Abstract
With its high incidence and mortality rate, lung cancer is one of the malignancies that poses the greatest threat to the health and life of the population. Mounting research found that the Membrane-spanning 4A (MS4A) family took part in tumor development. Nevertheless, the expression and prognostic value of the MS4A family genes in lung cancer remained unclear. Compared with normal tissues, the transcript levels of MS4A2, MS4A4A, MS4A4E, MS4A6A, MS4A6E, MS4A7, MS4A8, MS4A14, and MS4A15 were significantly decreased in lung cancer tissues, and there was a significant correlation between MS4A2, MS4A8, MS4A15, and the pathological stage of lung cancer patients. Survival analysis has shown that lung cancer patients with lower mRNA expression levels of MS4A2, MS4A4E, and MS4A15 had poorer prognosis. The function of MS4A family genes was primarily involved in immune-related pathways. Based on the results, it was indicated that the expression of the MS4A family was significantly correlated with the infiltration of immune cells (B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells). In conclusions, our research indicated that MS4A family might become a new immunotherapy target and prognostic biomarker for lung cancer.
Introduction
Lung cancer is one of the malignant tumors with the highest incidence and mortality around the world (Hirsch et al. Citation2017). Every year, 1.8 million people (11.6% of total cases) get diagnosed with lung cancer, and about 1.6 million people (18.4% of the total cancer deaths) die on account of this cancer. According to the stages and regional differences, 5-year survival rates vary from 4% to 17% (Bray et al. Citation2018). Comprehensive operative treatment is recommended for patients with early lung cancer (Vansteenkiste et al. Citation2014). The 5-year survival rate for pathological stage was 58–92% for stage I-II and 38–50% for stage III–IV in the previous study (Okami et al. Citation2019). In recent years, molecular targeted therapy has made tremendous progress. Although targeted therapy has a significant effect, it mainly targets young adenocarcinoma patients who have never smoked. This is far from enough, more new driver genes, therapeutic targets, and prognostic biomarkers must be discovered and used for targeted therapies for larger populations, more Accurate prediction of prognosis, and a better understanding of the mechanisms of lung carcinogenesis.
Membrane-spanning 4-domains subfamily A contains at least 16 family members, which are cell-surface proteins with four transmembrane regions. The MS4A family of genes are chromosomally close, encode structurally similar proteins, and are homologous in their amino acid sequences. MS4A1 (CD20) is an important marker of B-cell differentiation, regulates calcium ion inflow, and activates B-cell antigen receptors. It also is an important target of current lymphoma immunotherapy (van de Ven et al. Citation2012). MS4A2 (FcϵRIβ) has participated in the regulation of mast cell microtubule formation and degranulation in association with allergic reactions (Cruse et al. Citation2013). MS4A3 (HTm4) is a cell cycle regulator of hematopoietic cells and is associated with cell growth (Donato et al. Citation2002). In the past few years, an increasing number of researchers have begun to explore the nexus between MS4A gene family. It is reported that MS4A protein is specifically expressed in solid tumor tissue and is associated with tumorigenesis (Koslowski et al. Citation2008; Ye et al. Citation2014). Nevertheless, the potential characters of MS4A family proteins in the development or prognosis of lung cancer remain unknown.
This study thoroughly bioinformatics analyzed the transcriptional levels of MS4A family proteins and investigated their prognostic significance in lung cancer. Furthermore, we performed an analysis of the interaction network, functional enrichment, and immune infiltration of MS4A family proteins on several large public databases. This provides more data for clinicians to select proper therapeutic agents and more accurately predict long-term outcomes for lung cancer patients.
Materials and methods
ONCOMINE website analysis
Oncomine (https://www.oncomine.org/resource/login.html) (Rhodes et al. Citation2004) is a large tumor gene arrays database, covering over 480 million gene expression data, which can be used to analyze expression variances in genes, search for outliers, predict co-expression genes etc. In our study, we utilized Oncomine to analyze the expression level of MS4A family genes in pan-cancer and the threshold value was set at p-value < of 0.001, fold change > 2, and gene ranking of all.
GEPIA website analysis
Gene Expression Profiling Interactive Analysis (GEPIA) (http://gepia.cancer-pku.cn/index.html) (Tang et al. Citation2017) covers the RNA sequencing expression data of tumors and normal samples from the TCGA and the GTEx database. This study used GEPIA to seek out disparities in mRNA expression between tumors and normal tissues, disparities in pathological stages, and prognostic analysis related to MS4A family genes. Differential gene expression was analyzed by ANOVA, using pathological stage as a variable to calculate differential expression, and based on the differential results, GEPIA can perform overall survival (OS) or disease free survival (DFS), which uses the Log-rank test for hypothesis test and the p-value cutoff was set at <0.05.
Human Protein Atlas analysis
The Human Protein Atlas (HPA) (https://www.proteinatlas.org/) (Thul and Lindskog Citation2018) is a freely available web site, offering services to learn protein expression of human tissues and cells. On the basis of immunohistochemical images, a website tool, we assess the protein expression of the MS4A family between lung cancer and normal tissues.
Cbioportal website analysis
cBioPortal (www.cbioportal.org) (Gao et al. Citation2013) is a site simplifies molecular profiling data from cancer tissues and cell lines to make it more intuitive and straightway in analyzing multidimensional cancer genomics. Genetic alterations in the MS4A family genes from cBioPortal based on lung cancer data from the TCGA database. The mRNA expression z score (RNA Seq V2 RSEM) was obtained using a z score threshold of ± 2.0.
String website analysis
STRING (https://string-db.org/) (Szklarczyk et al. Citation2019) is a searchable database of functional protein association networks. We summarize the predicted association networks of MS4A family proteins using a network view.
Go and KEGG enrichment analysis
DAVID (https://david.ncifcrf.gov) (Huang et al. Citation2009) is an online analysis program with Gene Ontology functional annotations and Kyoto Encyclopedia of Genes and Genomes pathway enrichment. In our research, we utilized DAVID to perform GO enrichment analysis and KEGG pathway enrichment analysis of MS4A family genes and their relative genes.
TIMER website analysis
Tumor Immune Estimation Resource (TIMER) (http://timer.cistrome.org) (Li et al. Citation2017) is a compositive resource for systematically estimating the abundance of immune infiltrates, which is based on TCGA database, including over 10,897 samples and 32 cancer types. We take advantage of genetic module to analyze the correlation between MS4A family genes expression and immune infiltration abundance in lung cancer, including B cells, CD8+ T cells, CD4+ T cells, neutrophils, macrophages, and dendritic cells. Survival module was designed to find out the relationship among clinical outcomes, immune cells infiltration, and MS4A family genes expression.
Results
Abnormal expression of MS4A family in lung cancer patients
The ONCOMINE database was employed to search the transcriptional levels of 16 MS4A families in lung cancer tissues and normal lung tissues. The results were shown in Figure and Table . Transcription of MS4A2, MS4A3, MS4A4A, MS4A6A, MS4A7, MS4A8, MS4A14, and MS4A15 levels was significantly reduced. Seven datasets showed that MS4A2 expression was reduced in lung cancer tissues compared to paracancerous tissues (Beer et al. Citation2002; Stearman et al. Citation2005; Hou et al. Citation2010; Okayama et al. Citation2012; Selamat et al. Citation2012). In Arindam’s dataset, the fold change in MS4A3 expression in lung cancer cells was −4.89 (p = 3.97E-5) (Bhattacharjee et al. Citation2001). Hou et al. (Citation2010) and Garber et al. (Citation2001) found decreased MS4A4A levels in lung cancer cells. In the Garber and Selamat dataset, the transcription level of MS4A6A was significantly decreased in lung cancer cells than in normal tissues (Garber et al. Citation2001; Selamat et al. Citation2012). Okayama et al.’s Citation2012) data are accordant with Hou et al’s finding that MS4A7 was significantly down-regulation in lung cancer. Garber also showed that MS4A8 levels were significantly reduced in lung cancer (Garber et al. Citation2001). In addition, significantly lower levels of MS4A14 and MS4A15 were found in lung cancer tissues in Hou's data (Citation2010). The workflow diagram is shown in Figure .
Figure 2. mRNA levels of MS4A family genes in different cancer. The figure demonstrates the numbers of mRNA datasets of MS4A family genes in different cancers. Determined according to the following numerical thresholds: p-value ≤ 0.001, fold change ≥2, and gene ranking of all.
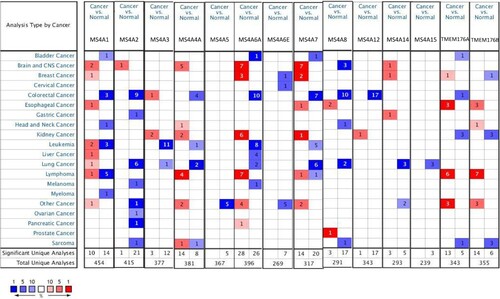
Table 1. Differential expression analyses of MS4A family in lung cancer.
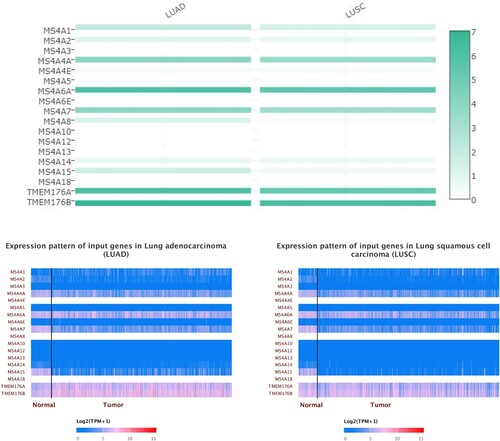
We also utilized GEPIA to evaluate the expression levels of the MS4A family of lung cancer tissues and normal lung tissues in the TCGA and GTEx data. In general agreement with the Oncomine results, MS4A2, MS4A4A, MS4A4E, MS4A7, MS4A14, and MS4A15 transcript levels were significantly reduced in lung adenocarcinoma (LUAD). Slightly different from the LUAD results, in lung squamous cell carcinoma (LUSC) MS4A2, MS4A4A, MS4A4E, MS4A6A, MS4A7, MS4A8, MS4A14, and MS4A15 transcript levels were decreased (Figure ). We also contrast the relative expression levels of MS4A family within LUAD and LUSC tissues and found that MS4A6A, TMEM176A, and TMEM176B had the highest relative expression among all MS4A families (Figure ).
Figure 3. The transcription levels of MS4A family genes in lung cancer. The transcriptional levels of MS4A2, MS4A4A4A, MS4A4E, MS4A7, MS4A14, and MS4A15 were significantly reduced in lung adenocarcinoma (LUAD). In lung squamous cell carcinoma(LUSC) MS4A2, MS4A4A, MS4A4E, MS4A6A, MS4A7, MS4A8, MS4A14, and MS4A15 transcript levels were decreased. The p value was set at 0.05.
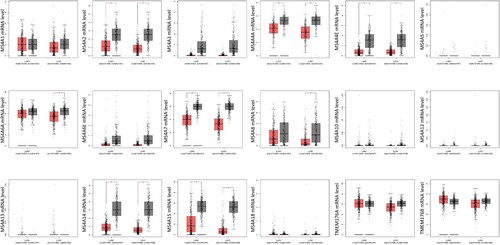
Figure 4. The expression matrix plot of MS4A family genes in lung cancer. MS4A6A, TMEM176A, and TMEM176B had the highest relative expression among all MS4A families.
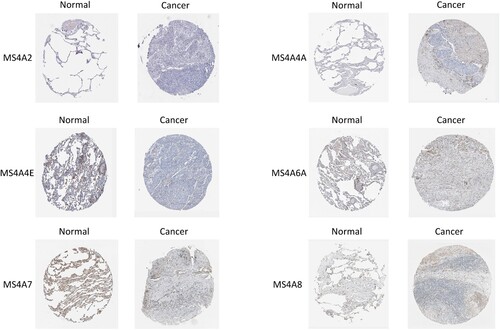
In order to further investigate the protein expression of MS4A family in lung cancer, we analyzed immunohistochemistry pictures from the HPA (Figure ). The result displays that MS4A2 and MS4A3 proteins were not expressed in both normal and lung cancer tissues. MS4A4A, MS4A6A, and MS4A7 proteins were not expressed in lung cancer tissues, while medium protein expressions were expressed in normal lung tissues. Furthermore, medium protein expressions of MS4A8 were observed in normal lung tissues while the low protein expressions were in lung cancer tissues. Unfortunately, datas for MS4A14, MS4A15 are not available in the database. In conclusion, we found that the expression of the proteins was mainly consistent with RNA expression data.
Figure 5. Immunohistochemistry pictures from HPA display the protein expression levels of MS4A family in lung cancer.
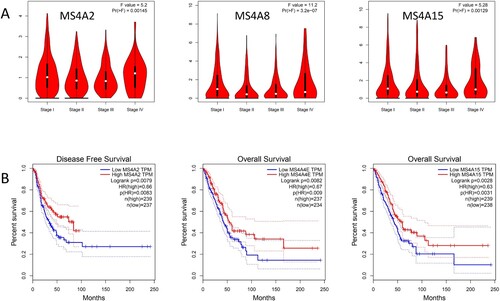
We then evaluated the association between differentially expressed MS4A family and the pathological stage of lung cancer patients and found that MS4A2 (p = 0.00145), MS4A8 (p = 3.2E-7), the expression of MS4A15 (p = 0.00129) was significantly correlated with the pathological stage (Figure (A)). The data in stages I, II, and III showed that as the oncology progressed, the expression of MS4A2, MS4A8, and MS4A15 reduced. These data suggest that these MS4A family genes act importantly in the early development of lung cancer. Interestingly, the expression of MS4A2, MS4A8, and MS4A15 was instead found to be elevated in the stage IV clinical data.
Analysis of the prognostic value of MS4A family in lung cancer patients
For the sake of assessing the prognostic value of differentially expressed MS4A families in lung cancer, we employed GEPIA to assess differential Correlation between expressed MS4A family and clinical data. High transcript levels of MS4A2 (p = 0.00083) were significantly related with longer DFS in lung cancer patients. We also evaluated the value of differentially expressed MS4A family in the OS of lung cancer patients and found that the higher levels of MS4A4E (p = 0.009) and MS4A15 (p = 0.0031) transcription were significantly associated with longer OS in lung cancer patients (Figure (B)).
Analysis of genetic alterations, neighboring gene networks, and protein interactions in the MS4A family in lung cancer patients
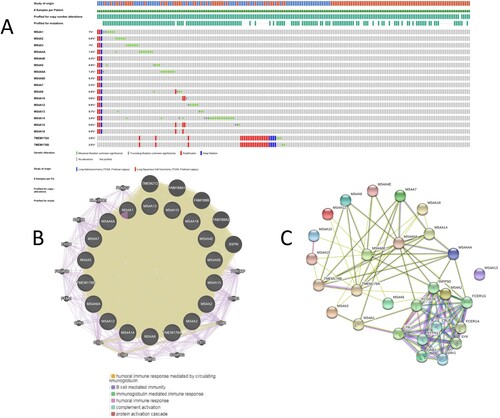
We used the TCGA dataset (LUAD, LUSC) to comprehensively analyze differentially expressed MS4A family genes Change. Results, MS4A1, MS4A2, MS4A3, MS4A4A, MS4A4E, MS4A6A, MS4A 6E, MS4A7, MS4A8, MS4A10, MS4A12, MS4A13. MS4A14, MS4A15, MS4A18, TMEM176A, and TMEM176B were altered 1%, 0.9%, 1%, 1.2%, 0.3%, 0.8%, 1.2%, 0.3%, 0.3%, respectively, 0.8%, 0.6%, 0.9%, 0.7%, 2.4%, 0.9%, 0.5%, 2.6%, and 2.5% in the lung cancer samples (Figure (A)). Amplification and missense mutation was the most common alteration in MS4A1, MS4A2, MS4A3, MS4A4A, MS4A6A, MS4A8, MS4A10, MS4A12, MS4A13, and MS4A14. MS4A4E, MS4A6E, MS4A7, MS4A15, MS4A18, TMEM176A, and TMEM176B were dominated by amplification. We then performed PPI network analysis on the differentially expressed MS4A family by using STRING to explore their interaction potential. We have constructed a 23-node, 63-edge PPI network (Figure (B)). The next results from the GeneMANIA protein interactions also showed that the differentially expressed MS4A family functioned mainly with humoral immune response mediated by circulating immunoglobulin, B cell mediated immunity, and protein activation cascade are related (Figure (C)).
Functional enrichment analysis of MS4A family in lung cancer
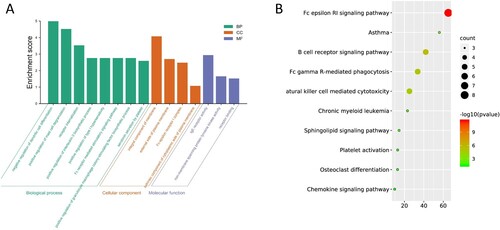
GO enrichment analysis and KEGG pathway analysis of the MS4A family and neighboring genes was performed online using DAVID. The top GO enrichment results are shown in (Figure (A,B)).
Association between MS4A family and immune cell infiltration in lung cancer patients
The MS4A family is participating in the human immunoreaction, in order to clarify its correlation with immune infiltration, we used TIMER database to analyze. There was a positive correlation between MS4A2 expression and the infiltration of B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells, with the most significant correlation with dendritic cells. Similar results were obtained in MS4A4A, MS4A6A, and MS4A7, with the difference that they correlated more strongly with monocytes, neutrophils, and dendritic cells. The results indicated that the expression of MS4A14A was positively correlated with the CD4+ T cell, macrophages, neutrophils and dendritic cells infiltration. However, there was a weak but insignificant correlation between MS4A8, MS4A14B, and immune cells (Figure ). The ‘Survival’ module was designed to explore the clinical relevance of immune subsets in lung cancer (LUAD, LUSC), which has the merit of amending multiple covariates in multivariate Cox proportional risk model. Covariates include individual factors (age, gender, ethnicity, tumor stages, etc.) and gene expression. B cells, CD4+ T cells were significantly associated with the clinical outcome of LUAD patients. And the clinical outcome of LUSC patients was significantly associated with MS4A4A, MS4A6A (Table ).
Figure 9. Correlation between differential expression of MS4A family genes and immune cell infiltration.
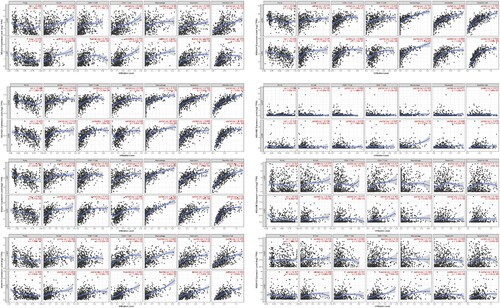
Table 2. The cox proportional hazard model of differentially expressed MS4A family and six tumor-infiltrating immune cells in LUAD and LUSC.
Discussion
lung cancer is one of the malignancies that pose the greatest threat to the health and life of the population all over the world due to its high incidence and mortality rate. In 2020, lung cancer is the primary cause of cancer death. According to the statistics, over two-thirds of death of lung cancer owe to tobacco (Sung et al. Citation2021). The population of smokers in China reached nearly 350 million in 2019, ranking first in the world. But there are other reasons that are considered to generate the disease, like outdoor air pollution and cooking oil fume which does harm to most of women and causes lung cancer given their low smoking prevalence. In most countries, the survival of lung cancer patients at 5 years is only 10–20% after being diagnosis (Allemani et al. Citation2018). Screening with low-dose computed tomography (CT) for high-risk people is considered to help diagnose cancer and under treatment earlier, but the disadvantage is obvious – expensive. Therefore, finding out a biomarker which is conducive to early and accurate diagnosis of lung cancer will be beneficial to control the death.
The MS4A family was originally found on the cell membrane surface of leukocytes. MS4A1 is a unique marker on the surface of B lymphocytes, which regulates calcium inflow and activates B cell antigen receptor function (van de Ven et al. Citation2012). MS4A2 is a signaling subunit of mast cell high-affinity IgE receptor and low-affinity IgG receptor, associated with allergic reactions and allergic diseases (Donnadieu et al. Citation2003). MS4A3 is found in the intracellular membranes of lymphocytes and plays a role in regulating the cell cycle (Kutok et al. Citation2011). MS4A4A is expressed in the plasma membrane of monocytes and has also been found to be present in lung tissue (Kutok et al. Citation2011; Zuccolo et al. Citation2013). In recent years, as the study of MS4A gene family has been much deeper, the MS4A family has been detected in other tissues with different expression patterns. MS4A5 is expressed only in testicular tissue (Hulett et al. Citation2001). MS4A6A is expressed in normal brain tissue and is associated with Alzheimer's disease (Martiskainen et al. Citation2015). MS4A12 is found to be expressed only in colonic epithelial cells and functions similarly to MS4A1, which also has a role in regulating calcium intracellular flow (Koslowski et al. Citation2008). Besides, the altered expression of MS4A protein has also been found in solid tumor tissues and is associated with tumorigenesis. MS4A8B was found to be strongly associated with prostate cancer in the research of Ye (Ye et al. Citation2014), and similarly, Michel proposed that the presence of reduced MS4A8B was also found in colon cancer (Michel et al. Citation2013). Drew et al. discovered that MS4A12 had obvious expression differences in normal colon, colon inflammatory polyps, and colon cancer tissues, and it was speculated that the expression may be related to the degree of differentiation of colon cells (Drew et al. Citation2014). Besides, there is a wealth of papers reporting the role of MS4A proteins in different immune cells under physiological or pathological conditions. For example, MS4A4A was highly expressed in human tumor-associated macrophages with lung cancer, colorectal carcinoma melanoma, and early-stage rheumatoid arthritis (Mattiola et al. Citation2019). In the research of Molgora et al. (Citation2020), it has already been confirmed that TREM2 emerged as a marker of tumor-infiltrating macrophages in all cancers. In Chiba’s (Citation2014) model of melanoma, macrophage activated cytotoxic NK cells to limit tumor burden and metastatic spread. However, the deficiency of MS4A4A triggered NK cell-mediated cytotoxic response, consequently impeding NK cells to control tumor metastasis (Mattiola et al. Citation2019). Evidence suggests that the MS4A family plays a crucial role in tumorigenesis and tumor metastasis. Nevertheless, the expression, prognostic value, and biological function of the MS4A family in lung cancer are still not been thoroughly investigated.
First, we compared the expression of the MS4A family in lung cancer with that in normal tissue using a public database and found that eight genes dissimilarly expressed in lung cancer compared to normal tissues (MS4A2, MS4A4A, MS4A4E, MS4A6A, MS4A7, MS4A8, MS4A14, and MS4A15 down-regulated). The abnormal expression of these genes indicates that they may contribute to the occurrence of lung cancer. Second, we evaluated the correlation between dissimilarly expressed MS4A family and pathological stage of lung cancer patients and found that MS4A2, MS4A8, MS4A15 were reduced with tumor progression in stage I–III patients. It is suggested that these genes may play a significant role in the development and progression of lung cancer. Next, we analyzed the prognostic value of differential genes in lung cancer and found that low expressions of MS4A2, MS4A4E, and MS4A15 were significantly associated with worse overall survival in lung cancer patients. These results imply that MS4A family members can be used as independent indicators of lung cancer prognosis.
Genetic alterations play an extremely significant role in tumorigenesis and progression, which are intricate and multifaceted. Since the MS4A family genes were considered to be significantly dissimilarly expressed in lung cancer, we then explored the factors that caused the differential expression in lung cancer. The results showed that MS4A family genes are frequently altered in lung cancer. Amplification and missense mutation was the most common alteration in the MS4A family.
Following this, we exploited GO and KEGG pathway enrichment analysis to focus on the function of the MS4A family. As predicted, we discovered that the functions of these genes are mainly related to immune-related pathways such as B cell receptor signaling pathway, Fc gamma R-mediated phagocytosis, Natural killer cell-mediated cytotoxicity. As the body has multiple immune surveillance mechanisms that exert their antitumor effects, including cellular and humoral immune mechanisms. Numerous studies have demonstrated there was a strong relationship between tumorigenesis and tumor immune escape (Liu and Cao Citation2016). From this, we hypothesized that reduced expression of the MS4A family in lung cancer was associated with tumor immune escape, leading to tumorigenesis.
There is growing evidence that complex interactions between tumors and immune cells influence tumor progression. Infiltration of immune cells can also serve as an important reference for immunotherapy and prognosis (Bindea et al. Citation2013). Bremnes indicates that tumor-infiltrating lymphocytes play a tumor-suppressive role in non-small cell lung cancer (Bremnes et al. Citation2016). Liu’s study showed that tumors with memory B cell deficiency or increased macrophage counts were related to a poor early prognosis in LUAD in the clinical course. In LUSC, T helper cells were associated with a good prognosis, whereas an increased number of neutrophils predicted a poor prognosis (Liu et al. Citation2017). In our current study, we discover that the expression of MS4A2, MS4A4A, MS4A6A, MS4A7, and MS4A14A was positively related with the infiltration of B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells. Among them, MS4A2 was most closely related to dendritic cells. MS4A4A, MS4A6A, and MS4A7 were significantly correlated with monocytes, neutrophils, and dendritic cells. All results indicated that the MS4A family could not only serve as a prognostic indicator but also reflect the tumor immune infiltration status.
Since our study was only a preliminary data analysis of the MS4A family, we did not analyze clinical specimens or in vivo and in vitro experiments. validation, thus our results have significant limitations, and their specific functions and mechanisms need to be further elucidated.
In conclusion, we systematically investigate the expression, functions, and prognostic of MS4A family of lung cancer, and provide a new insight between immune infiltration in lung cancer and the MS4A family, a rational prognostic marker for the clinic, and a new direction for immunotherapy.
Author contributions
Zheng Yang and Wuning Mo raised the concepts, design, definition of the study. Qisheng Su and Zunni Zhang contribute to literature search, data acquisition, and analysis. Jiameng Tang provided assistance for the above. Caize Cen is responsible for manuscript editing and modification. All the authors contribute to and approved the manuscript.
Data availability statement
The data that support the findings of this study are openly available in [Oncomine] at [https://www.oncomine.org/resource/login.html], reference number (Rhodes et al. Citation2004), [Gene Expression Profiling Interactive Analysis (GEPIA)] at [http://gepia.cancer-pku.cn/index.html], reference number (Tang et al. Citation2017), [The Human Protein Atlas (HPA)] at [https://www.proteinatlas.org/], reference number (Thul and Lindskog Citation2018), [cBioPortal] at [www.cbioportal.org], reference number (Gao et al. Citation2013), [STRING] at [https://string-db.org/], reference number (Szklarczyk et al. Citation2019), [DAVID] at [https://david.ncifcrf.gov], reference number (Huang et al. Citation2009), [Tumor Immune Estimation Resource(TIMER)] at [http://timer.cistrome.org], reference number (Li et al. Citation2017).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Allemani C, Matsuda T, Di Carlo V, Harewood R, Matz M, Nikšić M, Bonaventure A, Valkov M, Johnson CJ, Estève J, et al. 2018. Global surveillance of trends in cancer survival 2000-14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet. 391:1023–1075.
- Beer DG, Kardia SLR, Huang C-C, Giordano TJ, Levin AM, Misek DE, Lin L, Chen G, Gharib TG, Thomas DG, et al. 2002. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 8(8):816–824.
- Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, et al. 2001. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci U S A. 98(24):13790–13795.
- Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf A, Angell H, Fredriksen T, Lafontaine L, Berger A, et al. 2013. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 39(4):782–795.
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A, et al. 2018. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 68(6):394–424.
- Bremnes RM, Busund L-T, Kilvær TL, Andersen S, Richardsen E, Paulsen EE, Hald S, Khanehkenari MR, Cooper WA, Kao SC, Dønnem T. 2016. The role of tumor-infiltrating lymphocytes in development, progression, and prognosis of non-small cell lung cancer. J Thorac Oncol. 11(6):789–800.
- Chiba S, Ikushima H, Ueki H, Yanai H, Kimura Y, Hangai S, Nishio J, Negishi H, Tamura T, Saijo S, et al. 2014. Recognition of tumor cells by Dectin-1 orchestrates innate immune cells for anti-tumor responses. Elife. 3:e04177.
- Cruse G, Beaven MA, Ashmole I, Bradding P, Gilfillan AM, Metcalfe DD. 2013. A truncated splice-variant of the FcϵRIβ receptor subunit Is critical for microtubule formation and degranulation in mast cells. Immunity. 38(5):906–917.
- Donato J, Ko J, Kutok J, Cheng T, Shirakawa T, Mao X-Q, Beach D, Scadden DT, Sayegh MH, Adra CN. 2002. Human HTm4 is a hematopoietic cell cycle regulator. J Clin Invest. 109:51–58.
- Donnadieu E, Jouvin M-H, Rana S, Moffatt MF, Mockford EH, Cookson WO, Kinet J-P. 2003. Competing functions encoded in the allergy-associated FcεRIβ gene. Immunity. 18(5):665–674.
- Drew J, Farquharson A, Mayer C, Vase HF, Coates PJ, Steele RJ, Carey FA, Ashktorab H. 2014. Predictive gene signatures: molecular markers distinguishing colon adenomatous polyp and carcinoma. PloS one. 9:e113071.
- Gao J, Aksoy BA, Dogrusoz U, et al. 2013. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 6(269):pl1.
- Garber ME, Troyanskaya OG, Schluens K, Petersen S, Thaesler Z, Pacyna-Gengelbach M, van de Rijn M, Rosen GD, Perou CM, Whyte RI, et al. 2001. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci U S A. 98(24):13784–13789.
- Hirsch FR, Scagliotti GV, Mulshine JL, et al. 2017. Lung cancer: current therapies and new targeted treatments. The Lancet. 389(10066):299–311.
- Hou J, Aerts J, den Hamer B, et al. 2010. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One. 5(4):e10312–e10312.
- Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 4(1):44–57.
- Hulett MD, Pagler E, Hornby JR, Hogarth PM, Eyre HJ, Baker E, Crawford J, Sutherland GR, Ohms SJ, Parish CR. 2001. Isolation, tissue distribution, and chromosomal localization of a novel testis-specific human four-transmembrane gene related to CD20 and FcεRI-β. Biochem Biophys Res Commun. 280(1):374–379.
- Koslowski M, Sahin U, Dhaene K, Huber C, Türeci Ö. 2008. MS4A12 is a colon-selective store-operated calcium channel promoting malignant cell processes. Cancer Res. 68(9):3458–3466.
- Kutok JL, Yang X, Folkerth R, Adra CN. 2011. Characterization of the expression of HTm4 (MS4A3), a cell cycle regulator, in human peripheral blood cells and normal and malignant tissues. J Cell Mol Med. 15(1):86–93.
- Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, Li B, Liu XS. 2017. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 77(21):e108–e110.
- Liu X, Wu S, Yang Y, Zhao M, Zhu G, Hou Z. 2017. The prognostic landscape of tumor-infiltrating immune cell and immunomodulators in lung cancer. Biomed Pharmacother. 95:55–61.
- Liu Y, Cao X. 2016. Immunosuppressive cells in tumor immune escape and metastasis. J Mol Med. 94(5):509–522.
- Martiskainen H, Viswanathan J, Nykänen N-P, Kurki M, Helisalmi S, Natunen T, Sarajärvi T, Kurkinen KMA, Pursiheimo J-P, Rauramaa T, et al. 2015. Transcriptomics and mechanistic elucidation of Alzheimer’s disease risk genes in the brain and in vitro models. Neurobiol Aging. 36(2):1221.e15–1221.e28.
- Mattiola I, Tomay F, De Pizzol M, Silva-Gomes R, Savino B, Gulic T, Doni A, Lonardi S, Astrid Boutet M, Nerviani A, et al. 2019. The macrophage tetraspan MS4A4A enhances dectin-1-dependent NK cell-mediated resistance to metastasis. Nat Immunol. 20:1012–1022.
- Michel J, Schönhaar K, Schledzewski K, Gkaniatsou C, Sticht C, Kellert B, Lasitschka F, Géraud C, Goerdt S, Schmieder A. 2013. Identification of the novel differentiation marker MS4A8B and its murine homolog MS4A8A in colonic epithelial cells lost during neoplastic transformation in human colon. Cell Death Dis. 4(1):e469.
- Molgora M, Esaulova E, Vermi W, Hou J, Chen Y, Luo J, Brioschi S, Bugatti M, Omodei AS, Ricci B, et al. 2020. TREM2 modulation remodels the tumor myeloid landscape enhancing anti-PD-1 immunotherapy. Cell. 182:886–900.e17.
- Okami J, Shintani Y, Okumura M, Ito H, Ohtsuka T, Toyooka S, Mori T, Watanabe S-i, Date H, Yokoi K, et al. 2019. Demographics, safety and quality, and prognostic information in both the seventh and eighth editions of the TNM classification in 18,973 surgical cases of the Japanese joint committee of lung cancer registry database in 2010. J Thorac Oncol. 14(2):212–222.
- Okayama H, Kohno T, Ishii Y, Shimada Y, Shiraishi K, Iwakawa R, Furuta K, Tsuta K, Shibata T, Yamamoto S, et al. 2012. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res. 72(1):100–111.
- Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pander A, Chinnaiyan AM. 2004. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 6(1):1–6.
- Selamat SA, Chung BS, Girard L, Zhang W, Zhang Y, Campan M, Siegmund KD, Koss MN, Hagen JA, Lam WL, et al. 2012. Genome-scale analysis of DNA methylation in lung adenocarcinoma and integration with mRNA expression. Genome Res. 22(7):1197–1211.
- Stearman RS, Dwyer-Nield L, Zerbe L, Blaine SA, Chan Z, Bunn PA, Johnson GL, Hirsch FR, Merrick DT, Franklin WA, et al. 2005. Analysis of orthologous gene expression between human pulmonary Adenocarcinoma and a carcinogen-induced murine model. Am J Pathol. 167(6):1763–1775.
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. 2021. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 71(3):209–249.
- Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, et al. 2019. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47(Database issue):D607–D613.
- Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. 2017. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 45(Web Server issue):W98–W102.
- Thul PJ, Lindskog C. 2018. The human protein atlas: a spatial map of the human proteome. Protein Sci. 27(1):233–244.
- van de Ven AAJM, Compeer EB, Bloem AC, van de Corput L, van Gijn M, van Montfrans JM, Boes M. 2012. Defective calcium signaling and disrupted CD20–B-cell receptor dissociation in patients with common variable immunodeficiency disorders. J Allergy Clin Immunol. 129(3):755–761.e7.
- Vansteenkiste J, Crinò L, Dooms C, Douillard JY, Faivre-Finn C, Lim E, Rocco G, Senan S, Van Schil P, Veronesi G, et al. 2014. 2nd ESMO consensus conference on lung cancer: early-stage non-small-cell lung cancer consensus on diagnosis, treatment and follow-up. Ann Oncol. 25(8):1462–1474.
- Ye L, Yao X-D, Wan F-N, Qu Y-Y, Liu Z-Y, Shen X-X, Li S, Liu X-J, Yue F, Wang N, et al. 2014. MS4A8B promotes cell proliferation in prostate cancer. Prostate. 74(9):911–922.
- Zuccolo J, Deng L, Unruh T, Sanyal R, Bau JA, Storek J, Demetrick DJ, Luider JM, Auer-Grzesiak IA, Mansoor A, Deans JP. 2013. Expression of MS4A and TMEM176 genes in human B lymphocytes. Front Immunol. 4:195–195.