Abstract
Biodiversity of a gut microbiome has been shown as an important predictor of transplant-related outcomes and infections in allogeneic hematopoietic stem cell transplantation (HSCT). We conducted a single-center real-life clinical study and implemented a routine gut microbiome diversity monitoring across the course of allogeneic HSCT. Twelve patients (with ALL, AML, CML, biphenotypic leukemia and aplastic anemia) were enrolled in a stool samples collection protocol before the start of HSCT and during a 30-day post-transplant period. We have shown the feasibility of a gut microbiome monitoring in a real-life clinical setting and have proven that the alpha-biodiversity of the microbiome is significantly reduced with HSCT in comparison with the individual patient baseline level (Xdc 72.93; p < 0.001; multivariate Dirichlet analysis), what may be related to the antibiotic use and conditioning regimen. Overall, the gut microbiome biodiversity monitoring may be clinically used in a real-life HSCT setting to identify the high-risk groups for developing bloodstream infections and transplant-related negative outcomes.
Introduction
Over the past two decades, the microbial community, i.e. the human microbiome has been recognized as a fundamental determinant of host physiology and pathology. The associations of the spectrum and characteristics of the microbiome have already been proven in a number of infectious and noninfectious human diseases, and modern research has gone far beyond the classical understanding of the role of microorganisms in normal and pathological human physiology. One of the most studied roles of human microorganisms is to provide protection against bacterial pathogens. This is especially important in hospital settings to prevent health care-related infections caused by bacteria from the gastrointestinal tract. Commensal representatives of the microbiome are able to exclude these highly resistant pathogens from the microbial community of the gastrointestinal tract, as well as resist colonization de novo. Previously, the directions for studying the microbiome in modern clinical medicine were extensively discussed (Kim et al. Citation2017).
A differentiated microbiome and its role in protecting against infectious diseases are especially important in immunocompromized patients. It is known that the greatest depth and duration of damage to the immune system, as well as susceptibility to infectious agents, is observed in patients undergoing hematopoietic stem cell transplantation (HSCT, both peripheral and bone marrow). Current indications for HSCT include the following diseases: acute lymphoblastic leukemia, acute myeloid leukemia, chronic myeloid leukemia, MDS, nonHodgkin's lymphomas, Hodgkin's lymphoma, multiple myeloma. On the model of immunosuppression in HSCT, many fundamental data on the human microbiome have been obtained (Taur and Pamer Citation2016). It is important to note that the issue of microbiome biodiversity is of undoubted clinical significance, because an independent effect of microbiome diversity on the risk of transplant-associated mortality was shown in a large cohort of transplant patients for several years after allogeneic HSCT (Treatment by Cancer Type [Internet] Citation2021). Biodiversity reflects the number of different species/operating taxonomic units (OTUs) of bacteria in the microbiome. Importantly, alpha-diversity is the diversity of species within a biological sample, which is determined by the Chao 1, Shannon, Simpson indices. Beta-diversity is the diversity of species between two biological samples. Chao 1 is a measure of observed/latent diversity, while the Shannon index is a measure of the diversity and fluency of the microbiome, where a higher index corresponds to a greater diversity; Simpson's index is a measure of the dominance of a species in a specimen. Previously, we have proved taxonomic associations of the dominance of certain types in the microbiome and the development of bloodstream infections, which is true both for gram-negative bacteria (Yilmaz et al. Citation2014) and for individual gram-positive microorganisms (McMurdie Citation2013). Moreover, biodiversity before the HSCT procedure was a positive predictor of transplantation success (Taur et al. Citation2012). It is well known, that most of the sequence data is obtained with a substantial delay, and the associations are observed retrospectively. Meanwhile, we have implemented the pilot project of clinical microbiome monitoring, in small groups of patients being included in sequencing, almost ‘at the bedside’.
Aim of the study: to assess the dynamic parameters of the biodiversity of the intestinal microbiome in patients when performing HSCT in a real-life clinical practice.
Materials and methods
Study design and patient characteristics
Adult patients hospitalized for HSCT for the period 2019–2021 on the basis of the Minsk Scientific and Practical Center for Surgery, Transplantology and Hematology (Republic of Belarus) were included in the protocol of collection and low-temperature freezing of stool samples. The study protocol was approved by the ethical committee of the institution (protocol No. 10 for 2019). The study was registered in the Clinicaltrials.gov (NCT04281797). The study was conducted in accordance with the Declaration of Helsinki as well as the STROBE criteria for observational studies. Funding for the study was carried out at the expense of the state program ‘Science-intensive technologies and equipment’ for 2016–2020 (section ‘Medical biotechnology’), state registration number 20200047 dated 16 January 2020. Among the eligibility criteria for this clinical trial, the following were accepted: the presence of ≥ 2 consecutive high-quality sequenced biological samples in one patient. An important condition was that the first sample should be collected before the HSCT procedure in order to have a baseline comparison point for each of the participants. The other samples were taken in the early post-transplant period, up to 30 days after HSCT. The baseline sample and a first post-transplant sample were sequenced during the hospitalization period, overall there were three sequence groups formed among all the patients. Extensive clinical and laboratory data were collected from each patient prospectively in real clinical practice. The criteria for GVHD were defined as NCCN v. 1.2021 (Treatment by Cancer Type [Internet] Citation2021). Automatic bacteriological systems (VITEK from Biomerieux) in combination with MALDI-TOF mass spectrometry have been used for the rapid identification of pathogens. Among the clinical parameters in the analysis were used: age, sex of patients, underlying disease, conditioning regimen, source of hematopoietic stem cells and type of transplant, time, duration and choice of antibiotics, chemotherapy regimens.
Technical and laboratory procedures
After extraction and purification of DNA in each of the biological samples, PCR amplification of the V3–V4 region of the 16S rRNA gene was performed using modified universal bacterial primers (manufactured by Illumina, USA). After the index PCR, each of the DNAs was assigned unique indexes, allowing the identification of the amplicons of one sample. Isolation of total DNA from stool samples was performed using QIAamp® PowerFecal® DNA Kit (manufactured by Qiagen) on a QIAcube automated nucleic acid isolation station, Qiagen. Further processing of DNA samples for the preparation of the 16S metagenomic library was carried out in accordance with the instruction ‘16S Metagenomic Sequencing Library Preparation’ from Illumina. Sequencing of the prepared metagenomic library was performed on the MiSeq Illumina platform using MiSeq v3 sequencing reagents (600 cycles).
Mathematical modeling and statistical methods
Phylogenetic classification to the species level was carried out on the basis of the Bayesian model, as well as the SILVA ribosomal RNA database (Yilmaz et al. Citation2014). The sequences were grouped into operating taxonomic units (OTUs) based on 97% identity. The total number of 16S rRNA genes per gram of biological material, i.e. a measure of bacterial density, calculated using quantitative PCR based on the total amount of DNA isolated from each sample. The biodiversity indices used in the work included Shannon, Simpson, Chao 1, Simpson's inverse index. The processing and analysis of a vast array of clinical, laboratory and phylogenetic data were performed using the R (R Development Core Team, Vienna, Austria) along with Phyloseq package (McMurdie Citation2013). Bayesian nonparametric statistical methods, including the Dirichlet distribution (Dir (α)), a family of continuous multivariate probability distributions, were used to assess the independent nature of differences in microbiome biodiversity against the background of the HSCT procedure. UniFrac was used as a distance measure used to compare biological communities. As a measure of dissimilarity, the Bray–Curtis index was used to quantify the compositional dissimilarity between two different sites based on the counts at each site.
Results and discussion
A total of 12 patients receiving allo-HSCT were included in this real-life clinical practice prospective study. Table presents the baseline clinical and demographic characteristics of patients in the study.
Table 1. Basic characteristics of patients in the study and their transplantation period (MRD, minimal residual disease).
Thus, acute leukemia was the most common indication for transplantation. Both myeloablative and reduced intensity (predominantly) conditioning regimens were used. All of the included patients (except aplastic anemia) received the approved chemotherapy regimens to achieve remission prior to enrollment in HSCT program, while CML patient was in the chronic phase. The main source of HSCs was peripheral blood from related and unrelated donors. Mostly, transplantation was performed in patients in the first remission. Ex vivo depletion was not used, however, for transplantation from a completely compatible unrelated donor, the method of in vivo depletion with antithymocyte globulin was used, and for partially compatible transplantation in vivo, depletion with post-transplant cyclophosphamide.
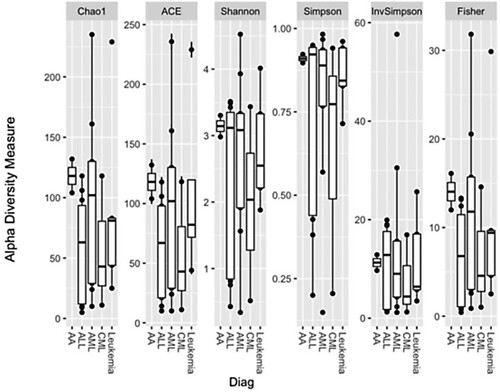
In an exploratory analysis of potential factors influencing the biodiversity of the microbiome, data for significant differences depending on the primary disease were not observed, Figure .
Figure 1. Biodiversity of intestinal microbiome samples depending on the main hematological disease (Leukemia - biphenotypic leukemia).
Another question was to study how the biodiversity of the intestinal microbiome changes during the HSCT procedure. In the graphical analysis, attention was drawn to the significant decrease in alpha-biodiversity in the second and third samples compared to baseline. Graphical analysis is shown in Figure .
Figure 2. Biodiversity of intestinal microbiome samples during HSCT, in dynamics, depending on the sample number (1,2,3).
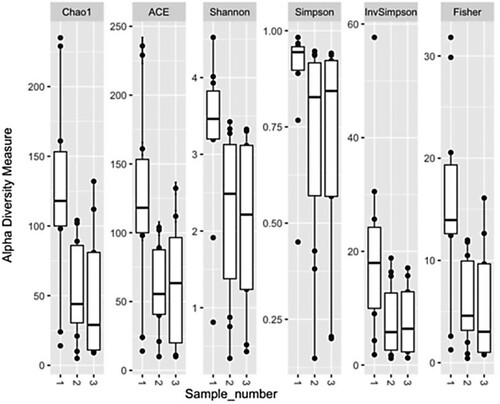
The nonparametric Bayesian Dirichlet model was used to mathematically confirm the hypothesis that the alpha-biodiversity of the intestinal microbiome is reliably and independently reduced during the HSCT procedure. According to the results of the analysis, it was proved that the alpha-biodiversity of the intestinal microbiome in samples 2 and 3 was significantly and independently lower than before HSCT in the first sample (Xdc = 72.93; p < 0.001). Median time interval between samples 1 and 2 was 8 days, while median interval between samples 2 and 3 was 5 days. Further, the study was carried out to analyze based on which representatives of the microbiome the loss of diversity occurs (Figure ).
Figure 3. Dynamics of the composition of the intestinal microbiome at the taxonomic level of the type before HSCT (samples 1) and after HSCT (samples 2, 3).
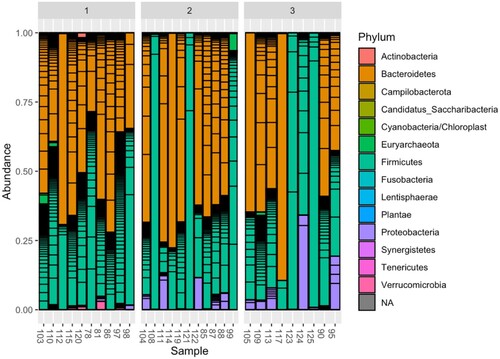
Further, to simplify understanding of the dynamics of changes in the intestinal microbiome during HSCT, the most common taxonomic phyla of bacteria (Bacteroidetes, Firmicutes, Proteobacteria) were selected, and a dynamic graph of their relative abundance in the microbiome before HSCT (sample 1) and after HSCT (samples 2 and 3) is shown in Figure .
Figure 4. Dynamics of the relative abundance of the 3 most common taxonomic phyla (Bacteroidetes, Firmicutes, Proteobacteria) of the intestinal microbiome before HSCT (samples 1) and after HSCT (samples 2, 3).
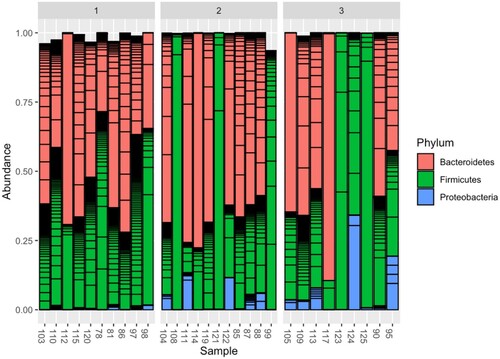
When graphically analyzing the dynamics of the microbiome composition, it should be noted that, in addition to the decrease in biodiversity, in some patients (samples 123, 124, 125) there is an almost complete replacement of the microbiome with one phylum. Earlier Taur et al. (Citation2012) have called this phenomenon as domination (replacement of more than 30% of the relative abundance of the microbiome with one taxonomic type). Moreover, in relation to Enterococci, this was an independent risk factor for generalized enterococcal infections. Later, in a collaboration with Memorial Sloan Kettering Cancer Center (USA) we have proven that the dominance of the Proteobacteria phylum is an independent risk factor for the development of Gram-negative bloodstream infections (Stoma et al. Citation2021 dec 6). Returning to Figure , it can be noted that, in comparison with the microbiome before HSCT, in a number of patients there is an increase in the relative abundance of the Proteobacteria phylum (samples 104, 111, 122, 87, 88 in the second group and samples 105, 109, 113, 124, 95 in the third group). Considering that, according to the literature, in a normal microbiome, the relative abundance of Proteobacteria, as a rule, is up to 1%, these changes can be called prognostically unfavorable (Turnbaugh et al. Citation2007; Gevers et al. Citation2012).
And in the case of sample 124, the patient after HSCT developed an episode of gut domination of the Proteobacteria phylum, which should be considered as a clinically relevant predictor of Gram-negative infections, and empirical antibiotic therapy for febrile neutropenia should be adjusted taking into account these data.
Conclusion
In this prospective real-life clinical study, we performed a dynamic assessment of the alpha-biodiversity of the gut microbiome in a cohort of adult patients undergoing HSCT. Graphic and mathematical analysis of sequence data in combination with clinical and laboratory variables made it possible to prove that the alpha-biodiversity of the microbiome is significantly reduced with HSCT in comparison with the baseline level (Xdc 72.93; p < 0.001; multivariate analysis of the Dirichlet distribution). Previously, it was indicated that patients with an initially lower level of biodiversity of the intestinal microbiome before HSCT have a higher level of 3-year post-transplant mortality, taking into account other variables (Taur et al. Citation2014). There were few studies that dynamically assesses the metagenomic features along the post-transplant period in HSCT (Kusakabe et al. Citation2020). While in the mentioned study authors analyzed both alpha- and beta-diversities, the results of our study represent a real-life clinical practice. It is well known, that most of the sequence data is obtained with a substantial delay, and the associations are observed mostly retrospectively. Meanwhile, we have implemented the pilot project of clinical microbiome monitoring, in small groups of patients being included in sequencing, with results obtained almost ‘at the bedside’.
In the performed study, in addition to the decrease in biodiversity, the dynamics of the ratio of the main taxonomic types in the microbiome during the HSCT procedure and the early post-transplantation period is shown. Namely, a decrease in the relative abundance of the Bacteroidetes, with an increase in Proteobacteria. In one of the cases in the study, the level of Proteobacteria phylum after HSCT reached a gut domination, i.e. more than 30%, which, according to the previously published data, is an independent predictor of bloodstream infections. Main study limitation is a relatively small sample size, but it is also important to assess the obtained results as first steps in a clinical real-life use of a next-generation sequencing techniques and microbiome analysis as a routine laboratory practice in HSCT-settings. Important and alarming observation is that Proteobacteria phylum is known for high prevalence of multiple antibiotic-resistance genes (ResistanceMap [Internet] Citation2021). Thus, the methodology for monitoring and assessing the indicators of microbiome biodiversity during the HSCT procedure has been worked out in a real-life clinical practice, which makes it possible to identify the risk groups for infections, GVHD, and even post-transplant mortality among the allogeneic HSCT recipients.
Author contributions.
Study conception and design: I.S., M.U., N.M. and A.U.; data collection: M.U., E.M., N.M. and T.G.; analysis and interpretation of results: I.S., V.S. and A.U.; draft manuscript preparation: I.S., V.S. and M.U. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgements
We acknowledge sponsorship for this study from the State Committee on Science and Technology of the Republic of Belarus. We acknowledge Andrey S. Babenko for important laboratory consultations. We acknowledge Oleg O. Rummo for an ongoing organizational support.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are available on request from the corresponding author (I.S.). The data are not publicly available due to containing information that could compromise the privacy of research participants.
Additional information
Funding
References
- Gevers D, Knight R, Petrosino JF, Huang K, McGuire AL, Birren BW, et al. 2012 Aug 14. The human microbiome project: a community resource for the healthy human microbiome. PLOS Biol. 10(8):e1001377.
- Kim S, Covington A, Pamer EG. 2017 Sep. The intestinal microbiota: antibiotics, colonization resistance, and enteric pathogens. Immunol Rev. 279(1):90–105.
- Kusakabe S, Fukushima K, Maeda T, Motooka D, Nakamura S, Fujita J, et al. 2020 Feb. Pre- and post-serial metagenomic analysis of gut microbiota as a prognostic factor in patients undergoing haematopoietic stem cell transplantation. Br J Haematol. 188(3):438–449.
- McMurdie PJ. 2013. Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PloS One. 8(4):e61217.
- ResistanceMap [Internet]. 2021. [cited 2021 Sep 4]. Available from: https://resistancemap.cddep.org/.
- Stoma I, Littmann ER, Peled JU, Giralt S, van den Brink MRM, Pamer EG, et al. 2021 Dec 6. Compositional flux within the intestinal microbiota and risk for bloodstream infection with gram-negative bacteria. Clin Infect Dis Off Publ Infect Dis Soc Am. 73(11):e4627–e4635.
- Taur Y, Jenq RR, Perales M-A, Littmann ER, Morjaria S, Ling L, et al. 2014 Aug 14. The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood. 124(7):1174–1182.
- Taur Y, Pamer EG. 2016 Apr 18. Microbiome mediation of infections in the cancer setting. Genome Med. 8(1):40.
- Taur Y, Xavier JB, Lipuma L, Ubeda C, Goldberg J, Gobourne A, et al. 2012 Jan 10. Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin Infect Dis. 55(7):905–914.
- Treatment by Cancer Type [Internet]. 2021. NCCN. [cited 2021 Sep 4]. Available from: https://www.nccn.org/guidelines/category_1.
- Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. 2007 Oct 18. The human microbiome project. Nature. 449(7164):804–810.
- Yilmaz P, Parfrey LW, Yarza P, Gerken J, Pruesse E, Quast C, et al. 2014 Jan 1. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 42(D1):D643–D648.
