Abstract
Nerve growth factor (NGF) plays a key role in controlling growth, maintenance and survival of neurons as well as nociceptor development. Many of its critical actions are believed to be mediated by activation of corresponding transcription factors. Here I attempt to reveal signaling cascades that relay NGF stimulation to NFAT, a family of Ca2+/calcineurin-dependent transcription factors that are implicated in playing a pivotal role in neuronal development and survival, and pain sensation. With a luciferase gene reporter assay, it was confirmed that NGF can trigger NFAT-dependent gene expression in PC12 cells. Subsequent research with pharmacological tools uncovers that this effect was mediated by the TrkA-MEK1/2 pathway. Furthermore, it was determined that NFATc3 is the predominant isoform by calculating an absolute copy number of each NFAT isoform with quantitative PCR. Taken together, it is proposed that elucidating cellular connections linking NGF and NFAT may help reveal novel therapeutic targets, which can be exploited to devise innovative remedies for the treatment of NGF and NFAT-associated clinical disorders such as anomalous neuronal development and hyperalgesia.
Introduction
Ever since its discovery in the 1950s, nerve growth factor (NGF) has been closely studied and universally identified as a pivotal player not only in controlling growth, maintenance, and survival of neurons but also in developing nociceptive primary neurons via its binding to high affinity TrkA and/or low affinity p75 receptors (Sofroniew et al. Citation2001; Aloe Citation2011). Among the consequences of the vast intracellular signaling triggered by NGF is the structural remodeling of neurons such as neurite sprouting, axon outgrowth, and synapse formation (Sharma et al. Citation2010; Dravid et al. Citation2020). From an anatomical point of view, those structural changes often require expression of proteins mediated by the recruitment, assembly and activation of transcriptional machinery.
Of many transcription factors potentially regulated by NGF, I examine the relationship between NGF and NFAT (nuclear factor of activated T-cells). Although NFAT was originally characterized as an essential factor for T-cell activation, a growing body of evidence strongly suggests that it plays critical roles in the nervous system as well (e.g. neuronal development and survival, axonal outgrowth, pain sensation) (Shaw et al. Citation1988; Graef et al. Citation2003; Groth et al. Citation2007; Jung and Miller Citation2008; Serrano-Perez et al. Citation2015; Huang et al. Citation2018). Interestingly, many of NFAT’s biological functions – particularly in the nervous system – are overlapped by those of NGF, implying that NFAT may be a vital mediator for relaying NGF signals. In support of this hypothesis, recent publications have provided evidence demonstrating that NFAT is activated by NGF under various experimental conditions (Groth et al. Citation2007; Stefos et al. Citation2013; Kim et al. Citation2014).
However, mechanisms by which NGF stimulates NFAT have not yet been fully elucidated. Considering their crucial roles in neurobiology, it is proposed that revealing the cellular components that relay signals initiated by NGF to NFAT for gene expression would not only uncover mechanisms behind the aforementioned neuronal functions but also lead to the development of novel agents that interrupt NGF signals in the treatment of clinically relevant disorders such as hyperalgesia. Therefore, the study described here was conducted to investigate how the cellular signaling triggered by NGF leads to NFAT-dependent gene expression in neurons using PC12 cells as a model system.
Materials and methods
PC12 cell culture
PC12 cells obtained from Korean Cell Line Bank (KCLB Cat# 21721) were plated on 100 ϕ dishes pre-coated with collagen (Collagen, Type 1 solution®, Sigma, USA, Cat#c3867, 1/100 diluted with 30% ethanol), and grown in RPMI1640 medium supplemented with 10% heat inactivated horse serum, 5% heat inactivated fetal bovine serum, and penicillin (100 units/ml) / streptomycin (100 µg/ml) (hereafter refer to as RPMI1640). Cells were grown to around 80% confluency, then cultures were split with trypsinization.
Transfection and NFAT reporter assay
NFAT-luciferase reporter (Clontech, Cat# K2052-1) and pGL4.74[hRluc/TK] (Promega, Cat#E6921) plasmids were transfected into PC12 cells using Amaxa NucleofectorTM II (Lonza, Germany). According to the manufacturer’s protocol, approximately 2 × 106 PC12 cells were harvested for a single nucleofection, then resuspended with 100 µl Cell Line Nucleofector® Solution V (Lonza, Germany), which was mixed with NFAT-luciferase reporter (0.9 µg, firefly luciferase) and pGL4.74[hRluc/TK] (1 µg, Renilla luciferase) plasmids. Cuvettes containing the reactant were inserted into the Nucleofector and electroporation was executed using Nucleofector® Program U-029. After that, 500 µl of RPMI1640 medium was immediately added to it, and the cells were gently transferred into a collagen precoated 96-well white cell culture plate (SPL, Korea) with an approximate density of 5 × 104 cells/well. The next day (around noon, approximately 20 h after transfection), cells were stimulated with NGF with or without inhibitors according to the protocols described in the Results section of this manuscript. Cells were pretreated with an inhibitor for 30 min before stimulation by replacing the original media with the one containing the appropriate inhibitor. To quantify NFAT transcriptional activity, the Dual-Glo Luciferase® Assay (Promega, USA) was performed according to the manufacturer’s protocol. Panomics-L100 luminometer (Affymetrix, USA) was used to detect luminescent signals from firefly and Renilla luciferases.
Measurement of NFAT isoforms
cDNA was synthesized from total RNA isolated from PC12 cells according to the manufacturer’s protocols (QuantiTect® Reverse Transcription Kit for cDNA synthesis, RNeasy Mini Kit for total RNA isolation, Qiagen, USA). SYBR green-based qPCR was performed with the cDNA from PC12 cells together with each NFAT plasmid DNA of known concentration (GFP-NFATc1, Addgene plasmid #11101; EGFP-NFATc2, provided by Dr. Rao; EGFP-NFATc3, originally provided by Dr. Iino, then modified by Dr. Usachev; EGFP-NFATc4, Addgene plasmid #10961). Standard curves for each NFAT isoform were constructed with serially diluted NFAT isoform. By relating cycle threshold (Ct) values of each NFAT plasmid with those of cDNA from PC12 cells, the absolute copy number of each NFAT isoform present in PC12 cells was calculated (Kim and Usachev Citation2009). The following NFAT primers were used for qPCR. NFATc1, F, 5′-CAT-CAA-CGC-CCT-GAC-CAC-3′, R, 5′-GTG-GTG-CCC-AGG-TCT-TCC-3′, NFATc2, F, 5′-TCT-GCT-GTT-CTC-ATG-GAT-GC-3′, R, 5′-TCA-GGA-CTG-GTC-TTC-CAT-ATC-3′, NFATc3, F, 5′-GAT-CAA-GCT-GCC-ATA-CTA-CCA-3′, R, 5′-CTG-AGA-TCC-AAG-GCC-ATC-ATC-3′, NFATc4, F, 5′-GAG-CAG-CTG-GAG-CTG-AGG-3′, R, 5′-TGT-AGC-CTA-GGA-GCT-TGA- C-3′. The qPCR program with Rotor-Gene Q Thermocycler (Qiagen, USA) was 95°C for 10 min for initial activation, then 95°C for 10 s, 57°C for 15 s, and 72°C for 15 s for 40 cycles. Finally, melting curve analysis was performed to determine the specificity of the amplicons.
Reagents
NGF (Cat# PMP04Z, AbD Serotec, USA), K252a (Cat#1683, Tocris), FK506 (Cat#3631, Tocris), Cyclosporin A (Cat#1101, Tocris), U73122 (Cat#1268, Tocris), LY294002 (Cat#440202, Calbiochem, USA), Wortmannin (Cat#681675, Calbiochem, USA), U0126 (Cat#1144, Tocris), RPMI1640 (Cat#22400, ThermoFisher, USA), KCl (Cat#P5405-500G, Sigma, USA), BayK8644 (Cat#1544, Tocris, USA), PMA(Cat#P1585, Sigma, USA), Horse serum (Cat#26-050-088, Gibco, USA), Fetal bovine serum (Cat#F0900-050, GenDEPOT, USA), and qPCR kit (Cat#RT501M, Enzynomics, Korea) were purchased. NFAT primers was synthesized from Biomedic (Korea).
Statistics
Student’s t-test for comparing two groups and One-Way ANOVA for comparing three or more groups were used to test for statistical significance. For ANOVA Bonferroni post-hoc test was employed since it is considered strict and conservative (Lee and Lee Citation2020).
Results
NGF increases NFAT-dependent gene expression
To test if NGF can activate NFAT in PC12 cells, a luciferase-based NFAT gene reporter assay was performed. A typical co-stimulation such as PMA (Phorbol 12-myristate 13-acetate, a PKC activator) and Ca2+ mobilization induces the binding of endogenous NFAT transcription factors to the enhance element with a help of loosely defined nuclear factors often referred to as NFATn, which initiates NFAT-dependent gene expression (Rao et al. Citation1997). Therefore, to validate the feasibility of the whole luciferase system, PC12 cells transfected with NFAT-luciferase reporter (firefly luciferase) and pGL4.74[hRluc/TK] (Renilla luciferase) plasmids were co-stimulated with PMA and 50 mM KCl for inducing Ca2+ influx by depolarization in the presence of 1 µM BayK8644, an L-type Ca2+ channel agonist to stabilize intracellular Ca2+ at the elevated levels, which resulted in a significant increase in a luciferase activity (Figure (B)). Next, the cells were stimulated with various concentrations of NGF (from 0.2 to 100 ng/ml) for 5 h. (Figure (A)) Although low levels of NGF (0.2 and 1 ng/ml) did not affect NFAT activity, high levels (5, 20, and 100 ng/ml) significantly activated NFAT (Figure (C)). Since it appeared that NFAT activation by NGF reached the plateau with 20 ng/ml NGF, this NGF concentration was adapted for further optimization. Next, to determine an optimal duration of NGF stimulation, cells were stimulated with or without 20 ng/ml NGF for 5, 10, or 24 h, followed by NFAT activity being measured at each time point. (Figure (A,D)) When the fold increase in NFAT activity was calculated, differences in NGF incubation time (5, 10 and 24 h) did not result in significant changes in NFAT-dependent gene expression. (Figure (D)) Therefore, it was decided to employ 20 ng/ml, 5 h NGF stimulation protocol for the remaining experiments.
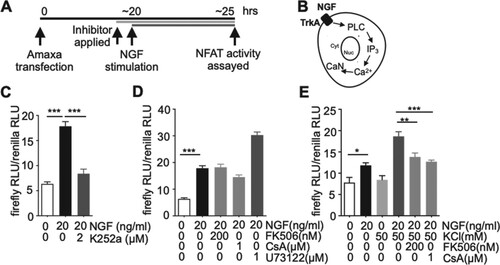
Figure 1. NGF activates NFAT-dependent gene expression in PC12 cells. A. Schematic diagram of depicting the experimental protocols. B. According to the protocol described in (A), PC12 cells transfected together with NFAT-luc (NFAT reporter, firefly luciferase) and pGL4.74[hRluc/TK] (constitutively active Renilla luciferase reporter) were co-stimulated with 20 ng/ml PMA (Phorbol 12-myristate 13-acetate, a PKC activator) and 50 mM KCl in the presence of 1 µM of BayK8644 for 5 h. After that, NFAT-dependent gene expression was determined by calculating ratios of firefly to Renilla luciferase activity. n = 7–8, **p < 0.01, Student’s t-test. C. Following the same protocol described in (B), the cells were stimulated by NGF (concentration ranged from 0 ng/ml to 100 ng/ml) for 5 h. NFAT-dependent gene expression was then determined by calculating ratios of firefly to Renilla luciferase activity. n = 6–12, ***p < 0.001 (vs 0 ng/ml NGF), One-Way ANOVA, Bonferroni post-hoc test. D. Either 0 ng/ml ((-)NGF) or 20 ng/ml NGF ((+)NGF) was applied for 5, 10, and 24 h to active PC12 cells transfected with the two luciferase reporters. NFAT-dependent gene expression was then assayed as described in (A). The ratio of NFAT-dependent gene expression in the presence of 20 ng/ml NGF to NFAT-dependent gene expression in the absence of NGF was calculated from (D), which was shown in the inset. n = 6–12. n stands for the number of well. Data are presented as mean ± S.E.M.
![Figure 1. NGF activates NFAT-dependent gene expression in PC12 cells. A. Schematic diagram of depicting the experimental protocols. B. According to the protocol described in (A), PC12 cells transfected together with NFAT-luc (NFAT reporter, firefly luciferase) and pGL4.74[hRluc/TK] (constitutively active Renilla luciferase reporter) were co-stimulated with 20 ng/ml PMA (Phorbol 12-myristate 13-acetate, a PKC activator) and 50 mM KCl in the presence of 1 µM of BayK8644 for 5 h. After that, NFAT-dependent gene expression was determined by calculating ratios of firefly to Renilla luciferase activity. n = 7–8, **p < 0.01, Student’s t-test. C. Following the same protocol described in (B), the cells were stimulated by NGF (concentration ranged from 0 ng/ml to 100 ng/ml) for 5 h. NFAT-dependent gene expression was then determined by calculating ratios of firefly to Renilla luciferase activity. n = 6–12, ***p < 0.001 (vs 0 ng/ml NGF), One-Way ANOVA, Bonferroni post-hoc test. D. Either 0 ng/ml ((-)NGF) or 20 ng/ml NGF ((+)NGF) was applied for 5, 10, and 24 h to active PC12 cells transfected with the two luciferase reporters. NFAT-dependent gene expression was then assayed as described in (A). The ratio of NFAT-dependent gene expression in the presence of 20 ng/ml NGF to NFAT-dependent gene expression in the absence of NGF was calculated from (D), which was shown in the inset. n = 6–12. n stands for the number of well. Data are presented as mean ± S.E.M.](/cms/asset/149be3f4-2341-426b-bdf9-cd3d6525724e/tfls_a_2034670_f0001_ob.jpg)
NFAT-dependent gene expression by NGF is mediated by TrkA receptor activation but not the PLC pathway
To exam if the observed NGF effect on increasing NFAT-dependent gene expression (Figure ) was mediated by the TrkA receptor, PC12 cells transfected with the two luciferase plasmids (firefly NFAT-reporter luciferase, Renilla pGL4.74[hRluc/TK]) were pretreated (30 min) with K252a, a potent tyrosine kinase inhibitor commonly used to study TrkA signaling, then NGF was applied to stimulate the cells (Figure (A)) (Knusel and Hefti Citation1992; Eibl et al. Citation2012; Kim et al. Citation2014). As shown in Figure (C), K252a essentially abolished the NGF effects, suggesting that TrkA responded to NGF to activate NFAT. TrkA receptor activation has shown to trigger three major intracellular signaling pathways – PLC, PI3 K, and MAPK pathways (Reichardt Citation2006). Therefore, to examine if TrkA triggered-PLC pathway was responsible for NFAT activation, the cells stimulated with NGF were pretreated (30 min) with U73122, a PLC inhibitor, which resulted in a rather significant increase of NFAT activity (Figure (D)). To further investigate the contribution of the PLC pathway, cells were pretreated (30 min) with calcineurin inhibitors (FK506 or CsA) since calcineurin, an NFAT phosphatase which triggers NFAT nuclear translocation, has been reported to be activated by a Ca2+ release from ER by TrkA-PLC-mediated IP3 generation. (Figure (B)) However, the increased NGF-mediated NFAT activity was not diminished following treatment with calcineurin inhibitors. Taken together, these observations implied that NGF did not rely on the TrkA-PLC pathway for NFAT activation (Figure (D)). Additionally, to verify the integrity of the calcineurin inhibitors (FK506, CsA) used in the experiment, the cells were co-stimulated with NGF together with 50 mM KCl (a mixture of RPMI1640 and 150 mM KCl stock solution) to depolarize the cells, thereby inducing Ca2+ influx via the opening of voltage-gated Ca2+ channels. BayK8644, an L-type Ca2+ channel agonist (1 µM), was added to the mixture to stabilize Ca2+ elevation during the stimulation period. In this case, the increased Ca2+ should stimulate NFAT by activating calcineurin. As expected, 50 mM KCl further increased NFAT activity and the calcineurin inhibitors essentially abolished the KCl effect, suggesting that the inhibitors used in this assay were intact (Figure (E)). A pharmacological approach to inhibit cellular targets often raises specificity issues. Therefore, previously validated commonly employed inhibitor concentration was carefully selected throughout the experiments (Kim and Usachev Citation2009; Kim et al. Citation2014).
Figure 2. NGF recruits TrkA but does not rely on the PLC pathway to activate NFAT-dependent gene expression. A. Schematic diagram of depicting the experimental protocols. B. A diagram describing the PLC pathway stimulated by NGF. C. According to the protocol described in (A), NGF (20 ng/ml, 5 h stimulation) was applied and its ability to activate NFAT-dependent gene expression in the presence or absence of K252a (a TrkA inhibitor) was assayed in PC12 cells as described in Figure . n = 6–7, ***p < 0.001, One-Way ANOVA, Bonferroni post-hoc test D. U73122 (a PLC inhibitor) or Ca2+-dependent phosphatase calcineurin (CaN) inhibitors (FK506, CsA) were applied to block TrkA-triggered PLC activation or PLC-IP3-Ca2+ mediated CaN activation in PC12 cells treated as explained in (A). n = 6–7 E. NGF-treated PC12 cells were co-stimulated with KCl to depolarize cells for inducing an intracellular Ca2+ increase. To verify the integrity of CaN inhibitors, the inhibitors were applied to PC12 cells prepared as described in (A) to check if they inhibit CaN which should be activated by KCl. All inhibitors were applied 30 min prior to NGF stimulation. n = 6–7, **p < 0.01, ***p < 0.001, One-Way ANOVA, Bonferroni post-hoc test. n stands for the number of well. Data are presented as mean ± S.E.M.
MEK1/2 but not PI3 K mediates NGF-dependent NFAT activation
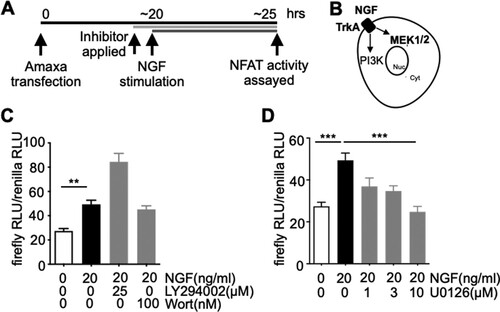
Next, the involvement of the PI3 K pathway for NGF-dependent NFAT activation was examined by treating the cells with PI3 K inhibitors (wortmannin, LY294002). Following essentially the same protocol used in Figure (D), NFAT activity was measured in cells stimulated with NGF in the presence of PI3 K inhibitors, which resulted in a significant increase in NFAT activity, a result that suggests that the TrkA-PI3 K pathway is not responsible for mediating NGF-dependent NFAT activation (Figure (C)). Lastly, the contribution of the MEK1/2 pathway stimulated by NGF for responding NGF-mediated NFAT activation was studied by incubating the cells with U0126, a MEK1/2 inhibitor (Duncia et al. Citation1998). As shown in Figure (D), U0126 reduced NGF-triggered NFAT-dependent gene expression in a dose-dependent manner (Figure (D)). Lastly, the cells were treated with U0126 in the absence of NGF, revealing insignificance changes in NFAT-dependent gene expression. This data implied that it was unlikely that the U0126-induced NFAT inactivation emerged as feedback signaling from basal-level MEK1/2 inhibition which was irrelevant to NGF (Supplementary Figure 1).
Figure 3. NFAT activation by NGF depends on MEK1/2 but not PI3 K pathway. A. Schematic diagram of depicting the experimental protocols. B. A diagram describing TrkA-mediated PI3 K and MEK1/2 activation by NGF. C-D. As described in Figure , NFAT reporter assays were performed with the exception that PI3 K inhibitor (LY294002, Wort (wortmannin) (C)) or MEK1/2 inhibitor (U0126, (D)) was applied to block TrkA-mediated PI3 K or MEK1/2 activation, respectively. All inhibitors were applied 30 min prior to NGF stimulation. n = 12–13 for (C), n = 6–12 for (D), **p < 0.01, ***p < 0.001, One-Way ANOVA, Bonferroni post-hoc test. n stands for the number of well. Data are presented as mean ± S.E.M.
NFATc3 is the predominant NFAT isoform in PC12 cells
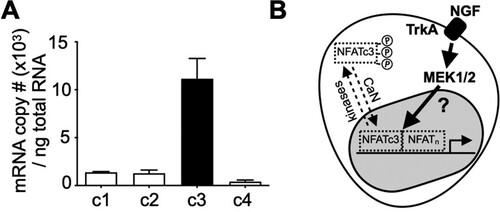
There are four classic members in the NFAT transcription factor family, which consists of NFATc1, c2, c3 and c4 (Rao et al. Citation1997). Each NFAT isoform has distinct cellular functions and tissue distributions. Therefore, it was decided to identify the main NFAT isoform responsible for mediating NGF signaling in PC12 cells. For this, quantitative real-time PCR was performed to estimate the absolute copy numbers of each NFAT, which revealed NFATc3 is most abundant in PC12 cells (Figure (A)). Based in these findings, I suggest that NGF stimulate NFATc3-dependent gene expression in PC12 cells via MEK1/2 (Figure (B)).
Figure 4. NFAc3 is the predominant NFAT isoform in PC12 cells. A. After constructing standard curves for each NFAT isoform, SYBR-based qPCR was performed with total RNA isolated from PC12 cells to calculate absolute copy number of each NFAT isoform. n = 3 B. Proposed model: NFATc3, a major NFAT isoform in PC12 cells shuttles in and out of the nucleus by CaN, an NFAT phosphatase and several kinases, respectively. An increase in NFAT-dependent gene expression by NGF depends on the MEK1/2 pathway via uncharacterized mechanisms in PC12 cells. Data are presented as mean ± S.E.M.
Discussion
In this research the novel signaling pathway initiated by NGF, which leads to NFAT-dependent gene expression in PC12 cells is demonstrated. Additionally, it is proposed that interrupting MEK1/2 pathway may exhibit the most prominent cellular consequence that halts NGF-related effects if it requires NFAT for exerting its biological functions. However, I believe this research also raises many intriguing questions worthy of being resolved in the future by ourselves or others who are enticed by this report.
First, NFAT needs to be dephosphorylated by calcineurin to be imported into the nucleus where it binds so-called NFAT consensus DNA sequences for initiating gene transcription with the help of loosely defined cofactors present in the nucleus generally referred to as NFATn. Likewise, NFAT can be re-phosphorylated by NFAT kinases such as GSK3β and p38, which antagonizes NFAT nuclear occupancy, thereby terminating its action (Crabtree and Olson Citation2002). Similar to the aforementioned NFAT kinases, MEK1/2 has the potential to oppose NFAT action so that inhibiting MEK1/2 activity by U0126 could increase NFAT-dependent gene expression by extending NFAT nuclear occupancy. However, this hypothesis was not confirmed in the study summarized here. The data presented here imply that MEK1/2 rather functions to augment NFAT’s transcriptional activity via uncharacterized mechanisms in PC12 cells. Importantly, Sanna et al. observed a similar phenomenon in cardiomyocytes and reported that (1) MEK1 helps to express AP-1 which was postulated to function as an NFATn and/or (2) MEK1 together with its substrate ERK1/2 forms a complex with calcineurin, which promotes NFAT phosphorylation near its C-terminal, strengthening NFAT’s DNA binding affinity instead of facilitating its nuclear export (Sanna et al. Citation2005). Therefore, although it should be scrutinized, it is plausible that similar molecular events may occur in PC12 cells. This open possibility needs to investigate in the future by such an alternative approach as Chip assay, which directly measures the strength of NFAT binding to its corresponding DNA sequence.
Second, among the three signaling pathways stimulated by NGF, a drastic increase of NFAT activity was observed when either the PLC or PI3 K pathways were blocked. (Figures and ). Since there were some reports suggesting the feedback activation of PI3 K upon MEK1/2 inhibition in cancer cells, it could be conceived as an open possibility that the blockade of PLC or PI3 K pathways could fortify the MEK1/2 pathway, which may give rise to the further increase in NFAT activity in neurons (Mirzoeva et al. Citation2009; Turke et al. Citation2012). However, there may likely be an existence of cell-type-specific signal transduction feedback mechanisms to optimally adapt to external stimuli. Therefore, further research needs to perform to uncover mechanisms underlying this interesting phenomenon caused by these inhibitors.
Third, the transcription factor family of NFAT consists of 4 major isoforms, named NFATc1, c2, c3, and c4. Each NFAT isoform has been reported to exert unique cellular functions with distinct tissue distributions. Ulrich et al. even demonstrated that the rate of nuclear import is quite distinct between NFATc3 and NFATc4 in neurons (Ulrich et al. Citation2012). Therefore, I believe it is important to identify a predominant NFAT isoform in a specific cell type when considering isolating drug targets to maximize clinical utility. Interestingly, NFATc3 has been reported as the main isoform in dorsal root ganglia (DRG) (Kim and Usachev Citation2009). Since PC12 cells are derived from a pheochromocytoma of the adrenal medulla, a structure considered as a modified ganglia of the postganglionic sympathetic nerve system, NFATc3 may be the most common NFAT isoform in neurons exhibiting properties of peripheral ganglia (i.e. DRG, adrenal medulla). Although sophisticated experiments such as replacing NFATc3 with other NFAT isoform may be necessary to reveal the reason why NFATc3 is heavily expressed in the peripheral neurons and what is the functional significance of small amount of other NFAT isoforms, uncovering the principal NFAT isoform in the peripheral neurons will definitely help usher drug development for targeting NGF-NFAT signaling in the nervous system. However, it should take into account that environmental stimuli could alter the level of each NFAT isoform. Actually, Cai et al. reported that nerve injury upregulated NFAT expression in an isoform- and neuronal cell-type-specific manner (Cai et al. Citation2013). Therefore, it is an open possibility that cells may regulate the expression levels of each NFAT isoform to adapt to environmental changes.
Finally, I want to emphasize ‘in our experimental settings’ the data support the hypothesis that NGF utilizes the MEK1/2 pathway for NFATc3 activation. However, the existence of other pathways that relay NGF signals to NFAT cannot be excluded at this time. It is an open question as to whether the main cellular components heavily stimulated by NGF for NFAT activation can be switched to other molecular elements depending on developmental and/or differentiation stages. Seybold et al. proposed that NGF alone triggered NFAT-dependent gene expression in a calcineurin-dependent manner in adult mouse DRG although they did not explain how NGF increases calcineurin activity (Groth et al. Citation2007). However, based on experiments using neonatal rat DRG, the Usachev group suggests that NGF should work in concert with electrical stimulation to activate NFAT via the TrkA-P13K-GSK3β pathway (Kim et al. Citation2014). Meanwhile, Stefos et al. reported NGF alone activates NFAT in a calcineurin-sensitive manner with in-house PC12-NFAT-Luc stable cell line (Stefos et al. Citation2013), which was different from what I observed in Figure . However, this discrepancy could be resolved when considering differences such as the transfection methods each group employed, detailed culture protocols for maintaining and splitting cells (i.e. composition of culture media, trypsinization), levels of differentiation, and passage number of PC12 cells used in the experiments, all of which could potentially affect intracellular signaling. Therefore, it can be postulated that considering the plethora of signaling cascades and dynamic aspect of cellular response to external stimuli, it is very likely that cells may trigger optimal intracellular signaling to best cope with environmental cues.
In conclusion, I hope this research revealing intracellular signaling triggered by NGF for NFAT activation helps to pave a new way to develop novel therapeutic agents, which can be exploited to devise innovative remedies for the treatment of NGF and NFAT-associated clinical disorders such as hyperalgesia and anomalous neuronal development.
Supplementary Material
Download MS Word (41.9 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in at 4TU.ResearchData repository [https://data.4tu.nl/portal] at https://doi.org/10.4121/16943956.v1.
Additional information
Funding
References
- Aloe L. 2011. Rita Levi-Montalcini and the discovery of NGF, the first nerve cell growth factor. Arch Ital Biol. 149(2):175–181.
- Cai Y-Q, Chen S-R, Pan H-L. 2013. Upregulation of nuclear factor of activated T-cells by nerve injury contributes to development of neuropathic pain. J Pharmacol Exp Ther. 345(1):161–168.
- Crabtree GR, Olson EN. 2002. NFAT signaling: choreographing the social lives of cells. Cell. 109(Suppl):S67–S79.
- Dravid A, Parittotokkaporn S, Aqrawe Z, O'Carroll SJ, Svirskis D. 2020. Determining Neurotrophin gradients in vitro to direct axonal outgrowth following spinal cord injury. ACS Chem Neurosci. 11(2):121–132.
- Duncia JV, Santella JB, 3rd, Higley CA, Pitts WJ, Wityak J, Frietze WE, Rankin FW, Sun J-H, Earl RA, Tabaka AC, et al. 1998. MEK inhibitors: the chemistry and biological activity of U0126, its analogs, and cyclization products. Bioorg Med Chem Lett. 8(20):2839–2844.
- Eibl JK, Strasser BC, Ross GM. 2012. Structural, biological, and pharmacological strategies for the inhibition of nerve growth factor. Neurochem Int. 61(8):1266–1275.
- Graef IA, Wang F, Charron F, Chen Lei, Neilson J, Tessier-Lavigne M, Crabtree GR. 2003. Neurotrophins and netrins require calcineurin/NFAT signaling to stimulate outgrowth of embryonic axons. Cell. 113(5):657–670.
- Groth RD, Coicou LG, Mermelstein PG, Seybold VS. 2007. Neurotrophin activation of NFAT-dependent transcription contributes to the regulation of pro-nociceptive genes. J Neurochem. 102(4):1162–1174.
- Huang W, Huang J, Jiang Y, Huang X, Xing W, He Y, Ouyang H. 2018. Oxaliplatin regulates chemotherapy induced peripheral neuropathic pain in the dorsal horn and dorsal root ganglion via the calcineurin/NFAT pathway. Anticancer Agents Med Chem. 18(8):1197–1207.
- Jung H, Miller RJ. 2008. Activation of the nuclear factor of activated T-cells (NFAT) mediates upregulation of CCR2 chemokine receptors in dorsal root ganglion (DRG) neurons: a possible mechanism for activity-dependent transcription in DRG neurons in association with neuropathic pain. Mol Cell Neurosci. 37(1):170–177.
- Kim M-S, Shutov LP, Gnanasekaran A, Lin Z, Rysted JE, Ulrich JD, Usachev YM. 2014. Nerve growth factor (NGF) regulates activity of nuclear factor of activated T-cells (NFAT) in neurons via the phosphatidylinositol 3-kinase (PI3K)-Akt-glycogen synthase kinase 3β (GSK3β) pathway. J Biol Chem. 289(45):31349–31360.
- Kim M-S, Usachev YM. 2009. Mitochondrial Ca2+ cycling facilitates activation of the transcription factor NFAT in sensory neurons. J Neurosci. 29(39):12101–12114.
- Knusel B, Hefti F. 1992. K-252 compounds: modulators of neurotrophin signal transduction. J Neurochem. 59(6):1987–1996.
- Lee S, Lee DK. 2020. What is the proper way to apply the multiple comparison test? Korean J Anesthesiol. 73(6):572.
- Mirzoeva OK, Das D, Heiser LM, Bhattacharya S, Siwak D, Gendelman R, Bayani N, Wang NJ, Neve RM, Guan Y, et al. 2009. Basal subtype and MAPK/ERK kinase (MEK)-phosphoinositide 3-kinase feedback signaling determine susceptibility of breast cancer cells to MEK inhibition. Cancer Res. 69(2):565–572.
- Rao A, Luo C, Hogan PG. 1997. Transcription factors of the NFAT family: regulation and function. Annu Rev Immunol. 15:707–747.
- Reichardt LF. 2006. Neurotrophin-regulated signalling pathways. Philos Trans R Soc Lond B Biol Sci. 361(1473):1545–1564.
- Sanna B, Bueno OF, Dai Y-S, Wilkins Benjamin J., Molkentin Jeffery D. 2005. Direct and indirect interactions between calcineurin-NFAT and MEK1-extracellular signal-regulated kinase 1/2 signaling pathways regulate cardiac gene expression and cellular growth. Mol Cell Biol. 25(3):865–878.
- Serrano-Perez MC, Fernandez M, Neria F, Berjón-Otero M, Doncel-Pérez E, Cano E, Tranque P. 2015. NFAT transcription factors regulate survival, proliferation, migration, and differentiation of neural precursor cells. Glia. 63(6):987–1004.
- Sharma N, Deppmann CD, Harrington AW, St. Hillaire C, Chen Z-Y, Lee F, Ginty DD. 2010. Long-distance control of synapse assembly by target-derived NGF. Neuron. 67(3):422–434.
- Shaw JP, Utz PJ, Durand DB, Toole J, Emmel E, Crabtree G. 1988. Identification of a putative regulator of early T cell activation genes. Science. 241(4862):202–205.
- Sofroniew MV, Howe CL, Mobley WC. 2001. Nerve growth factor signaling, neuroprotection, and neural repair. Annu Rev Neurosci. 24:1217–1281.
- Stefos GC, Soppa U, Dierssen M, Becker W, Gong C-X. 2013. NGF upregulates the plasminogen activation inhibitor-1 in neurons via the calcineurin/NFAT pathway and the down syndrome-related proteins DYRK1A and RCAN1 attenuate this effect. PLoS One. 8(6):e67470.
- Turke AB, Song Y, Costa C, Cook R, Arteaga CL, Asara JM, Engelman JA. 2012. MEK inhibition leads to PI3K/AKT activation by relieving a negative feedback on ERBB receptors. Cancer Res. 72(13):3228–3237.
- Ulrich JD, Kim M-S, Houlihan PR, Shutov LP, Mohapatra DP, Strack S, Usachev YM. 2012. Distinct activation properties of the nuclear factor of activated T-cells (NFAT) isoforms NFATc3 and NFATc4 in neurons. J Biol Chem. 287(45):37594–37609.
