Abstract
The Qinling mountains are one of the richest diversity hotspots in the world. This area is remote, and new species remain to be discovered, including macrofungi. In this study, Qinling Mountain of the Shaanxi province was surveyed for macrofungus, and one new species of macrofungi Microstoma ningshanica (Sarcoscyphaceae) was discovered and analyzed by light microscopy and scanning electron microscopy as well as DNA sequence analysis. The description, photographs, and comparisons with morphologically similar and phylogenetically related species are provided. Conclusions of its phylogenetic relationships with similar species are also drawn based on the sequence of the nuclear internal transcribed spacer region, as well as the large subunit rRNA.
KEYWORDS:
Introduction
The genus Microstoma is a member of the family Sarcoscyphaceae (Liu and Zhuang Citation2018). The characteristics of the genus are as follows: apothecia solitary to aggregate, deep-cupulate, and long stipitate generally; hymenium surface pink, orange, yellow to lavender; the margin and surface of the receptacle usually white and hairy with obtuse or sharp tips; ectal excipulums of textura angularis to globulosa; medullary excipulum two-layered, the outer layer of textura intricate and gelatinous, and the inner layer of textura porrecta to intricate; asci subcylindrical, J− in Melzer’s reagent; ascospores ellipsoidal to fusoidal, and smooth; paraphyses filiform (Liu and Zhuang Citation2018). Eight species of this genus have been reported (Kanouse Citation1948; Guy et al. Citation1997; Harada and Kudo Citation2000; Wang Citation2004; Douanlameli and Langer Citation2005; Kirk et al. Citation2008, Liu and Zhuang Citation2018), including M. aggregatum, M. apiculosporum, M. floccosum, M. insititium (≡ Cookeina insititia), M. macrosporum (≡ M. floccosum var. macrosporum), and M. radicatum. The characteristics, such as the color of apothecia, the distribution and shape of the hairs of the receptacle, or the shape of ascospores, are important bases for distinguishing species of the genus Microstoma. The key to species of the genus Microstoma has also been edited by Liu (Liu and Zhuang Citation2018).
Traditionally, macrofungal taxonomy was based on morphology and microscopic features of the fruiting bodies. However, due to the lack of a clear morphological distinction between closely related species and the extensive morphological variation among genera and sometimes even families, the identification of species based solely on morphology is difficult to apply (Grand et al. Citation2011; Park et al. Citation2013; Soo et al. Citation2017). DNA sequence analysis has become an important part of phylogeny and taxonomy of macrofungi (Ortiz-Santana et al. Citation2013; Park et al. Citation2014a; Olariaga et al. Citation2016; Wisitrassameewong et al. Citation2016). The internal transcribed spacer (ITS) region is universally used as a DNA barcode marker for fungal identification, and has been suggested to be the primary fungal barcode (Schoch et al. Citation2012). Translation elongation actor 1 (TEF1) gene, Large subunit (LSU) rRNA, RNA polymerase II subunit (RPB2) gene were also usually used as the DNA barcode marker for macrofungal identification (Moncalvo et al. Citation2006; Park et al. Citation2013; Wang et al. Citation2019). The combination of DNA sequence analysis and morphology studies has considerably improved the efficiency, accuracy, and rapidity of species identification of macrofungi (Park et al. Citation2014b; Cho et al. Citation2015).
Qinling Mountain, located in central China, is the geographical dividing line between the northern and southern sections of China, and is the transitional area of the warm temperate zone and the north subtropical zone. The special geographical location of Qinling Mountain provides a good foundation for the biodiversity of macrofungi. In order to add to our knowledge of macrofungus, we surveyed macrofungi of Qinling Mountain in the Shaanxi province, and a new species of Microstoma was discovered based on the combination of traditional morphology studies and DNA sequence analysis.
Materials & methods
Samples and morphological observations
Macrofungi were collected from Qinling Mountain of the Shaanxi province (China) in April 2020. Specimens were dried with silica gel and deposited in the Herbarium of the Shaanxi Institute of Microbiology (SXIM). Before drying, the specimens were preliminarily identified based on fruiting body morphology and microscopic characteristics using a field light microscope (Olympus BX43, Olympus, Tokyo, Japan) and a scanning electron microscope (SEM, Hitachi S-3400N, Hitachi, Tokyo, Japan). Dried tissue was rehydrated in 5% (w/v) KOH and stained with Melzer’s reagent for measurements and drawings. Asci (20 per sample) and ascospores (20 per sample) were measured. Spore size is presented as follows: (minimum–) mean±1σ (–maximum). The quotient (Q) refers to the length/width ratio of ascospores.
DNA extraction, PCR, and sequencing
Genomic DNA was extracted using the Ezup Column Fungi Genomic DNA Purification Kit (Sangon Biotech, Shanghai, China) according to the manufacturer’s instructions. PCR reactions targeting the ITS, LSU of the DNA samples were performed using the universal primers ITS5 (GGAAGTAAAAGTCGTAACAAGG) / ITS4 (TCCTCCGCTTATTGATATGC) and LR5 (TCCTGAGGGAAACTTCG) / LR0R (ACCCGCTGAACTTAAGC) respectively. PCR was carried out in a total reaction volume of 50 µL: 25 µL 2 × Rapid Taq Master Mix (Tsingke Biological Technology, Beijing, China), 1.5 µL DNA template (100 ng mL−1), 1.5 µL forward and reverse primers (10 mM), and 20.5 µL ddH2O. After an initial denaturation at 95°C for 3 min, an amplification was performed by 30 cycle incubations for 20 s at 95°C, 20 s at 55°C, and 70 s at 72°C, followed by a final extension at 72°C for 5 min. DNA sequencing was completed by Tsingke Biological Technology Co., LTD (Xi’an, China) using an ABI PRISM 3730XL Analyzer (Life Technologies, Gaithersburg, MD, USA) with the same primers used for PCR.
Phylogenetic analysis
DNA sequences were assembled, proofread, and edited using MEGA ver. 7.0 (Sudhir et al. Citation2016). BLASTn search were performed using the ITS and LSU sequences of Microstoma ningshanica. DNA sequences were deposited in GenBank (Accession Nos. MW718686 and MW718687), and reference sequences (Tables and ) were downloaded from GenBank. Multiple alignments were performed using ClustalW of the MEGA 7 software, and ambiguously aligned positions were adjusted manually. Neighbor-joining trees were constructed with MEGA 7 (Kimura 2-parameter model) with 1,000 bootstrap replicates (Saitou Citation1987). For ITS and LSU analyses, Xylaria subescharoidea (NR_171862) and Morchella esculenta (MH870872) were selected as an outgroup, respectively.
Table 1. Fungal species and ITS sequences used in phylogenetic analyses.
Table 2. Fungal species and LSU sequences used in phylogenetic analyses.
Results
Phylogenetic analyses
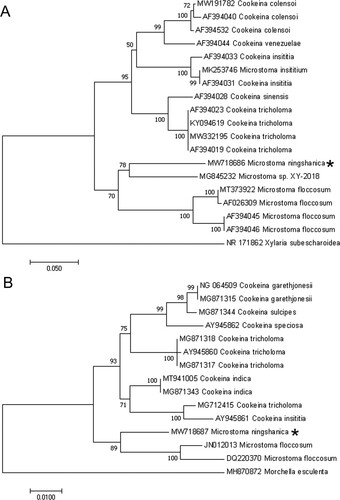
In a BLASTn search using the ITS sequence of Microstoma ningshanica (533 bp), Microstoma sp. XY-2018 (MG845232) was the closest hit with 82.72% sequence identity. Microstoma floccosum resulted in the closest hits (JN012013) with 95.49% sequence identity in a BLASTn search using the LSU sequence (907 bp) (Table ).
Based on an ITS dataset of 19 sequences, the NJ phylogenetic analysis generated a phylogenetic tree (Figure ) in which the new species grouped together with the Microstoma sp. XY-2018 forming a discrete clade with 78% bootstrap support. The results of the NJ phylogenetic analysis of the LSU dataset of 15 sequences showed that the new species grouped together with the similar species Microstoma floccosum with 89% bootstrap support (Figure ). These results indicated that Microstoma ningshanica was a new species of the genus Microstoma.
Taxonomy
Microstoma ningshanica Huo WY, Liu Y, Zhang LG & Li JZ, sp. nov. Figures 2–4.
Mycobank number: MB839332.
Etymology – the specific epithet “ningshanica” refers to NingShan County, China, the region where this species was first discovered.
Type – CHINA. Shaanxi province, AnKang City, NingShan County, HuangHua mountain, 33°46'24.4"N, 118°49'56.8"E, on rotten wood hidden in the ground in a broad-leaved forest [Holotype-SXIM20200016, GeneBank MW718686 (ITS) and MW718687 (LSU)].
Diagnosis – the species is well characterized with its cupulate, hairy and red apothecia, as well as its non-smooth ascospores. The apothecia are nearly spherical to pyriform when young.
Description – apothecia sparse, when young nearly spherical to pyriform, cupulate at maturity, stipitate, with margins dentate, 10−20 mm in diam.; stipes cylindrical, 4−6 cm long and 2−4 mm wide, solid; surface of apothecia and stipes covered with hairs; hairs straight, 0.2−8 mm long and 10-30 µm wide, with acute apices, thick-walled, septal, aggregated at the martin of apothecia and triangular in shape; hymenium surface and receptacle surface red, orange-red, or light orange-red; ectal excipulum of textura angularis, tissue gelatinous, 30−60 µm thick; medullary excipulum two-layered, the outer layer of textura intricata, highly gelatinous, 40−80 µm thick, the inner layer of textura porrecta, non-gelatinous, 70–110 µm thick; asci subcylindrical, 8-spored, J− in Melzer’s reagent, 449−517×16−20.5 µm; ascospores long ellipsoidal to cylindrical (Q = 2.83 ± 0.32), hyaline, non-smooth, (31−)40 ± 3(−45) (13−)14 ± 0.9(−17) µm, without appurtenance on both ends.
Discussion
The present study was conducted on the basis of molecular, light and scanning electron microscopy analyses. The apothecia of this species are nearly spherical to pyriform when young, cupulate at maturity, which indicates that this is a species of Microstoma (Liu and Zhuang Citation2018). BLAST searches and subsequent phylogenetic analysis (Figure and Tables and ) also confirm that it is indeed a species of Microstoma.
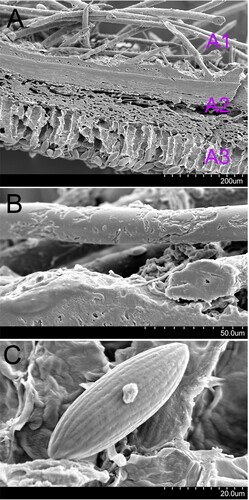
The hairs on the margin of apothecia of this species are aggregated, and are triangular in shape (Figure ), which distinguish M. ningshanica from M. aggregatum, M. floccosum, M. macrosporum, M. radicatum, M. apiculosporum, M. camerunense, and M. protractum (Liu and Zhuang Citation2018). There is no appurtenance on both ends of ascospores (Figure ), which is different from M. apiculosporum and M. camerunense (Liu and Zhuang Citation2018). The ascospore size is 31−45 × 13−17 µm (Figure ), which is larger than the spore of M. camerunense, and smaller than that of M. macrosporum (Liu and Zhuang Citation2018). There are hairs with acute apices for this species (Figure ), while the apices of hairs of M. aggregatum, M. protractum and M. radicatum are blunt (Liu and Zhuang Citation2018).
Figure 3. Light field light micrograph of Microstoma ningshanica. A: hymenium; B: ectal excipulum and medullary excipulum; C: asci and ascospore; D: paraphyses.
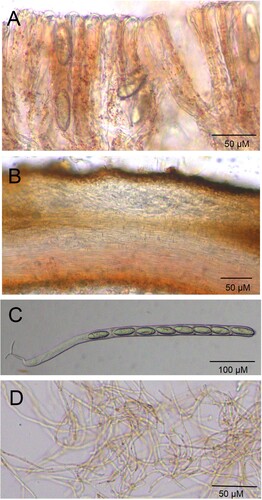
Figure 4. Scanning electron micrograph of Microstoma ningshanica. A: anatomic structure of portion of apothecium (A1, haris; A2, excipulum; A3, hymenium); B: hairs; C: ascospore.
The phylogenetic analysis of the ITS and LSU sequences showed that M. ningshanica was relatively related to M. floccosum (Figure ), but the identity of ITS sequences of the above two species was only about 80% (Table ), while the identity of LSU sequences of them was 95.48% (Table ).
Generally, the differences of morphological characters between M. ningshanica and other species of the genus Microstoma, as well as the less value than standard BLAST matched (less than 97% identity) and a well-differentiated position in the phylogenetic tree show that M. ningshanica is a new species of Microstoma.
Conflicts of interest
The authors report there are no competing interests to declare.
Acknowledgements
We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability
The data that support the findings of this study are openly available in figshare at https://figshare.com/s/caa32081ea13d82bb746, DOI 10.4121/16836547, and the ITS and LUS sequence information of M. ningshanica are also available in GenBank (https://www.ncbi.nlm.nih.gov/nuccore/MW718686.1 and https://www.ncbi.nlm.nih.gov/nuccore/MW718687.1/; Accession Nos. MW718686 and MW718687).
Additional information
Funding
References
- Cho HJ, Park MS, Lee H, Oh SY, Jang Y, Fong JJ, Lim YW. 2015. Four New species of amanita in inje county, Korea. Mycobiology. 43(4):408–414.
- Douanlameli C, Langer E. 2005. – notes on discomycetes (helotiales, pezizales): New species and new records from Cameroon. Mycotaxon. 92:223–237.
- Grand EA, Hughes KW, Petersen RH. 2011. Relationships within lentinus subg. lentinus (polyporales, agaricomycetes), with emphasis on sects. lentinus and tigrini. Mycol Prog. 10:399–413.
- Guy D, He Q, Chaumeton JP, Chauveau C. 1997. – cookeina insititia, a new discomycetes from China. Acta Bot Yunnanica. 19(2):128–130.
- Harada Y, Kudo SI. 2000. Microstoma macrosporum stat. nov., a new taxonomic treatment of a vernal discomycete (sarcoscyphaceae, pezizales). Mycoscience. 41:275–278.
- Kanouse BB. 1948. The genus plectania and its segregates in North America. Mycologia. 40:482–497.
- Kirk PM, Cannon PF, Minter DW, Stalpers JA. 2008. Dictionary of the fungi. Wallingford: Commonwealth Mycological Institute. 428 p.
- Liu TW, Zhuang WY. 2018. A new species of microstoma (sarcoscyphaceae) from China with a key to known species of the genus. Mycosystema. 37(5):555–558.
- Moncalvo JM, et al. 2006. The cantharelloid clade: dealing with incongruent gene trees and phylogenetic reconstruction methods. Mycologia. 98:937–948.
- Olariaga I, Moreno G, Manjón JL, Salcedo I, Buyck B. 2016. Cantharellus (cantharellales, basidiomycota) revisited in Europe through a multigene phylogeny. Fungal Divers. 83:1–30.
- Ortiz-Santana B, Lindner DL, Miettinen O, Justo A, Hibbett DS. 2013. A phylogenetic overview of the antrodia clade (basidiomycota, polyporales). Mycologia. 105(6):1391–1411.
- Park MS, Fong JJ, Lee H, Oh SY, Jung PE, Min YJ, Seok SJ, Lim YW. 2013. Delimitation of russula subgenus amoenula in Korea using three molecular markers. Mycobiology. 41:191–201.
- Park MS, Lee H, Oh SY, Jung PE, Seok SJ, Fong JJ, Lim YW. 2014a. Species delimitation of three species within the russula subgenus compacta in Korea: R. eccentrica, R. nigricans, and R. subnigricans. J Microbiol. 52(8):631–638.
- Park MS, Quan Y, Jung PE, Oh SY, Lim YW. 2014b. Re-evaluation of the genus antrodia(polyporales, basidiomycota) in Korea. Mycobiology. 42:114–119.
- Saitou N. 1987. – The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 4(4):406–425.
- Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W. 2012. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc Natl Acad Sci USA. 109:6241–6246.
- Soo PM, Jin CH, Kyu KN, Young PJ, Hyun L, Hyeong PK, Min-Ji K, Jae-Jin K, Changmu K, Woon LY. 2017. Ten New recorded species of macrofungi on ulleung island, Korea. Mycobiology. 45:286–296.
- Sudhir K, Glen S, Koichiro T. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Wang XC, Liu TZ, Chen SL, Li Y, Zhuang WY. 2019. A four-locus phylogeny of rib-stiped cupulate species of helvella (helvellaceae, pezizales) with discovery of three new species. MycoKeys. 60:45–67.
- Wang YZ. 2004. – A new species of microstoma from Taiwan. Mycotaxon. 89:119–122.
- Wisitrassameewong K, Looney BP, Le HT, De Crop E, Das K, Van dP K, Eberhardt U, Jiayu G, Stubbe D, Hyde KD. 2016. Lactarius subgenus russularia (basidiomycota, russulales): novel Asian species, worldwide phylogeny and evolutionary relationships. Fungal Biol. 120(12):1554–1581.