Abstract
Autophagy is important for cell survival during stress and nutrient recycle. However, the mechanisms involved in regulating autophagy in cucumber have not been determined. Here, we analyzed the transcript abundance of key autophagy genes in the conjugation pathway including ATG3, ATG4, ATG5, ATG7, ATG8, ATG10, ATG12 and ATG16 in response to nitrogen deficiency, pathogen infection and oxidative stress in Cucumis sativus. ATG8 protein abundance was analyzed using immunoblot analysis. Seven out of 13 autophagy genes studied have splice variances including ATG3, ATG4, ATG5, ATG8b, ATG8c, ATG8e and ATG10. All except ATG5 have changes in transcript variances abundance upon nitrogen starvation, indicating regulation via alternative splicing is pervasive in autophagy genes. This is the first report of alternative splicing in autophagy genes in cucumbers. However, transcription control is also important, especially in ATG4, and those without transcript variances. Moreover, different environment cues regulate autophagy genes through different mechanisms. The ATG8 protein tag was also regulated at the protein level through post-translational modification and blockage of degradation. This work shows that the regulation of autophagy in cucumber is complex and involves many mechanisms. Better understanding of autophagy regulation would thus help breeding plants that perform better even under environmental stresses.
Introduction
Autophagy, a nonspecific degradation and recycling of intracellular components, is important for cell survival, nutrient recycling and nutrient remobilization in various organisms (Mizushima Citation2007). In plants, it is particularly essential for mitigating the effects of stresses and environmental challenges (Liu and Bassham Citation2012). Various abiotic stresses including salt, drought, and hypoxia result in oxidative stress in plants. Autophagy is then induced to degrade proteins and cellular compartments damaged by the stress. Autophagy also plays an important role in the plant immune system to restrict pathogen invasion via hypersensitive response. Defects in the autophagy pathway cause plant to be sensitive to environmental stress, both biotic and abiotic, confirming the role of this process in stress responses (for a detail description of autophagy and plant stress response, see Signorelli et al. Citation2019 and Su et al. Citation2020). The transcriptional control of genes involved in the autophagy process in responses to salt, drought and dark treatments, as well as phytohormones was confirmed in various plants (Xia et al. Citation2011; Pei et al. Citation2014; Wang et al. Citation2020a). Given the role of autophagy under stress conditions, understanding the regulation of this process could provide a way to improve crop adaptation and tolerance against environmental challenges (Liu et al. Citation2009; Sun et al. Citation2018a; Huo et al. Citation2020).
Autophagy is mediated by AuTophaGy-related or ATG proteins. The cascade of activations leading to the autophagosome has been reviewed by Yang and Klionsky (Citation2009) and involves several elements. ATG12 is activated by ATG7 before transferred to ATG10, which catalyzes the conjugation of ATG12 to ATG5. The C-terminus of ATG8 is removed by ATG4, a protease. Processed ATG8 is then activated by ATG7 and conjugated to phosphatidylethanolamine (PE) lipid by ATG3 and ATG12-ATG5-ATG16 complex. ATG8-PE is then integrated into membranes to subsequently form autophagosomes (for a detailed description of these molecular mechanisms, see Yang and Klionsky (Citation2009)).
Alternative Splicing (AS) is a process in which more than one form of mRNA was produced as different splice sites were used. Up to 90% and 70% of all genes in human and Arabidopsis have splice variances (Pan et al. Citation2008; Calixto et al. Citation2018). This process alters the proteome landscape through changes in protein isoforms and quantity as well as introduces premature stop codons that induce nuclear-mediated decay and cis-regulatory elements targeted by miRNA and other regulatory machineries (Kazan Citation2003; Filichkin and Mockler Citation2012). In plants, splice variances were detected in association with differences in tissues, developmental stages and play an important role in plant response to environmental signals and stresses (Capovilla et al. Citation2018; Sun et al. Citation2018b; Zheng et al. Citation2019; Gao et al. Citation2021; John et al. Citation2021; Yan et al. Citation2021). Environmental stresses found to cause change in splicing in plants are pathogen infection, herbivore attack, extreme temperature, salinity, hypoxia, drought and deficiency of micronutrients (Laloum et al. Citation2018; Ganie and Reddy Citation2021; Lam et al. Citation2022). Genes targeted for AS upon stresses often have regulatory functions including transcription factors, splicing factors and protein kinase (Palusa et al. Citation2007; Rigo et al. Citation2019; Punzo et al. Citation2020). For example, a transcript variant encoding non-functional transcription factor was alternatively spliced to produce functional DREB2-type transcription factors in response to drought in rice (Matsukura et al. Citation2010). AS causes changes in protein structures of animal autophagy genes including ATG5, ATG7 and ATG8 (Liu et al. Citation2013; Ouyang et al. Citation2013; Park et al. Citation2016). Splice variances in autophagy genes were also detected in many plants including, Arabidopsis, maize, rice (Chung et al. Citation2009; Wang et al. Citation2020b). In addition, AS of ATG4 during grape berry development was recently reported (Ma et al. Citation2023). However, the extent of how AS regulates autophagy in plant responses to stresses is not yet well understood.
Cucumber (Cucumis sativus, Cucurbitaceae family) has been used as a model plant for research in various aspects of abiotic stresses, nutrient limitation and pathogen resistance (Wei et al. Citation2015; Xin et al. Citation2017; Dai et al. Citation2022). With the Cucumber genome available (Huang et al. Citation2009), autophagy gene orthologs were previously identified and their expression assessed (Han et al. Citation2021). Extensive transcriptome sequencing also provided abundant transcript data that could be explored by studying the role of AS in the responses to abiotic and biotic stress conditions.
Since AS regulates stress responses, we studied the regulation of AS upon autophagy genes and autophagy activity to establish if these processes take part in the stress response in cucumber. This work investigated the roles of AS, transcription and protein modifications that might affect functions of autophagy in cucumber. Here, we focus on key autophagy genes in the conjugation pathway, including ATG3, ATG4, ATG5, ATG7, ATG8 ATG10, ATG12 and ATG16. The expression of these autophagic genes and their splice variances under nitrogen starvation, oxidative stress and pathogen infection was analyzed, as well as ATG8 protein level.
Materials and methods
Plant treatments
The nitrogen starvation experiment was performed using 5-day-old seedlings of C. sativus grown hydroponically in liquid 1/2 MS medium with nitrogen before switching to 1/2 MS medium with and without nitrogen for 2, 4 and 6 days (Oka et al. Citation2012). The nitrogen source lacking in 1/2 MS medium without nitrogen was 21 mM NH4NO3 and 19 mM KNO3 with the supplement of 18 mM KCl. First true leaves were collected from seedlings before the treatment (day 0) as well as the other six time points (day 2 +N; day 2 -N; day 4 +N; day 4 -N; day 6 +N; day 6 -N) for subsequent analysis. Oxidative stress was conducted using 2-week-old seedlings grown on soil. These plants were sprayed once with 50 µM Methyl Viologen (MV) prepared in 0.05% (v/v) Tween 20 (Song et al. Citation2005), and the first true leaves were collected before treatment as well as at 3, 6, 9, 12 and 15 days after the treatment. For pathogen responses, seven-day-old seedlings of C. sativus grown on soil were inoculated with sporangia of the fungi causing downy mildew disease (Pseudoperonospora cubensis) with the concentration of 1 × 104 sporangia mL−1. Cotyledons were collected before inoculation and at 6, 12, 24, 72 and 168 h after inoculation. For each experiment, three replicates were collected for each time point before being frozen in liquid nitrogen and stored at −80°C.
RNA isolation and reverse transcription
RNA was isolated from cucumber samples using CTAB supplemented with 1% beta-mercaptoethanol and 4% sodium dodecyl sulfate and subsequent extraction using chloroform and trizol. The RNA was then precipitated using ethanol with 0.2 M NaCl. Total RNA was treated with DNaseI to remove genomic DNA. First-strand cDNA synthesis was performed using one microgram of RNA, 0.25 mM oligo (dT) primers and 2 mM dNTP (Sreeratree et al. Citation2022). The mixture was denatured for 5 min at 65°C before adding MMuLV Reverse Transcriptase (Biotechrabbit, Germany) and incubated for 1 h at 42°C.
Quantitative real-time PCR
The real-time PCR analysis was performed in duplicate assays in 96-well plates; each 10 µl reaction consisted of 2× QPCR Green Master Mix (Biotechrabbit, Germany), 5 µmol forward and reverse primers and 1 ng cDNA. The primers were designed to span the junction that differs between transcript variances and the sequences of primer are shown in Table . The data of Ct values obtained from two different experimental RNA samples were directly normalized to a housekeeping gene, CsActin. The fold-change in the expression of the gene of interest between each time point was then calculated as 2^ (−ΔΔCt). Statistical differences in the level expression compared to the expression at time point 0 or that of ATG7 were analyzed with student’s t-test with two-tailed distribution.
Table 1. Primers used in real-time RT-PCR.
Protein extraction and immunoblot analysis
Samples were ground and resuspended in extraction buffer (50 mM Tris–HCl pH 8.0, 150 mM NaCl, 2 mM EDTA, 10% (v/v) glycerol, 0.5% (v/v) IGEPAL CA-360, 1 mM phenylmethylsulfonyl fluoride) on ice. Crude proteins were separated on 18% SDS-PAGE before stained using Coomassie blue or transferred onto a PVDF membrane (GE Healthcare). Immunodetection of ATG8 was performed using anti-CrATG8 (Pérez-Pérez et al. Citation2010), with a working concentration of the primary antibody at 1:1000 in PBS containing 2.5% BSA. The Coomassie Brilliant Blue staining of the gel was used to estimate the amount of protein loaded.
Results
Splice variance in CsATG genes and its effects on the transcripts and protein-coding sequences
The splice variances of each ATG gene in the conjugation pathway were identified based on the Cucumber and NCBI databases. Only the splice variances detected by RT–PCR under any of the three conditions tested (nitrogen starvation, pathogen response and oxidative stress) were listed in Table , with the differences among splice variance described in detail as well as the condition that the splice variance was not detected. Out of 13 ATG genes assessed, seven genes contain multiple splice variances, including four variances for ATG3 and ATG8e, two variances for ATG4, ATG5, ATG8b and ATG8c and five variances for ATG10. Among these, only ATG4 and ATG5 have splice variances that affect the coding regions: N154H for CsATG4 × 1 and deletion at 98–127 amino acid position in CsATG5 × 2. The deletion is in the middle of conserved domain with Pfam ID of APG5 PF04106 and likely to affect the protein function. ATG3 variances alter 3’UTR after the stop codon, while the other four genes have splice variances that differ in 5’UTR with different forms of deletion. Because the splice variances of CsATG3 are clustered within 20 bp after the stop codons, this region of CsATG3 might have a regulatory function. That more than half of the genes in the cucumber conjugation pathway have multiple splice variances suggests the importance of AS in the autophagy regulation. In addition, various modes of regulation are likely to be displayed as the AS observed in these genes affects protein isoforms and UTRs. This result also suggests the presence of cis-acting elements in the UTRs that could affect mRNA stability or translation efficiency.
Table 2. Summary of transcript variances confirmed in this work.
Effects of environmental stresses on the abundance of the ATG splice variances
To understand the effects of environmental stress on the regulation of autophagy, we assessed the expression and the transcript variances of these ATG genes in seedlings under nitrogen starvation, and further investigated the key components (ATG7, ATG8 and ATG12) under pathogen infection and oxidative stress. The expression level in the control of all ATG8 genes was normalized to that of ATG7 to determine the most abundant isoforms, with ATG8f consistently has the highest level of expression (data not shown). Changes in transcript abundance upon treatments indicate the regulation through transcriptional control. For genes with multiple splice variances, different patterns of responses among splice variances suggest the role of AS in regulating the response to stresses.
Nitrogen starvation increased transcript abundance for all ATG genes without splice variances including ATG16, ATG7, ATG8a, ATG8d, ATG8f and ATG12, indicating the prominent role of transcription control on autophagy (Figure and Figure A). For ATG genes with multiple splice variances, all except ATG5 had different patterns of responses among splice variances. For example, only ATG3 × 1 and ATG3 × 3 abundance increased upon N starvation, but not the other two variances. In addition, a significant increase in abundance was detected for ATG10 × 1, ATG10 × 3 and ATG10 × 4 in all 3 samples under starvation, whereas ATG10 × 2 and ATG10 × 5 abundance decreased on day 4. The difference in response among splice variances of ATG8 is also detectable, as the increase in ATG8bx1 and ATG8cx1 were more prominent than their splice variances, and ATG8ex1 abundance significantly decreased upon starvation while the abundance of its variances increased. These results illustrate the pervasiveness of AS in the regulation of autophagy under nutrient stress.
Figure 1. Relative expression of ATG3, ATG4, ATG5, ATG10 and ATG16 genes and their splice variances under nitrogen starvation. * indicates statistically significant difference (p-value less than 0.05) from the expression at 0 h based on Student’s t-test.
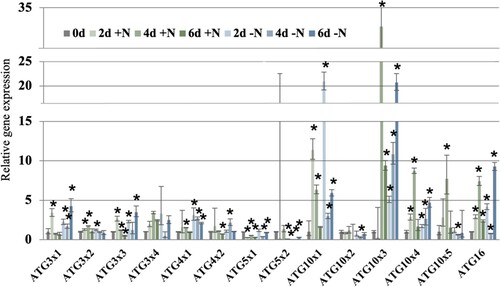
Figure 2. Relative expression of ATG7, ATG8 and ATG12 genes and their splice variances under nitrogen starvation (A), pathogen response (B) and oxidative stress (C). * indicates statistically significant difference (p-value less than 0.05) from the expression at Day 0 based on Student’s t-test.
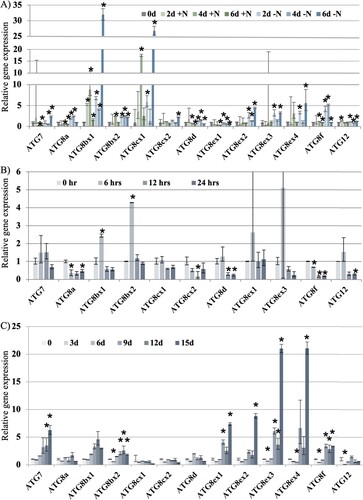
Downey mildew inoculation did not change the expression of ATG7, ATG8cx1, ATG8ex1 and ATG8ex3 (Figure B). However, both variances of ATG8b had increases in expression by 6 h before reducing to levels before inoculation by 12 h. Furthermore, ATG8a, ATG8cx2, ATG8d, ATG8f and ATG12 show a reduction in gene expression at 24 h after inoculation. This result indicates that the ATG8b might function as an early response to the pathogen infection, and this infection could block the accumulation of ATG transcripts potentially by inhibiting transcriptions or promoting transcript degradation. As a significance reduction in abundance among ATG8c transcripts was only detected for ATG8cx2, AS might also play a role in response to pathogen response.
Upon MV treatment, among the ATG genes without splice variances including ATG7, ATG8a, ATG8d, ATG8f and ATG12, only ATG7 and ATG8f increase in expression especially in days 12 and 15 (Figure C). In addition, in this group, a significance reduction in expression at 3 days after treatment was detected for ATG8f and ATG12. For ATG genes with splice variances including ATG8b, ATG8c and ATG8e, there was no significance changes in expression detected for both variances of ATG8c, indicating a lack of either transcriptional or splicing control for this gene under oxidative stress. However, different response patterns among splice variances were detected in ATG8b as only ATG8bx2 abundance was increased, but not ATGbx1. In addition, ATG8ex1 and ATG8ex3 increase in abundance happened earlier than the other variances. Therefore, the regulation of ATG expression was mediated through both transcription and AS. Moreover, ATG8f, ATG8bx2, and all four splice variances of ATG8e showed the highest transcript abundance increase under oxidative stress, suggesting that they are likely the main elements involved in the response to oxidative stress.
These results suggest that the transcriptional control as well as AS play a role in regulating ATG genes during abiotic and biotic stress response in cucumber, with different genes becoming targets of regulation by different mechanisms under each condition.
Effects of environmental stresses on ATG8 proteins
The level of ATG8 protein is often used as an indicator of autophagy levels since ATG8-PE conjugate is incorporated into the autophagosomal membrane (Klionsky et al. Citation2008). Therefore, assessing the effects of environmental stresses on ATG8 proteins allows better understanding of their effects on autophagy level in general. Here, the abundance and the modification of cucumber ATG8 proteins under the stress conditions were assessed through western blot using rabbit anti-CrATG8 antibody. The predicted size of free CsATG8 (fATG8) is around 14 kDalton, whereas the conjugated form of CsATG8 (cATG8) appeared slightly smaller than the free form (Chung et al. Citation2009). Bands with the predicted size of fATG8 were detected in crude extracts from seedlings grown with and without nitrogen (Figure D and Figure A). However, once normalized to the Coomassie gel used as a loading control, there is a slight increase in ATG8 protein abundance from 1 to 1.6-fold under nitrogen starvation.
Figure 3. Phenotype of cucumber and immunoblot analysis of ATG8 protein abundance under nitrogen starvation (A, D), pathogen response (B, E) and oxidative stress (C, F). CsATG8 was detected using antibodies specific to CrATG8 (upper panel). Coomassie staining was used as a loading control (lower panel). Black, red and blue arrowheads indicate free CsATG8, conjugated CsATG8 and the band used for estimating the amount of total protein loaded, respectively. The numbers indicate the abundance of CsATG8 normalized to a loading control compared to control.
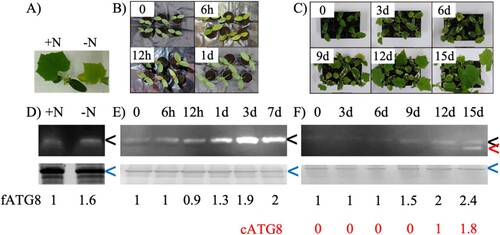
For the level of CsATG8 protein in response to pathogen infection, the sample for protein extraction was collected from both the early phase at 0, 6, 12, 24 h and the late phase at day 3 and day 7 after inoculation. Only one band at the predicted size of fATG8 was detected in all time points. The intensity of the band also increased along with time after inoculation, with a clear increase starting at 1 day (Figure E and Figure B). The ATG8 protein abundance increased from 1.3-fold after 1 day to 1.9-fold after 3 days and remained at a similar level after 7 days, suggesting that the accumulation occurred mostly between 3 and–7 days after inoculation. Upon treatment with MV, the band for fATG8 was detected with increased intensity starting from 9 days after treatment (1.5, 2 and 2.4 folds in days 9, 12 and 15, respectively) (Figure F and Figure C). In addition, another band for cATG8 was detected starting from 12 days after treatment and increase in intensity to 1.8-fold in day 15.
The increase in ATG8 protein abundance during these abiotic and biotic stresses indicated the roles of ATG8 protein and autophagy in cucumber responses to stress. In addition, the detection of the smaller size band suggested a role of post-translation modification in oxidative stress response.
Discussion
Our work shows that AS is frequent in the autophagy response, with 7 out of 13 cucumber autophagy genes studied here having multiple splice variances. In animals, AS of autophagy genes was reported to change the coding regions that affect protein structures of ATG5, ATG7 and ATG8 (Liu et al. Citation2013; Ouyang et al. Citation2013; Park et al. Citation2016). Here, AS of ATG5 allows changes in the coding sequence that potentially alter the protein function. However, as both splice variances of ATG5 have the same pattern of changes under N starvation, ATG5 was likely regulated mainly through transcriptional control under nutrient stress. In some cases, the regulations through the transcription control or AS could be selective depending on the stress conditions. AS affects ATG8b and ATG8e response to nitrogen starvation and oxidative stress but not during pathogen infection, whereas regulation through AS of ATG8c occurs during nitrogen starvation and pathogen infection but not under oxidative stress. In addition, for the ATG genes with splice variance, their expression was mainly regulated through both AS and transcription concurrently. These results illustrate the complex role of AS in regulated specific responses to stresses in plants (Wang et al. Citation2020b; Chaudhary and Kalkal Citation2021).
Transcription control is also important for cucumber response to abiotic and biotic stresses. Indeed, ATG8f, one of the genes with no transcript variance, has the highest expression level in the control group for all three experiments. Even though such high expression level might be partly due to the differences in the amplification efficiency of primers used, the increases of ATG8f expression in response to N starvation and oxidative stress suggest that ATG8f could be one of the key autophagy genes regulated by transcriptional controls during environmental stresses in cucumber. In rice, OsATG8a is the most abundant transcript and OsATG8d the most strongly induced upon N starvation and abiotic stress (Xia et al. Citation2011). The differences among the most abundant ATG8 isoforms and their role in response to environmental cues in different plant species illustrate the evolutionary divergence in the function of this gene. The reduction in ATG8f transcript abundance within a day after Downey mildew inoculation also indicates its significance as a target of transcription suppression in response to the pathogen. Wheat ATG8 also shows reduced expression upon pathogen infection in susceptible plants but is upregulated in resistant plants (Zhang et al. Citation2020). The expression of ATG12, another protein tag for the conjugative pathway, did not change significantly under the three treatments presented here. Given that ATG12 expression was previously shown to increase upon N starvation in rice and maize (Chung et al. Citation2009; Xia et al. Citation2011), this suggests a minor role of ATG12 under stress conditions in cucumber. In cucumber, ATG7 was responsive to abiotic stress, but not to biotic stress. ATG7 expression increases in maize under N starvation and carbon starvation, in Arabidopsis under heat treatment and sucrose starvation and in rice under salt treatment and nitrogen starvation (Rose et al. Citation2006; Chung et al. Citation2009; Xia et al. Citation2011; Zhou et al. Citation2013). These data indicate that ATG7 is transcriptionally regulated by abiotic stress across species.
An additional layer of control in autophagy is at the protein level. The increase in ATG8 protein abundance is consistent with the increases of transcripts of ATG8 gene family during N starvation, oxidative stress and pathogen infection, and this is quite conserved among plant species (Chung et al. Citation2009; Pérez-Pérez et al. Citation2010; Pérez-Martín et al. Citation2014). ATG8a and ATG8f are potentially responsible for the increases in ATG8 proteins during N starvation, and ATG8b, ATG8e and ATG8f are during oxidative stress. The selective induction of different ATG8 isoforms under different conditions was also observed in Arabidopsis, maize, rice and wheat (Yoshimoto et al. Citation2004; Chung et al. Citation2009; Xia et al. Citation2011; Pei et al. Citation2014). In contrast, upon pathogen infection, only ATG8b transcript increased, whereas the rest of the ATG8 family reduced in expression after only one day of inoculation. The increases in ATG8b transcript abundance seemed too low to be accounted for the increased protein level. This increase in ATG8 protein in contrast to the little increase in ATG8 transcripts may indicate either increase in translation or delay in protein degradation as a result of pathogen infection.
Post-translation modification of ATG8 through the conjugation of ATG8 onto PE before its integration into autophagosomal membrane is critical for ATG8 function (Yang and Klionsky Citation2009). In cucumber, a smaller band representing the PE-conjugated form of ATG8 became detectable after 12 days of oxidative stress confirming the modification of ATG8 and its role in oxidative responses, similar to reports in other organisms (Chung et al. Citation2009; Suttangkakul et al. Citation2011). In addition, the increase in transcript abundance of ATG7, the gene critical for the ATG8-PE conjugation, in days 12 and 15 also correlates with the detection of the PE-conjugated form of ATG8. Such increases in transcript and protein abundance in days 12 and 15 after MV treatment could also be due to a developmental process, namely senescence, since these first true leave samples were collected from 26- and 29-day-old plants at the age when cotyledon senescence can be detected (Kim Citation2004). ATG8-PE conjugate accumulation in cucumber in response to senescence is consistent with that detected in leaf senescence in maize (Chung et al. Citation2009). No detection of the conjugated form in N starvation and pathogen infection could imply a high turnover rate of the conjugated form or the lack of conjugation activity under these conditions. For N starvation, the first scenario is more likely as the conjugated form is normally present in the N starvation condition as shown in Arabidopsis, maize and Chlamydomonas (Chung et al. Citation2009; Pérez-Pérez et al. Citation2010; Suttangkakul et al. Citation2011). In pathogen infection, lack of conjugation could also be the case, based on the decrease in transcript abundance of most ATG8 genes. The targets of gene downregulation for Pseudoperonospora appeared to be ATG8s and ATG12, which encode protein tags, but not ATG7 (Zhang et al. Citation2020).
This work shows the regulation of autophagy genes in the conjugation pathway in cucumber under three different stress conditions. In addition to the transcriptional control, the autophagy process was also regulated post-transcriptionally both through AS and at the protein level, which can affect protein function as well as protein degradation. This work is also the first detailed analysis of splice variances in autophagy genes in plants. Further work will establish the exact mechanisms of protein accumulation and cis-regulatory elements in the 5’ and 3’UTRs that can deepen our understanding of cucumber tolerance under environmental stresses and suboptimal conditions.
Acknowledgement
We thank Dr. Sompid Samipak for mentoring and Chia Tai Co., Ltd. for technical support in Pseudoperonospora cubensis inoculation.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Author contributions
AS, PT and SV conceived the project and designed experiments. PT, TC, WP and PN performed the experiments. PT, PN, SV and AS analyzed the data. PT, SV, LG and AS interpreted the data, prepared Figures and Tables and wrote the manuscript. All authors have approved the final version.
Data availability
The data that support the findings of this study are openly available in Mendeley Data: http://doi.org/10.17632/v6f2rhv42z.2.
Additional information
Funding
References
- Calixto CP, Guo W, James AB, Tzioutziou NA, Entizne JC, Panter PE, Knight H, Nimmo HG, Zhang R, Brown JW. 2018. Rapid and dynamic alternative splicing impacts the Arabidopsis cold response transcriptome. Plant Cell. 30(7):1424–1444.
- Capovilla G, Delhomme N, Collani S, Shutava I, Bezrukov I, Symeonidi E, de Francisco Amorim M, Laubinger S, Schmid M. 2018. PORCUPINE regulates development in response to temperature through alternative splicing. Nat Plants. 4(8):534–539.
- Chaudhary S, Kalkal M. 2021. Rice transcriptome analysis reveals nitrogen starvation modulates differential alternative splicing and transcript usage in various metabolism-related genes. Life. 11(4):285.
- Chung T, Suttangkakul A, Vierstra RD. 2009. The ATG autophagic conjugation system in maize: ATG transcripts and abundance of the ATG8-lipid adduct are regulated by development and nutrient availability. Plant Physiol. 149(1):220–234.
- Dai Z, Dong S, Miao H, Liu X, Han J, Li C, Gu X, Zhang S. 2022. Genome-wide identification of TIFY genes and their response to various pathogen infections in cucumber (Cucumis sativus L.). Sci Hortic. 295:110814.
- Filichkin SA, Mockler TC. 2012. Unproductive alternative splicing and nonsense mRNAs: a widespread phenomenon among plant circadian clock genes. Biol Direct. 7(1):1–15.
- Ganie SA, Reddy AS. 2021. Stress-induced changes in alternative splicing landscape in rice: functional significance of splice isoforms in stress tolerance. Biology. 10(4):309.
- Gao R, Luo Y, Yun F, Wu X, Wang P, Liao W. 2021. Genome-wide identification, expression profile, and alternative splicing analysis of CAMTA family genes in cucumber (Cucumis sativus L.). Agronomy. 11(9):1827.
- Han Y, Yang Y, Wang Y, Elsheery NI, Ding G. 2021. Genome-wide identification and expression analysis of autophagy genes in cucumber. Res Sq. doi:10.21203/rs.3.rs-359244/v1.
- Huang S, Li R, Zhang Z, Li LI, Gu X, Fan W, Lucas WJ, Wang X, Xie B, Ni P, et al. 2009. The genome of the cucumber, Cucumis sativus L. Nat Genet. 41(12):1275–1281.
- Huo L, Guo Z, Jia X, Sun X, Wang P, Gong X, Ma F. 2020. Increased autophagic activity in roots caused by overexpression of the autophagy-related gene MdATG10 in apple enhances salt tolerance. Plant Sci. 294:110444.
- John S, Olas JJ, Mueller-Roeber B. 2021. Regulation of alternative splicing in response to temperature variation in plants. J Exp Bot. 72(18):6150–6163.
- Kazan K. 2003. Alternative splicing and proteome diversity in plants: the tip of the iceberg has just emerged. Trends Plant Sci. 8(10):468–471.
- Kim DJ. 2004. A study of cotyledon senescence in cucumber (Cucumis sativus L.) based on expressed sequence tags and gene expression. J Plant Biol. 47(3):244–253.
- Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, et al. 2008. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 4(2):151–175.
- Laloum T, Martín G, Duque P. 2018. Alternative splicing control of abiotic stress responses. Trends Plant Sci. 23(2):140–150.
- Lam PY, Wang L, Lo C, Zhu FY. 2022. Alternative splicing and its roles in plant metabolism. Int J Mol Sci. 23(13):7355.
- Liu C, Ma H, Wu J, Huang Q, Liu JO, Yu L. 2013. Arginine68 is an essential residue for the C-terminal cleavage of human Atg8 family proteins. BMC Cell Biol. 14(1):1–12.
- Liu Y, Bassham DC. 2012. Autophagy: pathways for self-eating in plant cells. Annu Rev Plant Biol. 63:215–237.
- Liu Y, Xiong Y, Bassham DC. 2009. Autophagy is required for tolerance of drought and salt stress in plants. Autophagy. 5(7):954–963.
- Ma SH, He GQ, Navarro-Payá D, Santiago A, Cheng YZ, Jiao JB, Li H-J, Zuo D-D, Sun H-T, Pei M-S, et al. 2023. Global analysis of alternative splicing events based on long-and short-read RNA sequencing during grape berry development. Gene. 852:147056.
- Matsukura S, Mizoi J, Yoshida T, Todaka D, Ito Y, Maruyama K, … Yamaguchi-Shinozaki K. 2010. Comprehensive analysis of rice DREB2-type genes that encode transcription factors involved in the expression of abiotic stress-responsive genes. Mol Genet Genomics. 283(2):185–196.
- Mizushima N. 2007. Autophagy: process and function. Genes Dev. 21(22):2861–2873.
- Oka M, Shimoda Y, Sato N, Inoue J, Yamazaki T, Shimomura N, Fujiyama H. 2012. Abscisic acid substantially inhibits senescence of cucumber plants (Cucumis sativus) grown under low nitrogen conditions. J Plant Physiol. 169(8):789–796.
- Ouyang DY, Xu LH, He XH, Zhang YT, Zeng LH, Cai JY, Ren S. 2013. Autophagy is differentially induced in prostate cancer LNCaP, DU145 and PC-3 cells via distinct splicing profiles of ATG5. Autophagy. 9(1):20–32.
- Palusa SG, Ali GS, Reddy AS. 2007. Alternative splicing of pre-mRNAs of Arabidopsis serine/arginine-rich proteins: regulation by hormones and stresses. Plant J. 49(6):1091–1107.
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. 2008. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 40(12):1413–1415.
- Park SM, Ou J, Chamberlain L, Simone TM, Yang H, Virbasius CM, Ali A, Zhu L, Mukherjee S, Raza A, Green MR. 2016. U2AF35 (S34F) promotes transformation by directing aberrant ATG7 pre-mRNA 3′ end formation. Mol Cell. 62(4):479–490.
- Pei D, Zhang W, Sun H, Wei X, Yue J, Wang H. 2014. Identification of autophagy-related genes ATG4 and ATG8 from wheat (Triticum aestivum L.) and profiling of their expression patterns responding to biotic and abiotic stresses. Plant Cell Rep. 33(10):1697–1710.
- Pérez-Martín M, Pérez-Pérez ME, Lemaire SD, Crespo JL. 2014. Oxidative stress contributes to autophagy induction in response to endoplasmic reticulum stress in Chlamydomonas reinhardtii. Plant Physiol. 166(2):997–1008.
- Pérez-Pérez ME, Florencio FJ, Crespo JL. 2010. Inhibition of target of rapamycin signaling and stress activate autophagy in Chlamydomonas reinhardtii. Plant Physiol. 152(4):1874–1888.
- Punzo P, Grillo S, Batelli G. 2020. Alternative splicing in plant abiotic stress responses. Biochem Soc Trans. 48(5):2117–2126.
- Rigo R, Bazin J, Crespi M, Charon C. 2019. Alternative splicing in the regulation of plant–microbe interactions. Plant Cell Physiol. 60(9):1906–1916.
- Rose TL, Bonneau L, Der C, Marty-Mazars D, Marty F. 2006. Starvation-induced expression of autophagy-related genes in Arabidopsis. Biol Cell. 98(1):53–67.
- Signorelli S, Tarkowski ŁP, Van den Ende W, Bassham DC. 2019. Linking autophagy to abiotic and biotic stress responses. Trends Plant Sci. 24(5):413–430.
- Song XS, Hu WH, Mao WH, Ogweno JO, Zhou YH, Yu JQ. 2005. Response of ascorbate peroxidase isoenzymes and ascorbate regeneration system to abiotic stresses in Cucumis sativus L. Plant Physiol Biochem. 43(12):1082–1088.
- Sreeratree J, Butsayawarapat P, Chaisan T, Somta P, Juntawong P. 2022. RNA-Seq reveals waterlogging-triggered root plasticity in mungbean associated with ethylene and jasmonic acid signal integrators for root regeneration. Plants. 11(7):930.
- Su W, Bao Y, Yu X, Xia X, Liu C, Yin W. 2020. Autophagy and its regulators in response to stress in plants. Int J Mol Sci. 21(23):8889.
- Sun X, Wang P, Jia X, Huo L, Che R, Ma F. 2018a. Improvement of drought tolerance by overexpressing MdATG18a is mediated by modified antioxidant system and activated autophagy in transgenic apple. Plant Biotechnol J. 16(2):545–557.
- Sun Y, Hou H, Song H, Lin K, Zhang Z, Hu J, Pang E. 2018b. The comparison of alternative splicing among the multiple tissues in cucumber. BMC Plant Biol. 18(1):1–12.
- Suttangkakul A, Li F, Chung T, Vierstra RD. 2011. The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell. 23(10):3761–3779.
- Wang P, Nolan TM, Yin Y, Bassham DC. 2020a. Identification of transcription factors that regulate ATG8 expression and autophagy in Arabidopsis. Autophagy. 16(1):123–139.
- Wang Y, Xu J, Ge M, Ning L, Hu M, Zhao H. 2020b. High-resolution profile of transcriptomes reveals a role of alternative splicing for modulating response to nitrogen in maize. BMC Genomics. 21(1):1–19.
- Wei L, Deng XG, Zhu T, Zheng T, Li PX, Wu JQ, Zhang DW, Lin HH. 2015. Ethylene is involved in brassinosteroids induced alternative respiratory pathway in cucumber (Cucumis sativus L.) seedlings response to abiotic stress. Front Plant Sci. 6:982.
- Xia K, Liu TAO, Ouyang J, Wang R, Fan T, Zhang M. 2011. Genome-wide identification, classification, and expression analysis of autophagy-associated gene homologues in rice (Oryza sativa L.). DNA Res. 18(5):363–377.
- Xin M, Wang L, Liu Y, Feng Z, Zhou X, Qin Z. 2017. Transcriptome profiling of cucumber genome expression in response to long-term low nitrogen stress. Acta Physiol Plant. 39(6):1–11.
- Yan X, Bai D, Song H, Lin K, Pang E. 2021. Alternative splicing during fruit development among fleshy fruits. BMC Genomics. 22(1):1–14.
- Yang Z, Klionsky DJ. 2009. An overview of the molecular mechanism of autophagy. Autophagy Infect Immunity. 1–32.
- Yoshimoto K, Hanaoka H, Sato S, Kato T, Tabata S, Noda T, Ohsumi Y. 2004. Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell. 16(11):2967–2983.
- Zhang J, Yang W, Yue J, Liu Y, Pei D, Wang H. 2020. The responses of wheat autophagy and ATG8 family genes to biotic and abiotic stresses. J Plant Growth Regul. 39(2):867–876.
- Zheng J, Liu F, Zhu C, Li X, Dai X, Yang B, Zou X, Ma Y. 2019. Identification, expression, alternative splicing and functional analysis of pepper WRKY gene family in response to biotic and abiotic stresses. PLoS One. 14(7):e0219775.
- Zhou J, Wang J, Cheng Y, Chi YJ, Fan B, Yu JQ, Chen Z. 2013. NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PLoS Genet. 9(1):e1003196.
