Abstract
Nosocomial infections are important causes of morbidity and mortality. There are inconsistent results among the antimicrobial resistance patterns of nosocomial infections. This study aimed to investigate the antimicrobial resistance patterns of bacteria isolated from the bloodstream and wound nosocomial infections among the inpatients in Hamadan, 2019–2020. In this study, the concurrent surveillance for nosocomial bloodstream and wound infections at three hospitals over 2 years detected and identified the nosocomial pathogens; then, they were surveyed antibiotic resistance. For this reason, we used API and disk diffusion methods. 2583 patients were enrolled in which 311 positive blood samples, 148 wound samples were isolated. In wound infections, the highest resistance rate of isolated bacteria to a wide range from the antibiotics was related to A. baumannii, P. aeruginosa, and E. coli, respectively. In blood infections, the highest resistance rate of isolated bacteria was related to A. baumannii, S. saprophyticus, and P. aeruginosa, respectively. These data could be helpful for the clinicians to decide better about target empirical therapy for the hospital-acquired cases of bacteremia and wound infections; however, it was concerned the isolates should properly monitor for a choice of antibiotics to be used as prophylaxis, and empiric treatment.
Introduction
In recent years, the resistant microorganisms against common antibiotics were a vast distribution worldwide. Nowadays, several reports indicate the rapid distribution of antibiotic resistance in the different parts of hospitals (Russotto et al. Citation2015), as well as in different regions of globe because of the inappropriate usage of antibiotics. On the other hand, the specific condition of patients, long-term hospitalization, immunodeficiency, immunosuppressive drugs, and rapid and invasive approaches, including urinary catheters, intravascular catheters, and tracheal tubes, is considered other causes of antibiotic resistance in various areas (Giannella et al. Citation2019). The four major categories which comprise the majority of healthcare-associated infections (HAIs) also called as Nosocomial infection are surgical-site infection (SSI), central line-associated bloodstream infection, catheter-associated urinary tract infection, and ventilator-associated pneumonia (Ranji et al. Citation2007). While about 15–30% of HAI in developing countries is related to blood infections, and it can increase the mortality rate in hospitals, so they are considered an important hospital infection not only in developing countries but also in the developed ones (Wenzel and Edmond Citation2001). However, SSIs are considered major problems in post-surgery and long-term hospitalized patients (Beig et al. Citation2021; Goodarzi et al. Citation2021). The emergence of resistant strains in diverse hospitals is caused by various health conditions and hospital infection management patterns. The emergence and spread of MDR infections have raised severe concerns about healthcare systems’ ability to treat drug-resistant illnesses.
There are inconsistent results among antimicrobial resistance patterns of nosocomial infections (Mahfouz et al. Citation2010). The epidemiology of emerging vancomycin-resistant isolates appeared like a complex mix of admitted cases and transmissions in small clusters, challenging infection control measures, but a decrease of methicillin resistance in S. aureus in nosocomial infections was reported in Germany. We investigated the antibacterial resistance patterns and the prevalence of bacteria isolated from different infectious sites in hospitalized patients in Hamadan, 2019–2020.
Materials and methods
Samples collection
This work was conducted as a descriptive cross-sectional study. To begin, all subjects signed a consent form, and all procedures were carried out in compliance with Hamadan’s ethics committee guidelines (IR.UMSHA.REC.1399.631). During this research, blood and wound samples, including bedsores, fistulae, blisters, and foot wounds, were isolated from patients admitted to the tertiary hospitals from 2019 to 2020 according to the centers for Disease Control and Prevention (CDC) standard criteria (Garner et al. Citation1988). Inclusion criteria, including people who had not used antibiotics in the past two weeks which was hospitalized for more than 48 h. The collected isolates were transferred to the microbiological laboratory for further confirmation. Two swabs were prepared for each wound sample, one for a smear test and another for culture. Ten-milliliter blood was prepared in Castaneda bottle for culturing, and it is repeated three times for each patient. The questionnaire forms, including personal information of each patient, such as sex, age, history of antibiotic consumption, sign and symptom, and the patient admission part, were prepared. Antibiotic consumption was considered an exclusion criterion.
Identification of isolated bacteria
After sampling, the wound samples were inoculated in TSB (Tryptic soy broth). A certain amount of this suspension was inoculated in differentiated and specialized media; then, it was incubated for 18 h at 37°C. Finally, the bacterial isolates were identified according to gram staining, morphological characteristics of the media cultures, and confirmed using an enzyme assay and standard biochemical tests. For this section, we used API method for differentiation.
Antimicrobial resistance patterns
Resistance pattern for each bacterial isolates against usual antibiotics, according to CLSI 2019 instructions, was carried out on Mueller Hinton agar using the disk diffusion method. Bacterial isolates were classified as sensitive, intermediate, or resistant according to antibiotic resistance patterns. Antibiotic disks (Mast Diagnostics, United Kingdom) which were used: CP, ciprofloxacin; CAZ, ceftazidime; FEP, cefepime; AN, amikacin; SAM, ampicillin/sulbactam; IMI, imipenem; CFM, cefixime; SXT, trimethoprim-sulfamethoxazole; FM, nitrofurantoin; CRO, ceftriaxone; LEV, levofloxacin; CZ, cefazolin; MEN, meropenem; PTZ, piperacillin/tazobactam; OFX; ofloxacin; PG, penicillin; E, erythromycin; FOX, cefoxitin; CL, clindamycin; VAN, vancomycin; AZM, azithromycin; NOR, norfloxacin; LZD, linezolid; NB, novobiocin; CTX, cefotaxime. In a 150-mm plate, including Mueller-Hinton agar, 10 disks were placed with a 24 mm gap between each of them. The plates were incubated at 37°C for 18 h, and inhibition zone diameters were measured to the millimeter.
Statistical analysis
The isolates’ frequency and antibiotic resistance rate were analyzed using SPSS (version 22) with a 0.05 significant level.
Results
Patients
In total, 2583 patients were enrolled during 2019–2020. Among them, 53% were female, and 47% were male in all of age range and there was no significant difference between males and females (p > 0.05). 1710 isolates were collected from Sina Hospital, 323 from Shahid Beheshti Hospital, and 550 from Be'sat Hospital. 311 positive blood samples, 148 wound samples, and other wound-related samples such as 12 bedsores, 2 fistulae, 4 blisters, and 3 wound samples were collected according to the CDC. Demographic findings of patient included in this study were presented in Table .
Table 1. Demographic findings of patient included in this study.
Bacterial isolation
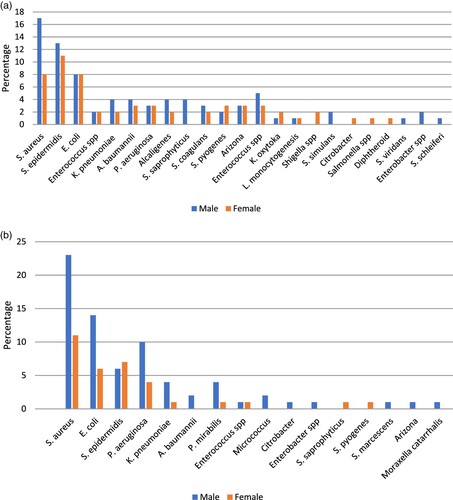
During this research, in blood specimen, we have distinguished S. aureus, S. epidermidis, E. coli, Enterobacter species, K. pneumoniae , A. baumannii, P. aeruginosa, Alcaligenes, S. saprophyticus, S. coagulans, S. pyogenes, Arizona, Enterococcus species, K. oxytoka, L. monocytogenesis, Shigella species, S. simulans, Citrobacter, Salmonella species , diphtheroid, and S. viridans concerning their frequency (Figure (a)). Moreover, in wound specimen, we have distinguished S. aureus, E. coli, S. epidermidis, P. aeruginosa, K. pneumoniae , A. baumannii, P. mirabilis, Enterococcus species, Micrococcus, Citrobacter, Enterobacter species, S. saprophyticus, S. pyogenes, S. marcescens, Arizona, Moraxella catarrhalis and, S. agalactiae concerning their frequency (Figure (b)). Figure (a,b) describes the prevalence of blood and wound isolates among men and women, respectively.
Figure 1. (a) Frequency of bacterial isolates of blood samples (b) Frequency of bacterial isolates of wound samples.
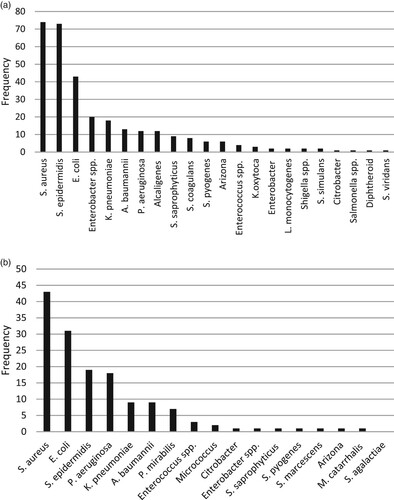
Figure 2. (a) Frequency of bacterial isolates of blood samples among patients (b) Frequency of bacterial isolates of wound samples among patients.
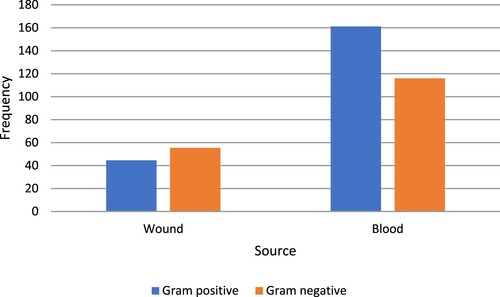
Our data showed that 24% of the blood isolates were S. aureus (Figure (a)) while, in the case of total wound isolates, 29% were S. aureus, and 21% were S. epidermidis (Figure (b)). In our study, the number of gram-positive bacteria in blood infections was higher than that of gram-negative bacteria, although the number of gram-negative bacteria in wound infections was higher (Figure ) and there was a significant difference between gram-positive and gram-negative bacteria obtained from blood samples (p < 0.05).
Antibiotic susceptibility assay
In total, in blood specimens, Klebsiella isolates from blood samples showed higher resistances rate (p < 0.05) proportions (94.4%) than Klebsiella isolates from wound infections (33.3%). We distinguished S. aureus isolates were more resistant to penicillin (98.6%), S. epidermidis resistance to norfloxacin (98.6%), E. coli resistance to cefotaxime (74.4%), Acinetobacter resistance to cefepime (100%), S. saprophyticus resistance to cefoxitin (100%), Enterobacter resistance to clindamycin (85%), P. aeruginosa resistance to imipenem (100%) isolated from blood specimens (Table ). Furthermore, in wound specimen, we have distinguished S. aureus was more resistant to ciprofloxacin (97.6%), E. coli resistance to ciprofloxacin (93.5%), Pseudomonas resistance to cefotaxime (100%), S. epidermidis resistance to oxacillin (94.7%), Acinetobacter resistance to ciprofloxacin, cefepime, ampicillin-sulbactam, imipenem, ceftriaxone and piperacillin + tazobactam (100%), Klebsiella resistance to piperacillin + tazobactam (77.7%). Beta-lactamase inhibitors can be used where other antibiotics are not used. The most resistance rate of piperacillin/tazobactam in blood samples is in order of A. baumannii (100%), S. aureus (95.9%%), E. coli (65.1%), P. aeruginosa (58.3%), and K. pneumonia (55.5%) and in wound samples is in order of A. baumannii (100%), E. coli (83.8%), K. pneumoniae (77.7%), S. epidermidis (73.6%), S. aureus (51.1%) were observed, according to their frequency. Tables and describe the antibacterial resistance patterns of different bacteria isolated from blood and wound specimens.
Table 2. The highest antibiotic resistance among bacteria isolated from blood samples.
Table 3. The highest antibiotic resistance among bacteria isolated from wound samples.
Discussion
Nowadays, nosocomial infections are very important, especially within surgical ICUs (Rosenthal et al. Citation2016). Antibiotic resistance has challenged ICU infection management strategies that focus on preventing and controlling nosocomial infections (Shokoohizadeh et al. Citation2019; Nouri et al. Citation2020). In developing countries, HAIs and antimicrobial use burden is very heavy on the government (Ling et al. Citation2015). The burden of HAIs has traditionally been measured using clinical and economic outcomes. The proper knowledge of antimicrobial resistance pattern in a region and the correct and effective treatment of the infections could affect the infection management strategy. Our study showed a high prevalence of nosocomial infections in the hospitals and a high rate of antimicrobial resistance among the causative pathogens isolated from the blood and wound infections complicate antibiotic treatment and its outcomes. Among the bacteria isolated from the studied hospitals, the highest resistance rate was related to A. baumannii and P. aeruginosa. In a study, in 2015, Kara et al. investigated bacterial bloodstream infections in patients with hematologic malignancies in Turkey (Kara et al. Citation2015). It was reported that gram-negative bacteria predominated (52.6%), with E. coli (17.3%) and Klebsiella species (11.0%) as the most frequent organisms, as in our study, we resulted that E. coli, Enterobacter, Klebsiella, as the most frequent gram-negative organisms, and in Kara study coagulase-negative staphylococci (10.4%) and corynebacterium species (6.3%), the most common gram-positive bacteria (35.8%), as in our study that we resulted in S. aureus, S. epidermidis as the most frequent gram-positive ones. It is be noted that S. epidermidis also often represents contamination in the sampling process and may not always be the causative infectious agent. In Kara study, the rate of extended-spectrum beta-lactamase (ESBL) production was 45% for E. coli and 58% for Klebsiella species. The quinolone resistance rate was 58% and 11% for E. coli and Klebsiella species, respectively. The overall frequency of ceftazidime resistance rate in MDR P. aeruginosa and Acinetobacter species were 28% and 87%, respectively (Kara et al. Citation2015).
Our study reported the resistance rate for levofloxacin in A. baumannii, S. saprophyticus, and E. coli bacteria as 100%, 88.8%, and 67.4%, respectively. Besides, in our previous study (Shokoohizadeh et al. Citation2019), the ciprofloxacin resistance rate of K. pneumoniae was reported as 38%, which in the present study was 50% in blood, and 33.3% in wound samples. It suggests that K. pneumoniae is becoming more resistant to fluoroquinolones, even though the bacteria are isolated from different sources. Despite the information mentioned, Trecarichi et al. investigated the bacterial bloodstream infections in the patients with hematologic malignancies in Italy and concluded the shift in the prevalence from gram-positive to gram-negative bacteria as causative agents of bloodstream infections which confirmed with our findings in the current study that showed a significant relationship between gram-positive and gram-negative bacteria isolated from blood samples (p < 0.05). Among 668 bacterial isolates from patients, 575 episodes were recorded. The susceptibility rates of gram-negative bacteria were 59.1% to ceftazidime, 20.1% to ciprofloxacin, 79.1% to meropenem, 85.2% to amikacin, 69.2% to gentamicin, and 69.8% to piperacillin/tazobactam. Among K. pneumoniae strains, 34.9% were resistant to carbapenems that are in contrast with the resistance observed to carbapenems (94.4%) in the current research that may be due to inappropriate administration or overuse of antibiotics (Trecarichi et al. Citation2015). Furthermore, Mohammad et al. (Citation2017) investigated bacterial isolates and their antimicrobial susceptibility in Ethiopian inpatients, and outpatients with wound infections. Among 115 bacterial isolates, 57% were gram-negative and 43% were gram-positive as S. aureus was the most predominant isolate 34% and then Klebsiella species (13%), coagulase-negative staphylococci species (12%), and P. aeruginosa. While in our study, S. aureus, E. coli, P. aeruginosa, S. epidermidis, Acinetobacter, and Klebsiella, were the most frequent, respectively (Mohammed et al. Citation2017). In our study, E. coli is in the second-order (20%) of frequency, demonstrating the importance of E. coli presence at many infection sites throughout the body. Besides, in our previous study on nosocomial infections obtained from the hospital, it was shown that in urinary tract infections and tracheal infections, the frequency of E. coli isolates was most higher than in other isolates (Nouri et al. Citation2020) which indicates the importance of this bacterium in causing various nosocomial infections.
In 2013, Bahador et al. in a study on A. baumannii showed high antibiotic resistance to ceftazidime, cefepime, and ceftriaxone which approximately indicates a resistance rate of 100% (Bahador et al. Citation2013). In our previous study on A. baumannii and K. pneumoniae, it was found that the highest frequency was related to the tracheal tube and throat cultures which also 95% of the isolates were MDR (Hazhirkamal et al. Citation2021; Karimi et al. Citation2021). Moreover, in 2020, Haghighifar et al. in a study on A. baumannii showed high antibiotic resistance to ceftazidime (95.6%), ciprofloxacin (92.8%) and, ceftriaxone (88.6%) that two studies are similar to this study's findings which approximately indicates a resistance rate of 95%. Besides, the patterns of antibiotic resistance to imipenem and amikacin in K. pneumoniae were similar to our study (Haghighifar and Kamali Dolatabadi Citation2020). The pattern of antibiotic resistance to E. coli in our study is higher than in Haghighifar study which may be due to differences in the site of bacterial isolation. Meanwhile, in Ethiopia study, gram-positive isolates were resistant to ampicillin (86.4%), amoxicillin (83%), penicillin (81.3%), oxacillin (74.6%), and tetracycline (59.4%), while gram-negative isolates were resistant to amoxicillin (97.4%), ampicillin (94.8%), tetracycline (72.7%), trimethoprim/sulfamethoxazole (66%), and chloramphenicol (54.5%). In both studies, S. aureus is the top leader among wound infections (Mohammed et al. Citation2017), similar to our results.
Piperacillin/tazobactam (PTZ) is a beta-lactamase inhibitor used for a wide range of gram-negative and gram-positive bacterial infections. The resistance rate of piperacillin/tazobactam in gram-negative and positive isolates was so much, as mentioned in the results. Hence, Antibiotic resistance and the prevalence of broad-spectrum beta-lactamase and metallobeta-lactamase in gram-negative strains in the world, especially in Iran, are of concern, and the need for infection control measures including antibiotic management and rapid identification of isolates produces beta-lactamase. A timely and correct diagnosis can assist physicians in prescribing antibiotics and avoiding expensive costs and long-term hospitalization. Bacteria-causing illnesses frequently indicate widespread antibiotic resistance in many regions of hospital, resulting in death and morbidity, particularly in ICUs. In addition to the direct impact on patients, the financial burden is remarkable. Increasing mortality and morbidity rates as a result of these infections, as well as therapeutic expenses as a result of long-term hospitalization, are two major reasons for antibiotic treatment failure, and they are a major source of concern in the medical community. Hence, finding and applying proper therapeutic approaches to control these infections and diminishing these microorganisms and antibiotic resistance is necessary in each community. Study of antibiotic resistance in larger sample sizes in other parts of country and identification of virulence genes can help our knowledge to use and prescribe antibiotics correctly which are among the limitations of this study. Finally, the following constructive comments are suggested, a continuous study about common type microorganisms isolated from the bloodstream and wound infections and their antimicrobial susceptibility for each geographical region due to empirical therapy.
Scientists are exploring novel antimicrobial components to eradicate and cure drug-resistant disease due to the emergence and spread of resistant infections worldwide. In conclusion, the high occurrence of antibiotic resistance in nosocomial infections in our hospital highlights the significance of better antibiotic use and infection control and management. These data could be helpful for clinicians to decide better about target empirical therapy for hospital-acquired cases of bacteremia and wound infections; however, it was concerned the isolates should properly monitor for the choice of antibiotics to be used as prophylaxis, and empiric treatment in the study area.
Ethical approval
This study was approved by the Ethics Committee of Hamadan University of Medical Sciences, Hamadan, Iran (Ethical approval No. IR.UMSHA.REC.1399.631).
Acknowledgements
We are thankful for the cooperation of the laboratory of the Be'sat, Beheshti, and Sina hospitals. F. N, M. T, and M. H designed the study. F. K and M. T contributed to the experimental studies and drafted the work. F. N, B. A and M. H contributed to the sample collections. All authors read and approved the final manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Due to the nature of this research, participants of this study did not agree for their data to be shared publicly, so supporting data is not available.
Additional information
Funding
References
- Bahador A, Bazargani A, Taheri M, Hashemizadeh Z, Khaledi A, Rostami H, et al. 2013. Clonal lineages and virulence factors among Acinetobacter baumannii isolated from Southwest of Iran. J Pure Appl Micribiol. 7:1559–1566.
- Beig M, Taheri M, Arabestani MR, Chikere CB. 2021. Comparison of different phenotypic tests versus PCR in the detection of carbapenemase-producing Pseudomonas aeruginosa isolates in Hamadan, Iran. Int J Microbiol. 2021:1–8.
- Garner JS, Jarvis WR, Emori TG, Horan TC, Hughes JM. 1988. CDC definitions for nosocomial infections, 1988. Am J Infect Control. 16(3):128–140.
- Giannella M, Pascale R, Gutiérrez-Gutiérrez B, Cano A, Viale P. 2019. The use of predictive scores in the management of patients with carbapenem-resistant Klebsiella pneumoniae infection. Expert Rev Anti-Infect Ther. 17(4):265–273.
- Goodarzi R, Farahani F, Roshani M, Taheri M, Asghari B. 2021. Comparative study of in vitro activities of polymyxin B commercial products on Pseudomonas aeruginosa isolated from hospitalized patients. Ars Pharmaceutica. 62(3):270–279.
- Haghighifar E, Kamali Dolatabadi R. 2020. Bacterial infections and antimicrobial resistance patterns of burn wound infections: a one year study from Burn Hospital, Isfahan, Iran. J Adv Med Biomed Res. 28(128):144–150.
- Hazhirkamal M, Zarei O, Movahedi M, Karami P, Shokoohizadeh L, Taheri M. 2021. Molecular typing, biofilm production, and detection of carbapenemase genes in multidrug-resistant Acinetobacter baumannii isolated from different infection sites using ERIC-PCR in Hamadan, west of Iran. BMC Pharmacol Toxicol. 22(1):1–7.
- Kara Ö, Zarakolu P, Aşçioğlu S, Etgül S, Uz B, Büyükaşik Y, Akova M. 2015. Epidemiology and emerging resistance in bacterial bloodstream infections in patients with hematologic malignancies. Infect Dis. 47(10):686–693.
- Karimi K, Zarei O, Sedighi P, Taheri M, Doosti-Irani A, Shokoohizadeh L, Chaves Lopez C. 2021. Investigation of antibiotic resistance and biofilm formation in clinical isolates of Klebsiella pneumoniae. Int J Microbiol. 2021:1–6.
- Ling ML, Apisarnthanarak A, Madriaga G. 2015. The burden of healthcare-associated infections in Southeast Asia: a systematic literature review and meta-analysis. Clin Infect Dis. 60(11):1690–1699.
- Mahfouz A, Al Azraqi T, Abbag F, Al Gamal M, Seef S, Bello C. 2010. Nosocomial infections in a neonatal intensive care unit in south-western Saudi Arabia.
- Mohammed A, Seid ME, Gebrecherkos T, Tiruneh M, Moges F. 2017. Bacterial isolates and their antimicrobial susceptibility patterns of wound infections among inpatients and outpatients attending the University of Gondar Referral Hospital, Northwest Ethiopia. Int J Microbiol. 2017:1–10.
- Nouri F, Karami P, Zarei O, Kosari F, Alikhani MY, Zandkarimi E, et al. 2020. Prevalence of common nosocomial infections and evaluation of antibiotic resistance patterns in patients with secondary infections in Hamadan, Iran. Infect Drug Resist. 13:2365–2374.
- Ranji SR, Shetty K, Posley KA, Lewis R, Sundaram V, Galvin CM, et al. 2007. Closing the quality gap: a critical analysis of quality improvement strategies. Vol. 6. Prevention of Healthcare–Associated Infections.
- Rosenthal VD, Al-Abdely HM, El-Kholy AA, AlKhawaja SAA, Leblebicioglu H, Mehta Y, Rai V, Hung NV, Kanj SS, Salama MF, et al. 2016. International nosocomial infection control consortium report, data summary of 50 countries for 2010–2015: device-associated module. Am J Infect Control. 44(12):1495–1504.
- Russotto V, Cortegiani A, Graziano G, Saporito L, Raineri SM, Mammina C, et al. 2015. Bloodstream infections in intensive care unit patients: distribution and antibiotic resistance of bacteria. Infect Drug Resist. 8:287.
- Shokoohizadeh L, Saniee M, Mirzaee M, Taheri M. 2019. Mutations in gyrA and parC genes in quinolone-resistant klebsiella pneumoniae isolates from borujerd hospitals. J Adv Med Biomed Res. 27(120):1–7.
- Trecarichi E, Pagano L, Candoni A, Pastore D, Cattaneo C, Fanci R, Nosari A, Caira M, Spadea A, Busca A, et al. 2015. Current epidemiology and antimicrobial resistance data for bacterial bloodstream infections in patients with hematologic malignancies: an Italian multicentre prospective survey. Clin Microbiol Infect. 21(4):337–343.
- Wenzel RP, Edmond MB. 2001. The impact of hospital-acquired bloodstream infections. Emerg Infect Dis. 7(2):174–177.