Abstract
Ectropis dentilineata Moore, 1868 has a wide geographic distribution while its genome data are largely unknown. In this study, we collected samples of E. dentilineata from a tea plantation in Guizhou province, China. Illumina sequencing showed that the mitochondrial genome of E. dentilineata is 15,356 bp in length, containing the entire set of 37 mitochondrial genes. Several tRNAs show gene arrangements compared with the ancestral gene order, mainly involving the tRNA cluster (M-I-Q). These data will facilitate a deeper understanding and exploitation of E. dentilineata.
Introduction
The superfamily Geometriodae is the most species-rich branch in the megadiverse Geometridae family and its taxonomy represents a challenge for researchers because of the taxonomic uncertainties around some species-rich genera (Sihvonen et al. Citation2011; Strutzenberger et al. Citation2012; Mitter et al. Citation2017). The mitochondrial genome is a useful tool for insect phylogenetics and has been widely applied to resolve uncertainty within this group (Timmermans et al. Citation2014). At present, the nucleotide database of the National Center for Biotechnology Information (NCBI) has only two published mitogenomes in the genus Ectropis (E. grisescens and E. obliqua) (Dai et al. Citation2018; Song et al. Citation2021). Ectropis dentilineata Moore, 1868 (Geometridae: Lepidoptera) was first described by Moore (Citation1867). The main characteristics of E. dentilineata adult are as follows: in the fore wing, there are three or four tiny dark brown spots displayed in the subbasal transverse oblique row, dentiform markings in discal that are double interrupted, and two dark brown streaks at the terminal; in the hind wing, there is a small dark brown discal spot and a line running down the abdominal border (Moore Citation1867). The species has a wide geographic distribution, discovered in Thailand, Bhutan (Ratnasingham and Hebert Citation2007), Pakistan (Ashfaq et al. Citation2017) and China, while no mitochondrial data of E. dentilineata are available. In this study, we got the mitochondrial genome of E. dentilineata by next-generation sequencing and the results will facilitate a deeper understanding and exploitation of this species.
Materials and methods
Sample collection and insect rearing
Larvae of E. dentilineata were collected from a tea plantation in Guizhou Province (25.67N, 107.52E), China. To understand its biology, the collected larvae were reared on fresh tea leaves grown at 25 ± 1°C, 60% relative humidity, and 14 h light: 10 h dark photoperiod.
The voucher specimens were stored in ethanol absolute and deposited at the Tea Research Institute, Chinese Academy of Agricultural Sciences (contact Xiao-Gui Zhou, [email protected]) under the voucher number ZJU-1 (Figure ). Three individuals of E. dentilineata (ZJU-1: female adult; ZJU-2: male adult; ZJU-3: pupa) were separately used as sequencing samples.
DNA extraction and library construction
The abdomen of adult samples was cut off to avoid the disturbance by symbiotic bacteria. The carcass was grounded in the lysis solution, and then placed in a water bath at 56°C for 5 h. For pupa, it was placed directly into the lysis solution and the same treatment was subsequently performed.
The whole genomic DNA was extracted using a DNeasy tissue kit (Qiagen, Hilden, Germany). The library was constructed by VAHTS™ Universal DNA Library Prep Kit for Illumina® v9.1, and sequenced by the Illumina NovaSeq 6000 platform (150 bp paired-end) at Grandomics (Wuhan, China).
Genome sequencing and annotation
FastQC was used to check the quality of the data, and Trimmomatic was used to trim adaptors and indexes with default parameters (Bolger et al. Citation2014; Wingett and Andrews Citation2018). The target mitochondrial reads were retained using BLAST (BLASTn with E value: 1 × 10−5) against a reference data set containing Geometridae mitochondrial genomes via the FastqExtract script (Crampton-Platt et al. Citation2015). The mitochondrial genome was assembled by IDBA-UD (Peng et al. Citation2012) and SPAdes version 3.13.1 (Bankevich et al. Citation2012). Sequence annotation, visualization and GC skew calculation are completed with Geneious Prime version 2019.1.3 with default parameters. In addition, the start and stop codons of protein-coding genes (PCGs) were corrected manually using the genomes of Ectropis oblique (KX827002.1) (Dai et al. Citation2018), and Ectropis grisescens (MN792921.1) (Song et al. Citation2021) as references.
Data description
Gene annotation and base composition
The assembly results showed that the mitochondrial genomes of the three samples were almost the same (with an identity of 97.24%∼99.59%) and we take ZJU-1 as an example. The mitochondrial genome of E. dentilineata is 15,356 bp in length (GenBank accession OP009386), containing the entire set of 37 mitochondrial genes and an A + T-rich region, but part of the control region (D-loop) failed to be sequenced or assembled (Figure ).
Figure 2. Mitogenome annotation of Ectropis dentilineata. CDS, tRNAs, and rRNAs are shown with standard abbreviations. Transcription directions are indicated by arrows, inner circles indicating GC Skew, and legends are shown in the upper right corner of the picture.
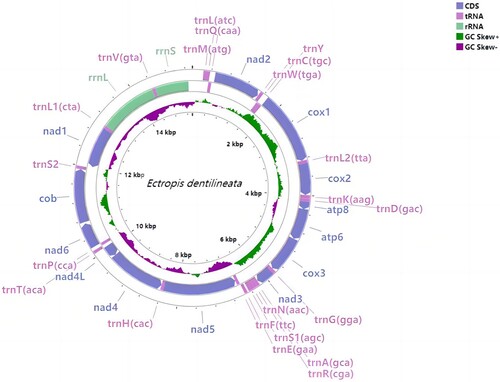
The overall base composition of the mitogenome was as follows: A (41.3%), T (38.5%), C (12.1%), and G (8.1%), with a high A + T-content of 79.8%. The mitogenome exhibits obvious negative GC-skews (−0.198) and positive AT-skews (0.035), while the protein-coding genes (PCGs) show the negative GC-skews (−0.005) and negative AT-skews (−0.049). 12 of the 13 PCGs have the conventional ATN start codons for invertebrate mitochondrial PCGs (two ATA, three ATT, two ATC and five ATG). The start codon of nad1 is TTG, which is different from other PCGs. About the stop codon, 10 of the 13 PCGs terminate with TAA. However, cox2, nad5, and nad4 terminate with the special stop codon T. The order of PCGs in E. dentilineata is the same with Drosophila yakuba, which is thought to be the ancestral arthropod mitogenome (Clary and Wolstenholme Citation1985) and the tRNA cluster of E. dentilineata rearranged to trnM – trnI – trnQ compared with the trnI – trnQ – trnM in D. yakuba (Figure ).
Author contributions statement
Conceptualization, Zhizhi Wang, Xiaogui Zhou and Xuexin Chen; software, Ruizhong Yuan, Pu Tang and Zhaohe Lu; writing – original draft preparation, Zhaohe Lu and Ruizhong Yuan; writing – review and editing, Zhizhi Wang, Pu Tang and Xuexin Chen; investigation, Xiaogui Zhou and Ruizhong Yuan; project administration, Zhizhi Wang, Pu Tang and Xuexin Chen; funding acquisition, Zhizhi Wang and Xuexin Chen. All authors have read and agreed to the published version of the manuscript.
Ethical approval
Ectropis dentilineata is a widely distributed Lepidoptera pest and is not a protected species. Therefore, the collection and research process are conventional and do not need to follow strict standards. This study did not involve ethical approval, and the field investigation was supported by the Tea Research Institute, Chinese Academy of Agricultural Sciences.
Acknowledgements
The authors would like to thank Xiao-Han Shu and Xi-QianYe of Institute of Insect Sciences, Zhejiang University for their help with the preparation of figures in this paper.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. OP009386. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA869498, SRR21047336, SAMN30308888 SAMN35736986, and SAMN35736987, respectively.
Additional information
Funding
References
- Ashfaq MA, Rafi MAS, Mansoor S, Hebert PDN. 2017. Mapping global biodiversity connections with DNA barcodes: Lepidoptera of Pakistan. PLoS ONE. 12:e0174749. doi:10.1371/journal.pone.0174749.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Son P, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477. doi:10.1089/cmb.2012.0021.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120. doi:10.1093/bioinformatics/btu170.
- Clary DO, Wolstenholme DR. 1985. The mitochondrial-DNA molecule of Drosophila-yakuba - nucleotide-sequence, gene organization, and genetic-code. J Mol Evol. 22:252–271. doi:10.1007/BF02099755.
- Crampton-Platt A, Timmermans MJTN, Gimmel ML, Kutty SN, Cockerill TD, Khen CV, Vogler AP. 2015. Soup to tree: the phylogeny of beetles inferred by mitochondrial metagenomics of a bornean rainforest sample. Mol Biol Evol. 32:2302–2316. doi:10.1093/molbev/msv111.
- Dai LS, Kausar S, Abbas MN, Wang TT. 2018. Complete sequence and characterization of the Ectropis oblique mitochondrial genome and its phylogenetic implications. Int J Biol Macromol. 107(Pt A):1142–1150.
- Mitter C, Davis DR, Cummings MP. 2017. Phylogeny and evolution of lepidoptera. Annu Rev Entomol. 62:265–283. doi:10.1146/annurev-ento-031616-035125.
- Moore F. 1867. On the Lepidopterous insects of bengal. Proc Zool Soc Lond. 35(1):612–686.
- Peng Y, Leung HCM, Yiu SM, Chin FYL. 2012. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 28:1420–1428. doi:10.1093/bioinformatics/bts174.
- Ratnasingham S, Hebert PDN. 2007. BOLD: the barcode of life data system (www.barcodinglife.org). Mol Ecol Notes. 7:355–364.
- Sihvonen P, Mutanen M, Kaila L, Brehm G, Hausmann A, Staude HS. 2011. Comprehensive molecular sampling yields a robust phylogeny for Geometrid moths (Lepidoptera: Geometridae). PLoS ONE. 6:e20356. doi:10.1371/journal.pone.0020356.
- Song XH, Yang TB, Xu XQ, Yan XH, Zhou CQ. 2021. Characterization of the complete mitochondrial genome of Ectropis grisescens (Lepidoptera, Geometridae). Mitochondrial DNA B Resour. 14;6(7):1953–1955. doi:10.1080/23802359.2021.1923423.
- Strutzenberger P, Brehm G, Fiedler K. 2012. DNA barcode sequencing from old type specimens as a tool in taxonomy: a case study in the diverse genus Eois (Lepidoptera: Geometridae). PLoS ONE. 7:e0049710. doi:10.1371/journal.pone.0049710.
- Timmermans MJTN, Lees DC, Simonsen TJ. 2014. Towards a mitogenomic phylogeny of Lepidoptera. Mol Phylogenet Evol. 79:169–178. doi:10.1016/j.ympev.2014.05.031.
- Wingett SW, Andrews S. 2018. Fastq Screen: a tool for multi-genome mapping and quality control. F1000Res. 7:1338–1338. doi:10.12688/f1000research.15931.1.

