Abstract
Previous studies revealed that both Helicobacter pylori infection and some cytokine gene polymorphisms are risk factors for gastric diseases. The association between H. pylori and gene polymorphisms is worth exploring. Here we conducted a case–control study and systematically evaluate on association between H. pylori infection and IL4, TNFA, and IL10 polymorphisms in a Chinese Han population in northwestern China, where gastric cancer is the major burden on the health and the leading cause of death in local community. The Sequenom MassArray platform was used to screen the genotypes and 14C-urea breath test was employed to determine H. pylori infection status of 705 participants of the local rural residents. Significant association between H. pylori infection and the IL4 rs34142320 polymorphism was observed in the dominant model (OR = 0.57, 95% CI = 0.40–0.80, P = 0.030). No significant association was found between IL4 rs2243250, TNFA rs1800630, TNFA rs1800629, TNFA rs1799964, IL10 rs1800871, IL10 rs1800896 and H. pylori infection. Haplotype analysis showed that T-DEL and G-C haplotypes of the observed IL4 SNPs were significantly associated with H. pylori infection. Our analysis demonstrated that the IL4 rs34142320 polymorphism has a protective effect on H. pylori infection in Chinese Han population.
Introduction
Helicobacter pylori is one of the most common bacteria worldwide (Lee et al. Citation2016). The spread of H. pylori infection is largely related to lifestyle factors and sanitary conditions, especially in developing countries (Marshall Citation2003; Axon Citation2014). The bacteria can cause various gastroduodenal diseases, including gastritis and peptic ulcer disease (PUD), and is closely related to the occurrence of gastric cancer (GC) and mucosa-associated lymphoid tissue (MALT) lymphoma. It has been reported as a class I carcinogen (IARC Citation1994). The discovery of H. pylori dramatically changes the awareness of the occurrence of upper gastrointestinal diseases. The infection rate of H. pylori in the population shows regional differences around the world (Axon Citation2014). Moreover, people in the same area infected with H. pylori also have different outcomes. A study of twins found that monozygotic twin pairs had higher H. pylori infection risk than dizygotic twin pairs, and confirmed that genetic effects influence the acquisition of H. pylori infection (Malaty et al. Citation1994). This demonstrated that the genetic factors of the host were closely linked with the infection and outcomes of the bacteria. H. pylori virulence factors could accumulate and activate neutrophils and mononuclear cells by inducing the local production of cytokines. Several cytokines including members of the interleukin (IL) family and tumor necrosis factor (TNF)-α, etc. had been reported to be involved in H. pylori infection (Gonzalez-Hormazabal et al. Citation2014; D'Elios and Czinn Citation2014). These host cytokines and their polymorphisms may be responsible for the outcomes of H. pylori infection in terms of protection, evasion, or pathology (Datta De and Roychoudhury Citation2015).
Wuwei City is located in Gansu Province in northwestern China. Gastric cancer is a major burden on the health and the leading cause of death in the local population. The incidence and mortality rates of gastric cancers in this area are 104.6 and 67.7 per 100,000 persons-years (Liang et al. Citation2016). The latest survey shows that the infection rate of H. pylori among the adults from Wuwei City is 53.0% (Zhang et al. Citation2021). It is very worthwhile to carry out and explore possible related mechanisms between the host's genetic background and H. pylori infection.
The IL4 gene is located on the long arm of human chromosome 5 (5q31.3), with a cluster of T help 2 (Th2) cytokine genes (Brown and Hural Citation2017). IL-4 is an immune regulatory cytokine that suppresses T help 1 (Th1) response while stimulating Th2 cells (Brown and Hural Citation2017; Yan et al. Citation2014). Two single nucleotide polymorphisms (SNPs), IL4 rs2243250 (also known as – 590T>C) and IL4 rs34142320 within the regulatory sequences of IL4 are considered to have influence on its expression (Brown and Hural Citation2017). TNFA gene, encoding the TNF-α, is located on the short arm of human chromosome 6 (6p21.3). There are several SNPs in the regulatory sequences of TNFA gene that may participate in its expression, including TNFA rs1800629 (−308G>A), TNFA rs1800630 (−863C>A) and TNFA rs1799964 (−1031T>C) (Hellmig et al. Citation2008; Lu et al. Citation2005). IL-10 is a potent inhibitor to the immunity and inflammatory responses (Trifunović et al. Citation2015). This cytokine is encoded by the IL10 gene, which is located in the long arm of human chromosome 1 (1q31–q32) (Qi et al. Citation2019). IL10 rs1800871 (−819T>C) and IL10 rs1800896 (−1082A>G) are two common polymorphisms in the IL10 gene (Qi et al. Citation2019).
Many previous studies conducted on the relationship between the IL4, TNFA, and IL10 gene polymorphisms and gastroduodenal diseases also discussed H. pylori infection simultaneously (Achyut et al. Citation2008; Murphy et al. Citation2009; Pavithra et al. Citation2018). Sun et al. (Citation2014) reported that IL4 rs2243250 polymorphism could reduce gastric cancer risk in Caucasians. However, a meta-analysis from Zhang et al. found that there was no significant association between IL4 rs2243250 polymorphism and gastric cancer (Zhang et al. Citation2016). Though the relationship between IL4, TNFA and IL10 gene polymorphisms and H. pylori related gastroduodenal diseases was explored by some previous studies, few were conducted directly on the association between these gene polymorphisms and H. pylori infection. Our previous meta-analysis discovered that TNFA rs1800629 and TNFA rs1799964 might be protective factors, while TNFA rs1800630 might be a risk factor against H. pylori infection, especially in Asian populations (Sun et al. Citation2016). However, there are still inconsistencies in the present literature (Santos et al. Citation2012; Queiroz et al. Citation2009). Since H. pylori infection, IL4, TNFA, and IL10 gene polymorphisms are risk factors for gastric cancer, we selected some SNPs which were indicated to be associated with gastric cancer (Sun et al. Citation2014; Yu et al. Citation2014; Liu et al. Citation2018), and focused on the association between the candidate IL4, TNFA, and IL10 gene polymorphisms and H. pylori infection in a Chinese Han population in Wuwei City in northwestern China. Our work provides new insights into the population genetic polymorphism and the occurrence of H. pylori infection.
Materials and methods
Study area and communities
All study participants were Chinese Han population and rural residents who were volunteers to attend China's National Science and Technology Program for Public Wellbeing in Wuwei City of Gansu Province in northwestern China (Ji et al. Citation2021). People, age ranging from 35 to 70 years, who signed informed consent, did not have mental illness and were not pregnant, were included. The participants included 705 people, and the mean age was 49.86 years. None of the participants had been found to have gastric cancer. Drinking was defined as consuming at least 150 g of wine or hard liquor, or 1000 g of beer once per week during the past 1 year; smoking was defined as smoking at least one cigarette per day over a 6-month period at any time in the past years. Furthermore, family history of gastric cancer was defined as at least one of the first-degree relatives having suffered from gastric cancer in the past years. A total of 2-milliliter peripheral venous blood was collected on EDTA anticoagulant tubes, for DNA extraction and genotyping of the candidate gene polymorphisms. All participants gave informed consent, and the study was approved by the Medical Ethics Committee of the First Hospital of Lanzhou University (ethical approval number: LDYYLL2012001, date: 2012.01.02).
Measurement of H. pylori infection
14C-urea breath test (UBT) was used to detect H. pylori infection (Skrebinska et al. Citation2018). UBT was performed according to the manufacturer's instructions (Headway, China). In brief, the participants took 14C-urea capsule orally. After waiting for 25 min, the exhaled CO2 of the participants were collected into a special bottle and tested by a H. pylori detector. A disintegration per minute (DPM) value greater than or equal to 100 per mmol CO2 was considered as H. pylori positive, while less than 100 per mmol CO2 was considered as negative. H. pylori positive participants were selected as case group, and negative participants were selected as control group.
Genotyping the candidate gene polymorphisms
Genomic DNA was extracted from 200 μL of peripheral blood by the Genomic DNA kit according to the manufacturer's instructions (Tiangen Corporation, Beijing, China). A DNA concentration of 20 ng/uL was examined by a NanoDrop 2000 system, and the 260/280 nm value between 1.6 and 2.0 was accepted for concentration and purity. The candidate gene polymorphisms involved in this study including IL4 rs2243250, IL4 rs34142320, TNFA rs1800630, TNFA rs1800629, TNFA rs1799964, IL10 rs1800871 and IL10 rs1800896. AssayDesigner 3.1 software (Sequenom, San Diego, CA, USA) was used to design the primers for locus-specific PCR, and the sequences of the primers were listed in Table . Sequenom's MassArray platform (Sequenom, San Diego, CA, USA) was used for genotyping the candidate gene polymorphisms according to the manufacturer's instructions. In brief, 10 ng of genomic DNA of each sample was amplified by multiplex PCR reactions, and the PCR products were then used in single base extension reactions. All the products were transferred to a 384-well SpectroCHIP, and a MALDI-TOF (Matrix-Assisted Laser Desorption Ionization-Time of Flight) mass spectrometer was used for allele detection. Finally, the spectrograms were analyzed using the MassArray Typer Analyzer software 4.0 (Sequenom, San Diego, CA, USA).
Table 1. The sequences of primers used in this study.
Statistical analysis
Allele and genotype frequencies of the candidate SNPs were obtained by direct counting. Sex and genotype frequencies were reported as percentages. A χ2-square test was used to examine differences in distribution by sex, age, the family history of gastric cancer, drinking and smoking status according to prevalence of H. pylori infection. The association between the cytokine gene polymorphisms and H. pylori infection was examined by comparing the odds ratios (ORs) and 95% confidence intervals (CIs) among different allele carriers using unconditional logistic regression test. Bonferroni method was used for multiple-comparison correction. Hardy–Weinberg equilibrium (HWE) was assessed by a χ2-square test. P values less than 0.05 were considered to be statistically significant. SNPStats (Solé et al. Citation2006) and SPSS 19.0 (IBM, Chicago, USA) software were used to perform all the above statistical analyses. Linkage disequilibrium (LD) and haplotype analysis were performed by HaploView 4.2 software (Barrett et al. Citation2005).
Results
Demographic characteristics of participants
Among the 705 participants, 448 samples were H. pylori infection positive while 257 samples were negative (Table ). No significant differences existed between H. pylori infection positive and negative groups by sex, age, the family history of gastric cancer, drinking or smoking status (Table ). The genotype frequencies of the investigated polymorphisms were in accordance with HWE for the H. pylori infection negative group (Table ).
Table 2. Main characteristics of the participants.
Table 3. Status of Helicobacter pylori infection among participants of different main characteristics.
Table 4. Association analysis between IL4, TNFA and IL10 genes polymorphisms and H. pylori infection.
The association between the IL4, TNFA and IL10 gene polymorphisms and H. pylori infection
To analyze the association between IL4, TNFA, and IL10 polymorphisms and H. pylori infection in the Chinese Han population in northwestern China, the status of the H. pylori infection and the candidate genotype frequencies were analyzed by unconditional logistic regression test. Significant association was found between overdominant model (OR = 0.71, 95% CI = 0.51–0.98, P = 0.039) of IL4 rs2243250, codominant model (OR = 0.53, 95% CI = 0.37–0.76, P = 0.003), dominant model (OR = 0.57, 95% CI = 0.40–0.80, P = 0.001) and overdominant model (OR = 0.63, 95% CI = 0.46–0.87, P = 0.005) of IL4 rs34142320 polymorphisms and H. pylori infection. However, after multiple correction, only the result of dominant model between H. pylori infection and IL4 rs34142320 was still significant (OR = 0.57, 95% CI = 0.40-0.80, P = 0.030). There was no significant association between H. pylori infection and TNFA rs1800630, TNFA rs1800629, TNFA rs1799964, IL10 rs1800871, IL10 rs1800896 polymorphisms (Table ).
Linkage disequilibrium (LD) and haplotype analysis
LD analysis was conducted based on the two polymorphisms of IL4 gene, three polymorphisms of TNFA gene and two polymorphisms of IL10 gene. LD was observed from the three SNPs of TNFA gene and the two SNPs of IL10 gene, respectively (Figure ). Haplotype analysis showed that T-DEL and G-C haplotypes of the two IL4 SNPs were significant associated with H. pylori infection (T-DEL, OR = 0.64, 95% CI = 0.48-0.86, P = 0.003; G-C, OR = 0.52, 95% CI = 0.30-0.90, P = 0.021) (Table ).
Figure 1. LD analysis of the candidate gene SNPs. The results of LD analysis for IL4 (A), TNFA (B) and IL10 (C) genes were described. The color shift from white to red represented that the results of D’ and r2 changed from 0 to 1. A deep red block means the result is 1. The closer the value is to 1, the higher degree of LD is. Kb represented the distance between two SNPs in the genome. LD was observed from the three SNPs of TNFA gene and the two SNPs of IL10 gene, respectively.
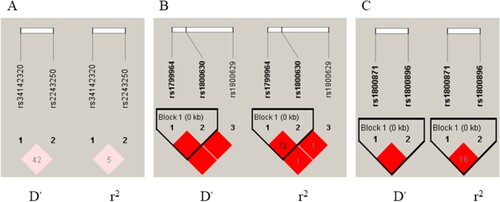
Table 5. Results of haplotype analysis.
Discussion
It had been confirmed that the genetic background of the host could influence H. pylori infection (Clyne and Rowland Citation2019). A more complete understanding of how the genetics of the host contribute to complex diseases will improve the exploration of the susceptibility factors that contribute to H. pylori infection and the possible mechanisms underlying its pathogenicity. The infection rate of H. pylori among the adults from Wuwei City is 53.0% (Zhang et al. Citation2021), and this area is also a high-incidence area for gastric cancer (Liang et al. Citation2016; Li et al. Citation2016; Liu et al. Citation2021). The prevalence of H. pylori infection may play an important role in the occurrence and development of gastric cancer in this area.
Our results indicated that there was a significant association between the IL4 rs34142320 polymorphism and H. pylori positive subjects. No significant association was found between the other candidate gene polymorphisms and H. pylori infection. Haplotype analysis showed that T-DEL and G-C haplotypes of the observed IL4 SNPs were significantly associated with H. pylori infection. The genotypes of candidate genes in our study were similar to previous research in the Chinese Han population (Zeng et al. Citation2012; Su et al. Citation2010).
IL-4 belongs to the Th2 cytokine family. Besides Th2 being the main producer of IL-4, Th1, CD8+ T cells, B lymphocytes, mast cells, macrophages, etc. also could secrete lower levels of IL-4 (Iwaszko et al. Citation2021; Paul Citation2015; Liang et al. Citation2012). IL-4 plays an important role in activating B cells, influencing cell differentiation, and regulating antibody production (Brown and Hural Citation2017). IL-4 is the key cytokine that regulates the transformation of immunoglobulin types to IgE and IgG4 (Pène et al. Citation1988), and participates in the human's allergic disorders and anti-parasitic immunity (Finkelman et al. Citation2004; Gour and Wills-Karp Citation2015). Furthermore, IL-4 can promote Th2 differentiation and antagonize Th1-driven proinflammatory immune response (Iwaszko et al. Citation2021). Studies had confirmed that Th1 immune response was suggested to be predominant in H. pylori infection (Montecucco and Rappuoli Citation2001; Larussa et al. Citation2015). Decreased activity of IL-4 in the gastric mucosa can promote the Th1-driven immune response, which may activate more protective immunity mechanisms against H. pylori infection (Larussa et al. Citation2015). In addition, IL-4 was suggested to be related with gastric mucosal tolerance, which could be beneficial to the settlement and proliferation of H. pylori in the gastric mucosa (Orsini et al. Citation2007). In brief, the IL4 rs34142320 polymorphism protects against H. pylori infection, which may contribute to explore the association between the IL4 polymorphisms and upper gastrointestinal diseases including gastritis, peptic ulcer and even gastric cancer (Gonzalez-Hormazabal et al. Citation2014).
Previous literature indicated that TNFA, IL10 polymorphisms and H. pylori were associated with gastric cancer (Guo et al. Citation2013). Our meta-analysis suggested that TNFA rs1800629, TNFA rs1800630 and TNFA rs1799964 polymorphisms were significantly correlated with H. pylori infection (Sun et al. Citation2016). TNF-α could inhibit gastric acid secretion and amplify inflammatory responses (Zhao et al. Citation2013), and IL-10 is a potent inhibitor to some inflammatory cytokines, which might be correlated with H. pylori infection and gastric cancer (Hamajima Citation2003). But no significant association was found between the candidate TNFA, IL10 polymorphisms and H. pylori infection in the Chinese Han population in northwestern China. As the results were inconclusive, further studies with larger sample sizes are needed to verify the association between TNFA, IL10 polymorphisms and H. pylori infection.
There were some limitations to our study. The sample size of this study is relatively small. We only analyzed seven polymorphisms of IL4, TNFA and IL10, and other polymorphisms or haplotypes in these genes might be also implicated in persistent H. pylori infection. We only used UBT method to detect H. pylori infection, while various measurements of H. pylori infection may impact the results (Sabbagh et al. Citation2019). We only used MALDI-TOF method to detect genetic polymorphisms, while other SNP genotyping methods and monitoring mRNA levels of cytokines are meaningful to this work. Future studies with larger sample sizes and various study populations should be conducted to clarify the association between the IL4, TNFA, and IL10 polymorphism and H. pylori infection.
In conclusion, our analysis suggests the presence of an association between the IL4 rs34142320 variants and risk of H. pylori infection in a Chinese Han population in northwestern China, which provides insights into the functions of IL4 SNPs, and new clues for the prevention of gastric cancer in this area.
Author contributions
Conceived and designed the experiments: XDS JHZ TJ YNZ JH.
Performed the experiments: XDS SJZ FHZ XHL LLH LW.
Analyzed the data: XDS FHZ.
Contributed reagents/materials/analysis tools: XDS FHZ SJZ JHZ XHL YNZ JH.
Wrote the paper: XDS JH.
Acknowledgements
We thank villagers, the rural health workers and the staff of Liangzhou District Hospital of Wuwei who participated in this study.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in figshare at https://doi.org/10.6084/m9.figshare.21563523.
Additional information
Funding
References
- Achyut BR, Tripathi P, Ghoshal UC, Moorchung N, Mittal B. 2008. Interleukin-10 (−819 C/T) and Tumor Necrosis factor-α (−308 G/A) Gene variants influence Gastritis and Lymphoid Follicle development. Dig Dis Sci. 53:622–629. doi:10.1007/s10620-007-9925-y.
- Axon A. 2014. Helicobacter pylori and public health. Helicobacter. 19(1):68–73. doi:10.1111/hel.12155.
- Barrett JC, Fry B, Maller J, Daly MJ. 2005. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 21:263–265. doi:10.1093/bioinformatics/bth457.
- Brown MA, Hural J. 2017. Functions of IL-4 and control of Its expression. Crit Rev Immunol. 37:181–212. doi:10.1615/CritRevImmunol.v37.i2-6.30.
- Clyne M, Rowland M. 2019. Advances in experimental medicine and biology. Adv Exp Med Biol. 1149:151–172. doi:10.1007/5584_2019_364.
- Datta De D, Roychoudhury S. 2015. To be or not to be: The host genetic factor and beyond in helicobacter pylori mediated gastro-duodenal diseases. World J Gastroenterol. 21:2883–2895. doi:10.3748/wjg.v21.i10.2883.
- D'Elios MM, Czinn SJ. 2014. Immunity, inflammation, and vaccines for Helicobacter pylori. Helicobacter. 19(1):19–26. doi:10.1111/hel.12156.
- Finkelman FD, Shea-Donohue T, Morris SC, Gildea L, Strait R, Madden KB, Schopf L, Urban JF, 2004. Interleukin-4- and interleukin-13-mediated host protection against intestinal nematode parasites. Immunol Rev. 201:139–155. doi:10.1111/j.0105-2896.2004.00192.x.
- Gonzalez-Hormazabal P, Musleh M, Bustamante M, Stambuk J, Escandar S. 2014. Host polymorphisms and virulence genotypes of helicobacter pylori as risk factors for gastric cancer. Anticancer Res. 34:5927.
- Gour N, Wills-Karp M. 2015. IL-4 and IL-13 signaling in allergic airway disease. Cytokine. 75:68–78. doi:10.1016/j.cyto.2015.05.014.
- Guo XF, Wang J, Yu SJ, Song J, Ji MY. 2013. TNF-α−308polymorphism and risk of digestive system cancers: A meta-analysis. World J Gastroenterol. 19:9461–9471. doi:10.3748/wjg.v19.i48.9461.
- Hamajima N. 2003. Persistent helicobacter pylori infection and genetic polymorphisms of the host. Nagoya J Med Sci. 66:103–117.
- Hellmig S, Bartscht T, Fischbach W, Fölsch UR, Schreiber S. 2008. Interleukin-10 (−819 C/T) and TNF-A (−308 G/A) as risk factors for H. pylori-associated gastric MALT-lymphoma. Dig Dis Sci. 53:2007–2008. doi:10.1007/s10620-008-0231-0.
- Iwaszko M, Biały S, Bogunia-Kubik K. 2021. Significance of interleukin (IL)−4 and IL-13 in inflammatory arthritis. Cells. 10:3000. doi:10.3390/cells10113000.
- Ji R, Zhang Z, Zhang J, Wu Z, Li M, Ye Y, An F, Xu C, Lu L, Fan P, et al. 2021. Cohort profile: A population-based cohort for the study of gastric cancer in northwest area of China (Wuwei Cohort). Int J Epidemiol. 50:1433–1442. doi:10.1093/ije/dyab083.
- Larussa T, Leone I, Suraci E, Imeneo M, Luzza F. 2015. Helicobacter pylori and T helper cells: mechanisms of immune escape and tolerance. J Immunol Res. 2015:1. doi:10.1155/2015/981328.
- Lee YC, Chiang TH, Chou CK, Tu YK, Liao WC, Wu M-S, Graham DY. 2016. Association between Helicobacter pylori eradication and gastric cancer incidence: A systematic review and meta-analysis. Gastroenterology. 150:1113–1124.e5. doi:10.1053/j.gastro.2016.01.028.
- Li CY, Ye YC, Liang GY, Zhang WH, Zhang ZY, Liu X-Q, Xu F-L, Xiang J-L. 2016. Cancer incidence and mortality survey in Wuwei, Gansu Province, Northwestern China from 2003 to 2012. Chin Med J. 129:636–644. doi:10.4103/0366-6999.177969.
- Liang HE, Reinhardt RL, Bando JK, Sullivan BM, Ho IC, Locksley RM. 2012. Divergent expression patterns of IL-4 and IL-13 define unique functions in allergic immunity. Nat Immunol. 13:58–66. doi:10.1038/ni.2182.
- Liang Y., Ye YC., Zhang WH., Zhang ZY., Liu XQ., Li CY., Xu FL. 2016. An analysis of stomach cancer from 2008 to 2012 in Liangzhou district Wuwei city, Gansu province. China Cancer. 25:519–523.
- Liu K, Song S, Fu T, Liu Y, Zhang H, Yan M, Su H, Li Z, Ji Z, Shao Z, et al. 2021. Spatiotemporal trends in the incidence of Gastrointestinal Neoplasms in Wuwei City of northwestern China from 1995 to 2016: A hospital-based retrospective observational study. Front Oncol. 11:712857. doi:10.3389/fonc.2021.712857.
- Liu S, Liu JW, Sun LP, Gong YH, Xu Q, Jing J-J, Yuan Y. 2018. Association of IL10 gene promoter polymorphisms with risks of gastric cancer and atrophic gastritis. J Int Med Res. 46:5155–5166. doi:10.1177/0300060518792785.
- Lu CC, Sheu BS, Chen TW, Yang HB, Hung KH, Kao A-W, Chuang C-H, Wu J-J. 2005. Host TNF-alpha-1031 and −863 promoter single nucleotide polymorphisms determine the risk of benign ulceration after H. pylori infection. Am J Gastroenterol. 100:1274–1282. doi:10.1111/j.1572-0241.2005.40852.x.
- Malaty HM, Engstrand L, Pedersen NL, Graham DY. 1994. Helicobacter pylori infection: genetic and environmental influences: A study of twins. Ann Intern Med. 120:982–986. doi:10.7326/0003-4819-120-12-199406150-00002.
- Marshall B. 2003. Helicobacter pylori: past, present and future. Keio J Med. 52:80–85. doi:10.2302/kjm.52.80.
- Montecucco C, Rappuoli R. 2001. Living dangerously: how Helicobacter pylori survives in the human stomach. Nat Rev Mol Cell Biol. 2:457–466. doi:10.1038/35073084.
- Murphy G, Thornton J, McManus R, Swan N, Ryan B, Hughes DJ, O'Morain CA, O'Sullivan M. 2009. Association of gastric disease with polymorphisms in the inflammatory-related genes IL-1B, IL-1RN, IL-10, TNF and TLR4. Eur J Gastroenterol Hepatol. 21:630–635. doi:10.1097/MEG.0b013e3283140eea.
- Orsini B, Vivas JR, Ottanelli B, Amedei A, Surrenti E, Galli A, Milani S, Pinzani P, Del Prete G, Surrenti C, et al. 2007. Human Gastric Epithelium produces IL-4 and IL-4δ2 Isoform only upon Helicobacter pylori infection. Int J Immunopathol Pharmacol. 20:809–818. doi:10.1177/039463200702000417.
- Paul WE. 2015. History of interleukin-4. Cytokine. 75:3–7. doi:10.1016/j.cyto.2015.01.038.
- Pavithra D, Gautam M, Rama R, Swaminathan R, Gopal G, Ramakrishnan AS, Rajkumar T. 2018. TGFβ C-509T, TGFβ T869C, XRCC1 Arg194Trp, IKBα C642T, IL4 C-590T genetic polymorphisms combined with socio-economic, lifestyle, diet factors and gastric cancer risk: A case control study in south Indian population. Cancer Epidemiol. 53:21–26. doi:10.1016/j.canep.2018.01.004.
- Pène J, Rousset F, Brière F, Chrétien I, Bonnefoy JY, Spits H, Yokota T, Arai N, Arai K, Banchereau J, et al. 1988. Ige production by normal human lymphocytes is induced by interleukin 4 and suppressed by interferons gamma and alpha and prostaglandin E2. Proc Natl Acad Sci USA. 85:6880–6884. doi:10.1073/pnas.85.18.6880.
- Qi Y, Kong J, He J. 2019. Genetic relationship between IL-10 gene polymorphisms and the risk of clinical atopic dermatitis. BMC Med Genet. 20:83. doi:10.1186/s12881-019-0817-8.
- Queiroz DM, Saraiva IE, Rocha GA, Rocha AM, Gomes LI, Melo FF, Bittencourt PFS. 2009. IL2-330G polymorphic allele is associated with decreased risk of Helicobacter pylori infection in adulthood. Microbes Infect. 11:980–987. doi:10.1016/j.micinf.2009.07.008.
- Sabbagh P, Mohammadnia-Afrouzi M, Javanian M, Babazadeh A, Koppolu V, Vasigala VR, Nouri HR, Ebrahimpour S. 2019. Diagnostic methods for helicobacter pylori infection: ideals, options, and limitations. Eur J Clin Microbiol Infect Dis. 38:55–66. doi:10.1007/s10096-018-3414-4.
- Santos JC, Ladeira MS, Pedrazzoli J, Ribeiro J, L M. 2012. Relationship of IL-1 and TNF-α polymorphisms with helicobacter pylori in gastric diseases in a Brazilian population. Braz J Med Biol Res. 45:811–817. doi:10.1590/S0100-879X2012007500099.
- Schistosomes, liver flukes and Helicobacter pylori. 1994. IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Lyon, 7-14 June 1994. IARC Monogr Eval Carcinog Risks Hum 61: 1-241.
- Skrebinska S, Mégraud F, Bessède E. 2018. Diagnosis of Helicobacter pylori infection. Helicobacter. 23(1):e12515. doi:10.1111/hel.12515.
- Solé X, Guinó E, Valls J, Iniesta R, Moreno V. 2006. SNPStats: a web tool for the analysis of association studies. Bioinformatics. 22:1928–1929. doi:10.1093/bioinformatics/btl268.
- Su SP, Yang ZB, Tian YL. 2010. Relationship between polymorphisms of IL-1β−31, IL-10-819 and TNF-α−1031 genes and susceptibilities to H. pylori infection-associated gastric ulcer and cancer. Chinese J Biol. 23:517–520.
- Sun X, Xu Y, Wang L, Zhang F, Zhang J. 2016. Association between TNFA gene polymorphisms and helicobacter pylori infection: A meta-analysis. PLoS One. 11:e0147410.
- Sun Z, Cui Y, Jin X, Pei J. 2014. Association between IL-4 −590C>T polymorphism and gastric cancer risk. Tumor Biol. 35:1517–1521. doi:10.1007/s13277-013-1209-x.
- Trifunović J, Miller L, Debeljak Ž, Horvat V. 2015. Pathologic patterns of interleukin 10 expression – A review. Biochem Med (Zagreb). 25:36–48. doi:10.11613/BM.2015.004.
- Yan Y, Weng H, Shen ZH, Wu L, Zeng XT. 2014. Association between interleukin-4 gene −590 C/T, −33 C/T, and 70-base-pair polymorphisms and periodontitis susceptibility: A meta-analysis. J Periodontol. 85:e354–e362. doi:10.1902/jop.2014.140317.
- Yu T, Lu Q, Ou XL, Cao DZ, Yu Q. 2014. Clinical study on gastric cancer susceptibility genes IL-10-1082 and TNF-α. Genet Mol Res. 13:10909–10912. doi:10.4238/2014.December.19.12.
- Zeng X, Li Y, Liu T, Zhang J. 2012. Diverse H. pylori strains, IL-10 promoter polymorphisms with high morbidity of gastric cancer in Hexi area of Gansu province, China. Mol Cell Biochem. 362:241–248. doi:10.1007/s11010-011-1149-y.
- Zhang C, Huang JY, He ZQ, Weng H. 2016. Genetic association between interluekin-4 rs2243250 polymorphism and gastric cancer susceptibility: evidence based on a meta-analysis. Onco Targets Ther. 9:2403–2408.
- Zhang F, Pu K, Wu Z, Zhang Z, Liu X, Chen Z, Ye Y, Wang Y, Zheng Y, Zhang J, et al. 2021. Prevalence and associated risk factors of Helicobacter pylori infection in the Wuwei cohort of north-western China. Trop Med Int Health. 26:290–300. doi:10.1111/tmi.13517.
- Zhao Y, Wang JW, Tanaka T, Hosono A, Ando R, Zhao Y, 2013. Association between TNF-α and IL-1β genotypes vs helicobacter pylori infection in Indonesia. World J Gastroenterol. 19:8758–8763. doi:10.3748/wjg.v19.i46.8758.
