ABSTRACT
The blue-crowned laughingthrush Pterorhinus courtoisi (Ménégaux, 1923) is listed as a critically endangered species on the International Union for Conservation of Nature Red List. This species occupies an extremely small known breeding range in China. In this study, we obtain the complete mitochondrial genome of the blue-crowned laughingthrush by using Illumina NovaSeq sequencing. The total length of the genome is 17,875 bp, containing 13 protein-coding genes, 22 transfer RNA genes, 2 ribosomal RNA genes and 2 control regions, with a slight A-T preference (52.84%) in base composition.
Introduction
Blue-crowned laughingthrush (Pterorhinus courtoisi, formerly as: Garrulax courtoisi Ménégaux, 1923) is a small bird of the family Leiothrichidae which is listed as critically endangered species on the International Union for Conservation of Nature (IUCN) Red List (BirdLife International Citation2018). It is endemic to China and occupies an extremely small known breeding range in Wuyuan county of Jiangxi province and Simao county of Yunnan Province (He et al. Citation2017). A wild population of 323 individuals was estimated in 2016 (BirdLife International Citation2018). The main food for the blue-crowned laughingthrush is insects and invertebrates. It also eats the fruits of plants such as loquat. Its upper body is brown, with a rich blue crown and nape, black eye patch and bright yellow throat. Its tail is black and has a white edge. The color of the belly and the undertail cover gradually changes from yellow to white (BirdLife International Citation2018). Studies on the blue-crowned laughingthrush have mostly focused on some ecological issues such as the home range and habitat use (Liu et al. Citation2020), breeding habitat characteristics and selection (Liao et al. Citation2007; Huang et al. Citation2018). Mitochondrial genomes are a valid molecular marker for studying the evolutionary status of the species. There are a few mitochondrial genomes of Pterorhinus animals reported, such as Pterorhinus lanceolatus (formerly as: Babax lanceolatus) (Qi et al. Citation2016a), Pterorhinus albogularis (formerly as: Garrulax albogularis) (Liu et al. Citation2018), and Pterorhinus poecilorhynchus (formerly as: Garrulax poecilorhynchus) (Qi et al. Citation2016b), etc. However, the mitochondrial genome of the blue-crowned laughingthrush has not been reported.
Methods
Sample collection
In this study, we collected about 3g muscle sample from the leg of a female individual of the subspecies of blue-crowned laughingthrush (Pterorhinus courtoisi courtoisi) in Wuyuan, Jiangxi province, China (Latitude: 29°18′15.45″N, Longitude:117°52′59.61″E; H: 83 m). The specimen was deposited at Jiangxi Agricultural University (Weiwei Zhang, email: [email protected]) under the voucher number GCWY01.
DNA extraction, PCR amplification, sequencing and analysis
This study complied with the IUCN policies about research involving species at risk of extinction and was approved by the Bioethics Committee of Qufu Normal University (No. 2021096). Total genomic DNA was extracted by using a modified cetyltrimethylammonium bromide (CTAB) method. 500 bp paired-end library was constructed by using the NEBNext Ultra DNA Library Prep Kit for Illumina sequencing. Bridge PCR amplification was performed on a cBot solid-phase vector to obtain DNA clusters based on the universal primer sequences of the avian mitochondrial genomes (Sorenson et al. Citation1999; Qi et al. Citation2016b; Liu et al. Citation2018). One end of the DNA fragment was complementary to the primer and fixed on the chip, and the other end was randomly complementary to other nearby primer to be fixed to form a “bridge”. Sequencing was carried out on the Illumina NovaSeq 6000 platform with a read length of 150 bp paired-end.
A total of about 3,896 Mb raw data were obtained from Illumina sequencing. The low quality and duplication reads were removed by filtering to obtain about 3,555 Mb clean data. De novo assembly with GetOrganelle v1.6.4 referencing mitochondrial genomes of closely related species of genus Pterorhinus produced contigs of mitochondrial genome. A number of potential mitochondrion reads were extracted from clean data using BLAST searches against mitochondrial genomes of related species of genus Pterorhinus and the contigs. The mitochondrion Illumina reads were obtained to perform cp genome de novo assembly using the SPAdes 3.13.1 package. The GetOrganelle assembly contigs were optimized by the scaffolds from SPAdes result. Finally, the assembled sequence was reordered and oriented according to the reference mitochondrial genomes, thus generating the final assembled mitochondrion genomic sequence of the blue-crowned laughingthrush. The starting location and direction of the mitochondrial assembly sequence were determined according to the reference genomes of Pterorhinus animals. The protein-coding genes, tRNA and rRNA genes of the mitochondrial genome were predicted by using MITOS software (Bernt et al. Citation2013). The start and stop codon positions of the genes were manually corrected to obtain a highly accurate conservative genome.
Description of the data
The complete mitochondrial genome sequence of the blue-crowned laughingthrush was submitted to NCBI GenBank with the accession number ON584558. The total length of the genome is 17,875 bp and it contains 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes and 2 control regions (D-loop regions, one is between the tRNA-Thr and tRNA-Pro, and the other one is between the tRNA-Glu and the tRNA-Phe) (Figure ). Most genes were encoded on the heavy strand except for ND6 and 8 tRNA genes. The individual base proportion of the blue-crowned laughingthrush mitochondrial genome amounts to 29.53% A, 23.31% T, 32.63% C, 14.53% G, with a weak A-T preference (52.84%). Six tRNA genes (tRNA-Gln, tRNA-Tyr, tRNA-Asp, tRNA-His, tRNA-Thr and tRNA-Pro) lack the DHU arms, and tRNA-Gly lacks the anticodon loop. The starting codons of 12 protein-coding genes are ATG, while the COX1 gene starts with GTG. The stop codons are: TAA for COX2, ATP8, ND4L and CYTB; TAG for ATP6 and ND6; AGA for ND1 and ND5; and AGG for COX1. In addition, the stop codons of 4 protein-coding genes are incomplete TA- (ND2, ND3) or T-- (COX3, ND4), which are complemented by Poly A at the 3′ end of mRNA to TAA (Table ).
Figure 1. The complete mitochondrial genome of blue-crowned laughingthrush Pterorhinus courtoisi (Ménégaux, 1923).
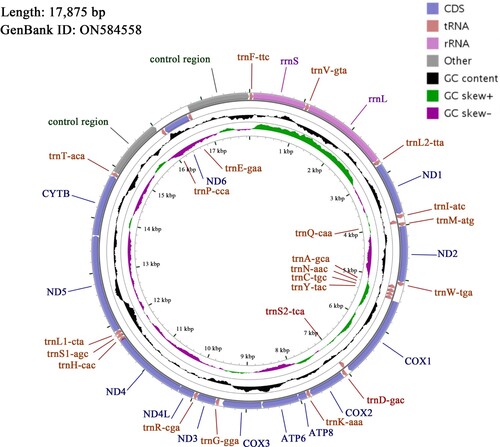
Table 1. Gene organization of the blue-crowned laughingthrush (Pterorhinus courtoisi Ménégaux, 1923) mitochondrial genome, H: heavy strand, L: light strand.
Disclosure statement
No potential conflict of interest was reported by the authors.
Author contributions statement
Lei Chen contributed to the conception and design of this paper substantially. Weiwei Zhang collected the experimental samples and acquired the data. Mengyao Sun, Di Xu and Zenghao Gao were involved in the analysis and interpretation of the data and drafting of the paper. Lei Chen and Weiwei Zhang revised the manuscript critically for important intellectual content. All authors approved the final version to be published and agreed to be accountable for all aspects of the work.
Data availability statement
The genome sequence data that support the findings of this study is openly available in GenBank of NCBI (https://www.ncbi.nlm.nih.gov/) under the accession number ON584558. The associated BioProject, SRA, and BioSample numbers are PRJNA837048, SRR19451413 and SAMN28188779, respectively.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- BirdLife International. 2018. Garrulax courtoisi. The IUCN Red List of Threatened Species 2018: e. T22732350A131890764.
- He FQ, Lin JS, Wen C, Lin Z, Shi QH, Huang HQ, Cheng SL, Xiao H. 2017. A preliminary study on the biology Garrulax courtoisi in Wuyuan. Chin J Zool. 52(1):167–175.
- Huang HQ, Liu T, Shi JZ, Liu P, Zhang WW. 2018. Habitat selection of the Blue-Crowned Laughingthrush during the breeding season. Acta Ecologica Sinica. 38(2):493–501.
- Liao WM, Hong YH, Yu SB, Ouyang XZ, He GX. 2007. A study on the propagation habitat of Garrulax galbanus courtoisi and the relationship of the birds with village forests in Wuyuan, Jiangxi Province. Acta Agricultruae Universitatis Jiangxiensis. 5:837–841+850.
- Liu T, Xu YT, Mo B, Shi JZ, Cheng Y, Zhang W, Lei FM. 2020. Home range size and habitat use of the blue-crowned laughingthrush during the breeding season. PeerJ. 8(2):e8785.
- Liu X, Xu HL, Zhou YY, Li DY, Ni QY, Zhang MW, Xie M, Wen AX, Wang Q, Wu JY, Yao YF. 2018. Complete characteristics and phylogenetic relationships of the Garrulax albogularis mitochondrial genome (Passeriformes: Timaliidae). Mitochondrial DNA Part B. 3(2):1272–1273.
- Qi Y, Zhou YY, Yao YF, Huan ZJ, Li DY, Xie M, Ni QY, Zhang MW, Xu HL. 2016a. The complete mitochondrial genome of Babax lanceolatus (Passeriformes: Timaliidae). Mitochondrial DNA Part A. 27(4):2925–2926.
- Qi Y, Zhou YY, Yao YF, Huan ZJ, Li DY, Xie M, Ni QY, Zhang MW, Xu HL. 2016b. The complete mitochondrial genome sequence of Garrulax poecilorhynchus (Aves, Passeriformes, Timaliidae). Mitochondrial DNA Part A. 27(5):3636–3637.
- Sorenson MD, Ast JC, Dimcheff DE, Yuri T, Mindell DP. 1999. Primers for a PCR-based approach to mitochondrial genome sequencing in birds and other vertebrates. Mol Phylogenet Evol. 12(2):105–114.
