCDK14 protects against osteoarthritis by sponging miR-125a-5p and promoting the expression of Smad2. Theranostics. 10(20):9113–9131.
- Shen S, Wu Y, Chen J, Xie Z, Huang K, Wang G, Yang Y, Ni W, Chen Z, Shi P, et al. 2019. CircSERPINE2 protects against osteoarthritis by targeting miR-1271 and ETS-related gene. Ann Rheum Dis. 78(6):826–836.
- Shen S, Yang Y, Shen P, Ma J, Fang B, Wang Q, Wang K, Shi P, Fan S, Fang X. 2021. circPDE4B prevents articular cartilage degeneration and promotes repair by acting as a scaffold for RIC8A and MID1. Ann Rheum Dis. 80(9):1209–1219.
- Shi L, Zhang H, Sun J, Gao X, Liu C. 2021. CircSEC24A promotes IL-1β-induced apoptosis and inflammation in chondrocytes by regulating miR-142-5p/SOX5 axis. Biotechnol Appl Biochem. Mar 9:1–13. Epub ahead of print.
- Sofat N, Kuttapitiya A. 2014. Future directions for the management of pain in osteoarthritis. Int J Clin Rheumtol. 9(2):197–216.
- Song S, Tan J, Miao Y, Li M, Zhang Q. 2017. Crosstalk of autophagy and apoptosis: involvement of the dual role of autophagy under ER stress. J Cell Physiol. 232(11):2977–2984.
- Sophia Fox AJ, Bedi A, Rodeo SA. 2009. The basic science of articular cartilage: structure, composition, and function. Sports Health. 1(6):461–468.
- Su X, Xing J, Wang Z, Chen L, Cui M, Jiang B. 2013. microRNAs and ceRNAs: RNA networks in pathogenesis of cancer. Chin J Cancer Res. 25(2):235–239.
- Sun M, Yang J, Jiang D, Bao G. 2021. Overexpression of hsa_circ_0094742 inhibits IL-1β-induced decline in CHON-001 cell viability by targeting microRNA-127-5p. Histol Histopathol. 36(2):207–216.
- Szabo L, Salzman J. 2016. Detecting circular RNAs: bioinformatic and experimental challenges. Nat Rev Genet. 17:679–692.
- Tao SC, Huang J-Y, Gao Y, Li Z-X, Wei Z-Y, Dawes H, Guo S-C. 2021. Small extracellular vesicles in combination with sleep-related circRNA3503: A targeted therapeutic agent with injectable thermosensitive hydrogel to prevent osteoarthritis. Bioact Mater. 6(12):4455–4469.
- Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U, Karreth F, Poliseno L, Provero P, Di Cunto F, et al. 2011. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 147(2):344–357.
- Tkach M, Théry C. 2016. Communication by extracellular vesicles: where we are and where we need to go. Cell. 164(6):1226–1232.
- Tu C, He J, Chen R, Li Z. 2020. The emerging role of exosomal non-coding RNAs in musculoskeletal diseases. Curr Pharm Des. 25(42):4523–4535.
- Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, Zhang Y, Wu Y-M, Dhanasekaran SM, Engelke CG, Cao X, et al. 2019. The landscape of circular RNA in cancer. Cell. 176(4):869–881.e13.
- Wang C, Li N, Liu Q, Su L, Wang S, Chen Y, Liu M, Lin H. 2021. The role of circRNA derived from RUNX2 in the serum of osteoarthritis and its clinical value. J Clin Lab Anal. 35(7):e23858.
- Wang J, Wang K, Huang C, Lin D, Zhou Y, Wu Y, Tian N, Fan P, Pan X, Xu D, et al. 2018. SIRT3 activation by dihydromyricetin suppresses chondrocytes degeneration via maintaining mitochondrial homeostasis. Int J Biol Sci. 14(13):1873–1882.
- Wang KF, Shi ZW, Dong DM. 2021. CircATRNL1 protects against osteoarthritis by targeting miR-153-3p and KLF5. Int Immunopharmacol. 96:107704.
- Wang Q, Luo S, Yang J, Li J, Huan S, She G, Zha Z. 2021. Circ_0114876 promoted IL-1β-induced chondrocyte injury by targeting miR-671/TRAF2 axis. Biotechnol Lett. 43(4):791–802.
- Wang T, Hao Z, Liu C, Yuan L, Li L, Yin M, Li Q, Qi Z, Wang Z. 2020. LEF1 mediates osteoarthritis progression through circRNF121/miR-665/MYD88 axis via NF-B signaling pathway. Cell Death Dis. 11(7):598.
- Wang Y, Wu C, Yang Y, Ren Z, Lammi MJ, Guo X. 2019a. Preliminary exploration of hsa_circ_0032131 levels in peripheral blood as a potential diagnostic biomarker of osteoarthritis. Genet Test Mol Biomarkers. 23(10):717–721.
- Wang Y, Wu C, Zhang F, Zhang Y, Ren Z, Lammi MJ, Guo X. 2019b. Screening for differentially expressed circular RNAs in the cartilage of osteoarthritis patients for their diagnostic value. Genet Test Mol Biomarkers. 23(10):706–716.
- Wu J, Tao Y, Shang A, Wang W, Zhang Y, Hu L, Wang J, Wang Y, Guo N. 2017. Effect of the interaction between MiR-200b-3p and DNMT3A on cartilage cells of osteoarthritis patients. J Cell Mol Med. 21(10):2308–2316.
- Wu Q, Kim K-O, Sampson ER, Chen D, Awad H, O’Brien T, Puzas JE, Drissi H, Schwarz EM, O’Keefe RJ, et al. 2008. Induction of an osteoarthritis-like phenotype and degradation of phosphorylated Smad3 by Smurf2 in transgenic mice. Arthritis Rheum. 58(10):3132–3144.
- Wu Q, Yuan Z-H, Ma X-B, Tang X-H. 2020. Low expression of CircRNA HIPK3 promotes osteoarthritis chondrocyte apoptosis by serving as a sponge of miR-124 to regulate SOX8. Eur Rev Med Pharmacol Sci. 24(15):7937–7945.
- Wu R, Zhang F, Cai Y, Long Z, Duan Z, Wu D, Zhou Y, Wang Q. 2021. Circ_0134111 knockdown relieves IL-1β-induced apoptosis, inflammation and extracellular matrix degradation in human chondrocytes through the circ_0134111-miR-515-5p-SOCS1 network. Int Immunopharmacol. 95:107495.
- Wu Y, Hong Z, Xu W, Chen J, Wang Q, Chen J, Ni W, Mei Z, Xie Z, Ma Y, et al. 2021. Circular RNA circPDE4D protects against osteoarthritis by binding to miR-103a-3p and regulating FGF18. Mol Ther. 29(1):308–323.
- Wu Y, Zhang Y, Zhang Y, Wang J-J. 2017. CircRNA hsa_circ_0005105 upregulates NAMPT expression and promotes chondrocyte extracellular matrix degradation by sponging miR-26a. Cell Biol Int. 41(12):1283–1289.
- Xi P, Zhang C, Wu S, Liu L, Li W, Li Y. 2021. CircRNA circ-IQGAP1 knockdown alleviates interleukin-1β-induced osteoarthritis progression via targeting miR-671-5p/TCF4. Orthop Surg. 13(3):1036–1046.
- Xiao J, Wang R, Zhou W, Cai X, Ye Z. 2021. Circular RNA CSNK1G1 promotes the progression of osteoarthritis by targeting the miR-4428/FUT2 axis. Int J Mol Med. 47(1):232–242.
- Xiao K, Xia Z, Feng B, Bian Y, Fan Y, Li Z, Wu Z, Qiu G, Weng X. 2019. Circular RNA expression profile of knee condyle in osteoarthritis by illumina HiSeq platform. J Cell Biochem. 120(10):17500–17511.
- Xu J, Ma X. 2021. Hsa_circ_0032131 knockdown inhibits osteoarthritis progression via the miR-502-5p/PRDX3 axis. Aging. 13(11):15100–15113.
- Yang Y, Shen P, Yao T, Ma J, Chen Z, Zhu J, Gong Z, Shen S, Fang X. 2021. Novel role of circRSU1 in the progression of osteoarthritis by adjusting oxidative stress. Theranostics. 11(4):1877–1900.
- Yao T, Yang Y, Xie Z, Xu Y, Huang Y, Gao J, Shen S, Ye H, Iranmanesh Y, Fan S, Ma J. 2021. Circ0083429 regulates osteoarthritis progression via the Mir-346/SMAD3 axis. Front Cell Dev Biol. 8:579945.
- Yu CX, Sun S. 2018. An emerging role for circular RNAs in osteoarthritis. Yonsei Med J. 59(3):349–355.
- Yu F, Xie C, Sun J, Feng H, Huang X. 2018. Circular RNA expression profiles in synovial fluid: a promising new class of diagnostic biomarkers for osteoarthritis. Int J Clin Exp Pathol. 11(3):1338–1346.
- Yu J, Qin Y, Zhou N. 2021. Knockdown of Circ_SLC39A8 protects against the progression of osteoarthritis by regulating miR-591/IRAK3 axis. J Orthop Surg Res. 16(1):170.
- Yuan X, Zhang Y, Cai C, Liu C, Xie J, Yi C. 2021. Circular RNA circZNF652 is overexpressed in osteoarthritis and positively regulates LPS-induced apoptosis of chondrocytes by upregulating PTEN. Autoimmunity. 54(7):415–421.
- Zhang H, Ge J, Lu X. 2021. CircFADS2 is downregulated in osteoarthritis and suppresses LPS-induced apoptosis of chondrocytes by regulating miR-195-5p methylation. Arch Gerontol Geriatr. 96:104477.
- Zhang J, Cheng F, Rong G, Tang Z, Gui B. 2020. Hsa_circ_0005567 activates autophagy and suppresses IL-1β-induced chondrocyte apoptosis by regulating miR-495. Front Mol Biosci. 7:216.
- Zhang L, Sui C, Zhang Y, Wang G, Yin Z. 2021. Knockdown of hsa_circ_0134111 alleviates the symptom of osteoarthritis via sponging microRNA-224-5p. Cell Cycle. 20(11):1052–1066.
- Zhang M, Mou L, Liu S, Sun F, Gong M. 2021. Circ_0001103 alleviates IL-1β-induced chondrocyte cell injuries by upregulating SIRT1 via targeting miR-375. Clin Immunol. 227:108718.
- Zhang P, Wu W, Chen Q, Chen M. 2019. Non-coding RNAs and their integrated networks. J Integr Bioinform. 16(3):20190027.
- Zhang Q, Qiao X, Xia W. 2020. CircSERPINE2 weakens IL-1β-caused apoptosis and extracellular matrix degradation of chondrocytes by regulating miR-495/TGFBR2 axis. Biosci Rep. 40(11):BSR20201601.
- Zhang W, Qi L, Chen R, He J, Liu Z, Wang W, Tu C, Li Z. 2021. Circular RNAs in osteoarthritis: indispensable regulators and novel strategies in clinical implications. Arthritis Res Ther. 23(1):23.
- Zhang W, Zhang C, Hu C, Luo C, Zhong B, Yu X. 2020. Circular RNA-CDR1as acts as the sponge of microRNA-641 to promote osteoarthritis progression. J Inflamm. 17:8.
- Zhang Z, Yang B, Zhou S, Wu J. 2021. CircRNA circ_SEC24A upregulates DNMT3A expression by sponging miR-26b-5p to aggravate osteoarthritis progression. Int Immunopharmacol. 99:107957.
- Zhang Z, Yang T, Xiao J. 2018. Circular RNAs: promising biomarkers for human diseases. EBioMedicine. 34:267–274.
- Zhao J, Li T, Luo W. 2021. Silencing of circ-PRKCH protects against lipopolysaccharide (LPS)-evoked chondrocyte damage and extracellular matrix loss by the miR-140-3p/ADAM10 axis. Gen Physiol Biophys. 40(2):89–101.
- Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, et al. 2016. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 7:11215.
- Zheng W, Hou G, Li Y. 2021. Circ_0116061 regulated the proliferation, apoptosis, and inflammation of osteoarthritis chondrocytes through regulating the miR-200b-3p/SMURF2 axis. J Orthop Surg Res. 16(1):253.
- Zhou J, Wang Y, Liu Y, Zeng H, Xu H, Lian F. 2019. Adipose derived mesenchymal stem cells alleviated osteoarthritis and chondrocyte apoptosis through autophagy inducing. J Cell Biochem. 120:2198–2212.
- Zhou JL, Deng S, Fang H, Du X, Peng H, Hu Q. 2021. Circular RNA circANKRD36 regulates Casz1 by targeting miR-599 to prevent osteoarthritis chondrocyte apoptosis and inflammation. J Cell Mol Med. 25(1):120–131.
- Zhou X, Jiang L, Fan G, Yang H, Wu L, Huang Y, Xu N, Li J. 2019. Role of the ciRS-7/miR-7 axis in the regulation of proliferation, apoptosis and inflammation of chondrocytes induced by IL-1β. Int Immunopharmacol. 71:233–240.
- Zhou X, Li J, Zhou Y, Yang Z, Yang H, Li D, Zhang J, Zhang Y, Xu N, Huang Y, Jiang L. 2020. Down-regulated ciRS-7/up-regulated miR-7 axis aggravated cartilage degradation and autophagy defection by PI3K/AKT/mTOR activation mediated by IL-17A in osteoarthritis. Aging. 12(20):20163–20183.
- Zhou Z, Du D, Chen A, Zhu L. 2018. Circular RNA expression profile of articular chondrocytes in an IL-1β-induced mouse model of osteoarthritis. Gene. 644:20–26.
- Zhou Z, Ma J, Lu J, Chen A, Zhu L. 2021. Circular RNA CircCDH13 contributes to the pathogenesis of osteoarthritis via CircCDH13/miR-296-3p/PTEN axis. J Cell Physiol. 236(5):3521–3535.
- Zhou ZB, Du D, Huang G-x, Chen A, Zhu L. 2018. Circular RNA atp9b, a competing endogenous RNA, regulates the progression of osteoarthritis by targeting miR-138-5p. Gene. 646:203–209.
- Zhou ZB, Huang G-x, Fu Q, Han B, Lu J-j, Chen A-m, Zhu L. 2019. circRNA.33186 contributes to the pathogenesis of osteoarthritis by sponging miR-127-5p. Mol Ther. 27(3):531–541.
- Zhu C, Shen K, Zhou W, Wu H, Lu Y. 2021. Exosome-mediated circ_0001846 participates in IL-1β-induced chondrocyte cell damage by miR-149-5p-dependent regulation of WNT5B. Clin Immunol. 232:108856.
- Zhu H, Zhu S, Shang X, Meng X, Jing S, Yu L, Deng Y. 2021. Exhausting circ_0136474 and restoring miR-766-3p attenuate chondrocyte oxidative injury in IL-1β-induced osteoarthritis progression through regulating DNMT3A. Front Genet. 12:648709.