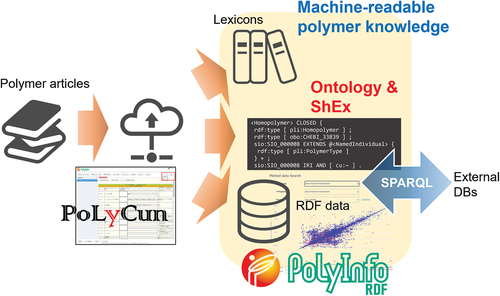
ABSTRACT
Herein, we propose a machine-readable knowledge representation of PoLyInfo, the polymer database of National Institute for Materials Science (NIMS) of Japan with more than half a million data points. Through the use of the Shape Expressions (ShEx) language, PoLyInfo was made entirely machine-readable, allowing the formulation of polymer chemistry concepts. In particular, the following tasks were accomplished: (1) the relationships between homopolymer – copolymer – polymer blends were described at the molecular level, replicating the essential data management of PoLyInfo; (2) the relationships between composites, compounds, and neat resins were formulated, and the positions of additives, such as fillers, were clarified; (3) the synthesis process was made machine-readable, and the roles of materials, such as catalysts and initiators, were described according to international standards; (4) a post-synthesis forming process was formulated, and its relationship with the final polymer sample was concretized; and (5) all properties in PoLyInfo were modeled and semantically related to the measurement conditions.
Impact statement
More than half a million data points and data structures in the NIMS polymer database PoLyInfo were machine-readable by ontology and data normalization based on global standards.
1. Introduction
As data-driven science is becoming more popular, the polymer database PoLyInfo of National Institute for Materials Science (NIMS) of Japan [Citation1], comprising more than half a million polymer data points manually collected over several decades, is changing from its originally expected use as a reference for polymer chemistry to a variety of unprecedented uses, including training data for machine learning and material exploration of unexplored areas [Citation2,Citation3]. This paper is the second half of a series of research papers, of which the first half is called PoLyInfo (I), and the second half, i.e. this paper, is called PoLyInfo (II). It should be noted that the PoLyInfo (I) [Citation4] and (II) papers are twins: PoLyInfo (I) details the dataset behind the graphical user interface (GUI) version of PoLyInfo, which is published for human-readable views of the data, whereas PoLyInfo (II) describes an international common-knowledge expression for machine-readable views of the polymer chemistry inherent in the data. These papers cover the same topics as much as possible. Thus, it is recommended to read through PoLyInfo (I) before reading PoLyInfo (II) to understand PoLyInfo as a database. As mentioned therein, PoLyInfo includes systematized multi-scale polymer structures, which, together with the various properties determined by the resulting higher-order structures, have produced over half a million data points. Whereas the PoLyInfo GUI enhances the storytelling and visibility of the data by presenting the stored data redundantly, we discussed the outline of the curated original dataset from an editor’s perspective in PoLyInfo (I). We believe that this overview will help users understand PoLyInfo as a whole, going beyond the limitations of a GUI dedicated to daily use. However, if this overview were further refined and each piece of data were documented in a human-readable manner, it would likely result in a massive manual that would be difficult to read through. We believe that what is currently needed is to normalize the PoLyInfo data structure and create a machine-readable schema to accurately represent polymer chemistry knowledge. By creating this schema, we expect to accomplish the following:
Presentation, sharing, and preservation of items necessary for the representation of polymer chemistry knowledge.
Accurate understanding of the conceptual relationships among these items.
Use for validation of new polymer dataset.
Linkage of this knowledge with external non-polymer databases, i.e. huge knowledge construction.
PoLyInfo (II) consists of the following:
1. Introduction
2. PoLyInfo ontology overview
3. Designed namespace architecture
4. Semantic search using Resource Description Framework (RDF) and schema generalization
5. Contents of PoLyInfo schema
5.1. Master information
5.2. Material information
5.3. Fabrication information
5.4. Formation information
5.5. Properties information
6. Challenges and prospects
7. Conclusion
The Semantic Web technology used in this study, standardized by the World Wide Web Consortium (W3C) [Citation5], enables automatically findable and sophisticated processing of web content on various sites, normally read by humans, by providing it with machine-readable semantics. The underlying technologies are the Web Ontology Language (OWL) [Citation6], RDF [Citation7], and the SPARQL Protocol and RDF Query Language (SPARQL) [Citation8]. The W3C technical papers [Citation9–11] provide more information on these specifications. Twenty years have already passed since RDF became a W3C recommendation in 1999 and its basic specification was finalized in 2004. During this time, RDF has become a fundamental technology for adding semantics to data and for realizing advanced search. RDF enables integrated retrieval by defining the vocabulary in an ontology and then writing all information in machine-understandable sentences called ‘triples’, which are subject – predicate – object combinations. The idea of linking data using RDF has already been established, and thus, there is no need to reiterate it. The main topic of discussion herein is a schema that determines what conceptual definitions of vocabulary are needed to express polymer data, what predicates are used to represent polymer chemistry, and how the triples are graphically networked. The schema aggregates the knowledge of the polymer domain, from which PoLyInfoRDF is ultimately created.
Domain knowledge aggregation schemas have been developed outside the field of materials science, particularly in biology [Citation12], and indeed, there is a long history of semantic database-to-database linkages using RDF. Herein, we describe the current status of schema and data linkages, focusing on PubChemRDF [Citation13] and NikkajiRDF [Citation14], which are widely used in the field of organic small-molecule chemistry, which is the closest to the field of polymer chemistry, under which PoLyInfo belongs. PubChemRDF is the most current with its version 1.8.0 beta, released in February 2023 [Citation15]. It has been updated every year since the release of its first version, 1.0.0 beta, in January 2014 and is already 10 years old [Citation16]. The current total number of triples in PubChemRDF is 238,429,023,647, of which the namespace ‘Compound’ (https://rdf.ncbi.nlm.nih.gov/pubchem/Compound/), the namespace most closely related to PoLyInfoRDF, has triple counts of
Non-neighboring links: 2,764,650,647
2D neighboring links: 85,958,021,112
3D neighboring links: 134,282,846,831
Practical documents on the content and examples of SPARQL are summarized in the PubChemRDF official documentation [Citation17].
The semantic use of RDF, i.e. cross-domain integration with external resources, is made possible by the Chemical Information Ontology (CHEMINF) [Citation18] certified by the Open Biological and Biomedical Ontologies (OBO) Foundry [Citation19], and its importance in the linkage with PoLyInfo is described later. Although the Compound namespace in PubChemRDF contains mainly computational results that are different from the experimental data in PoLyInfo, it still has many similarities with PoLyInfo’s schema, such as the relationship between individual sample names and normalized material names, and the notation of property names and values.
NikkajiRDF, managed by the National Bioscience Database Center (NBDC) of Japan, was first published in August 2015 and has since been expanded to a database of small molecules similar to PubChem, with 128,704,657 triples in the main namespace (http://rdfportal.org/dataset/nikkaji/main). The Nikkaji number, which can be mapped to PubChem compound identifiers (CIDs), is also structured based on CHEMINF, and general information specific to molecular structures, such as the International Chemical Identifier (InChI) [Citation20], canonical simplified molecular-input line-entry system (SMILES) [Citation21], and molecular weight, are related as attributes [Citation22]. Of particular importance to this NBDC initiative is the integration of the Nikkaji number with the IDs of PubChem and UniChem (an integrated resource for small molecules, including the European Molecular Biology Laboratory – European Bioinformatics Institute, EMBL-EBI [Citation23]. The Nikkaji number serves as a gateway to external resources. Linkages with PubChem are provided via RDF for 34,671,881 triples and with UniChem for 18,910,927 triples (both as of 2022).
However, these data schema provision methods are in human-readable format referring to several examples, which is thought to be a bottleneck in the actual automatic understanding of data structures by machines. Once machine-readable schemas are created, Semantic Web technologies will become more empowered. This is the main objective of this study from a Semantic Web perspective.
2. PoLyInfo ontology overview
PoLyInfo (I) provides an overview of the more than half a million data points displayed on the PoLyInfo GUI. Prior to creating machine-readable schemas for these data, we defined the target materials (material entity class) such as Homopolymer, Copolymer, and Monomer, the processes used (process class) such as Polymerization, Measurement, and Molding, and other concepts necessary for polymer chemistry. In addition, these defined classes were organized hierarchically using OWL to create a PoLyInfo ontology. Although ‘ontology’ is originally a philosophical term, the ontology described here is an informatics method that classifies the subject to be discussed (known as the ‘Universe of Discourse’) into concept classes, annotates them, clarifies the definitions, and provides the relationships and constraints among the classes and the named individuals that are types of each class. The characteristics of the PoLyInfo ontology are as follows:
Use of underlying concepts based on the well-known international top ontology Basic Formal Ontology (BFO) [Citation24] designed by the OBO Foundry
Use of middle-class ontologies from OBO, such as Information Artifact Ontology (IAO) [Citation25], if necessary for polymer chemistry representation
Use of predicates from well-known external ontologies such as RDF Schema (RDFS) [Citation26] and semanticscience integrated ontology (SIO) [Citation27] whenever possible to describe relationships between entities, minimizing domain-specific predicates to be defined in the PoLyInfo ontology
Use of definitions of polymer chemistry terms from PoLyInfo Help [Citation28], the International Union of Pure and Applied Chemistry (IUPAC) Gold book [Citation29], etc., to ensure the interface between machine readability and human readability
The BFO conforms to the requirements of ISO/IEC (International Organization for Standardization; International Electrotechnical Commission) 21838–1 for a top-level ontology, is based on ontological realism and neutrality, and is designed to cover the concepts of modern physics. Details on the BFO concept have been published in a book [Citation24]. Although cross-domain SPARQL queries that directly include BFO classes are rare, the consistency of the upper concepts ensures that there are no discrepancies among the domains, even homonyms. That PubChem and Nikkaji are based on the OBO family ontology strengthens their linkages with PoLyInfo. The PoLyInfo ontology uses the following five BFO classes, which are basic concepts used in science:
process (obo:BFO_0000015)
role (obo:BFO_0000017)
specifically dependent continuant(obo:BFO_0000020)
generically dependent continuant(obo:BFO_0000031)
material entity (obo:BFO_0000040)
Here in, obo is a prefix, and with the list of prefix/URI correspondences summarized in Appendix A, the definitions of and detailed information on each class can be obtained on the web (e.g. for obo:BFO_0000040 of material entity, http://purl.obolibrary.org/obo/BFO_0000040). It is important to emphasize that there are more than 100 ontologies in the OBO definition site Ontobee [Citation30] alone that share these class concepts, and that there is no conceptual discrepancy between each of these domains and the PoLyInfo ontology. Perhaps, we should detail how the aforementioned five BFO conceptual classes are broken down into polymer concepts (via a middle-class ontology, if necessary). However, to avoid complicating the article, we refer the reader to the published ontology [Citation31] for more details, and note two points in particular that should be emphasized.
The acronym PSPP, which represents process, structure, property, and performance, is a well-known term used to summarize material concepts [Citation32]. One concept not included in the PSPP but is included among the listed five classes is (2) role (obo:BFO_0000017). This means that the PoLyInfo ontology can explicitly describe cases in which one material has more than one role. In other words, a role is defined within an action (process) and is not an attribute intrinsic to a material. A representative process in PoLyInfo is polymerization, whereas suitable roles include reactants, catalysts, and initiators, among others. This definition enables searches for polymerization roles in PoLyInfoRDF that are not searchable in the PoLyInfo GUI.
Another important BFO conceptual class for the PoLyInfo data structure is (5) material entity (obo:BFO_0000040). This class has the subclasses ‘Chemical substance’ and ‘Sample structured material’. ‘Chemical substance’ defines substances lexicographed in PoLyInfo, such as polyethene with the IUPAC structure-based name ‘poly(methylene)’ and polymer ID (PID) P010001 in PoLyInfo, and other general chemicals that are not identified as individuals, such as decahydronaphthalene as solvent. On the other hand, ‘Sample structured material’ refers to individual polymers using the PoLyInfo sample ID. Thus, this class reproduces the two major features of PoLyInfo described in PoLyInfo (I):
Required PoLyInfo registration conditions: molecular structure determination and normalization by IUPAC name
Data curation per sample
The statistics of the PoLyInfo ontology (version 1.0) are as follows:
Number of classes (owl:Class): 1,417
Number of predicates (owl:ObjectProperty): 7
Number of named individuals (owl:NamedIndividual): 907
Total triples: 9,691
Hereafter, the prefix of the PoLyInfo ontology will be ‘pli’, and the actual URI of pli is indicated in Appendix A.
3. Designed namespace architecture
The key idea in the schema determining the PoLyInfoRDF data structure is ‘static data management’. Polymers have multi-scale structures, ranging from the molecular-level constitutional unit (CU) determined by synthesis to the macroscopic shape determined by the molding process, etc. If we trace the sample preparation process, the polymer scale will evolve over time from micro- to meso- to macro-scale. Even if we consider only the synthesis, the structure and composition will change dynamically as elementary processes related to polymerization and additives progress. However, PoLyInfo does not maintain all the dynamic information. Although it sometimes partially records time-series information in literal form, it has a static data structure consisting of typical material roles, such as reactants and additives, and typical experimental conditions of the processes. This may not be sufficient to produce a large collection of polymer sample preparation procedures; however, it is superior in that it organizes items commonly dealt with in many research studies as metadata and makes them semantically and cross-domain searchable in the RDF.
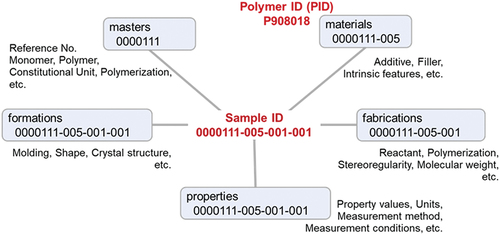
The idea of static data management made us realize that the polymer and sample ID systems discussed in PoLyInfo (I) can be reconstituted in the schema; PoLyInfo manages the processes of polymer sample preparation from primary to higher-order structures with rules by sequentially increasing the ID branch numbers. However, it is possible to reproduce database functions with another data architecture centered on the sample and arranged radially in terms of the following namespace groups ():
Figure 1. Machine-readable data architecture of PoLyInfo. Namespace groups are arranged radially around the polymer sample.
Master information, which normalizes the chemical substances, CUs of polymers, and polymerization (masters)
Materials information, which summarizes additives, fillers, etc. (materials)
Fabrication information, which summarizes synthetic processes (fabrications)
Formation information, which summarizes the higher-order structures of samples (formations)
Properties information of the resulting sample (properties)
With a radial data architecture, the attributes and properties of a sample can be accessed directly and multiplied with short SPARQL queries. If this information were stacked to mimic a dynamic process, it would require long SPARQL queries to follow the data links from the materials to the properties namespaces. In this figure 0000111-005-001-001 is an example of a sample ID, which is normalized to PID P908018 in the materials namespace group, leading to the normalization of polymer sample structures based on SMILES and the IUPAC name. For this example, the IUPAC name of the normalized polymer is poly{1-carboxyethylene/1-[(ethane-1,1,2-triyl-1-carbonyl)oxyzincanediyloxycarbonyl]ethylene/ethylene/1-(methoxycarbonyl)ethylene}, as is described in the masters namespace. As shown in the figure, for each namespace group, the ID branches are organized from the primary to higher-order structures.
These namespace groups actually have many connections with the central polymer sample with predicates (graph edges) of RDF. The predicate statistics are summarized in detail in for the predicates that make RDF objects into polymer samples and in 1(b) for those that make subjects. All of the predicates in the RDF are defined by external ontologies, and there are three types for the objects and two for the subjects. These specific types and numbers are also outlined in the table. The predicate with the largest number of triples is the ‘attribute’ relation (sio:SIO_000008, has attribute) in item #1, which is consistent with the large number of properties in PoLyInfo that act as attributes (the details will be discussed in Section 5.5). Specifically, there are 104 types of objects with 1,453,968 triples present in PoLyInfoRDF. On the other hand, the predicate with the smallest number of triples is the obo:BFO_0000051 (has part) in item #2. This is closely related to the management policy for polymer blends in PoLyInfo; specifically, PoLyCun curates the polymer components in each polymer blend sample as other nested samples. This precise component management is reflected in its 49,355 triples as part of the radial connection.
Table 1. (a) Predicates that make RDF objects into polymer samples and (b) those that make subjects.
4. Semantic search using RDF and schema generalization
Each namespace group discussed in Section 3 will be subdivided into actual PoLyInfoRDF. The machine-readable schema discussed in Section 5 will be based on the PoLyInfoRDF, but it will not be created by automatically normalizing all relevant RDFs; instead, it will be refined through a process of manual elaboration, including conceptual design (discussed as follows), modularization of common items, and finally, release as optimal for representing polymer knowledge. During the schema-creation process, RDF that is not well described may be revised. Through a series of reviews, both the schema and RDF issues will converge. Naturally, the RDF and schema become consistent, and thus, the PoLyInfoRDF validation check with the schema passes successfully.
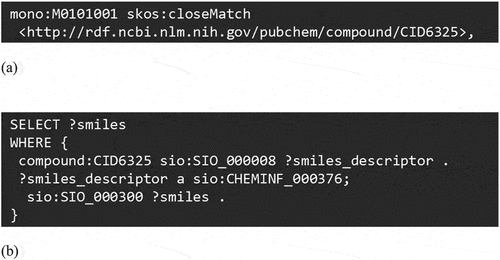
PoLyInfoRDF provides machine-readable data, including concepts, attributes, and semantic relations, for all instances described in PoLyInfo (I). These are defined in the PoLyInfo ontology in ways consistent with the BFO, resulting in a semantically integrated search of all PoLyInfo data in external databases. This means that the simultaneous use of external endpoints allows a seamless connection to data outside the polymer domain akin to traversing only a single database, thus creating a huge knowledgebase. A representative example of an integrated search is presented as follows: PoLyInfoRDF declares that ethene with a monomer ID of M0101001 in PoLyInfo is equivalent to a chemical with an ID of CID6325 in PubChem (). Herein, ‘mono’ refers to the monomer namespace in the master information (masters) shown in . To retrieve the canonical SMILES of ethene (C=C) from PubChemRDF, we can execute a SPARQL query, as shown in . It should be noted here that canonical SMILES (sio:CHEMINF_000376) is a descriptor for chemicals (in this case ethene) in PubChem: Other computational descriptors similar to canonical SMILES, such as structure complexity (sio:CHEMINF_000390) and hydrophobicity/hydrophilicity (sio:CHEMINF_000395), can be retrieved with the similar query, thus seamlessly connecting PoLyInfo experimental data with the various computational data.
Figure 2. Integrated search schemes. (a) Example of ID linkage description with PubChem in PoLyInfoRDF. (b) SPARQL query to retrieve canonical SMILES from PubChemRDF.
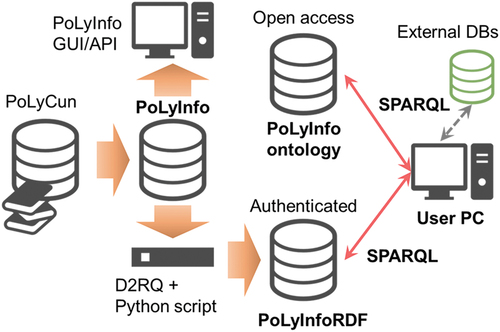
PoLyInfo is a relational database (RDB), and its data are collected by the PoLyInfo Curation System (PoLyCun). The RDB is the core of PoLyInfo and provides users with data in various formats via GUI, application programming interface (API), and RDF, as conceptually shown in . The RDB is converted to RDF using D2RQ [Citation33] and an original Python script and fed to a triple store. The latest version of PoLyInfoRDF has 24,593,403 triples. There are two types of triple stores: open-access and authenticated. The open-access type contains ontologies and other highly public items that can be freely used to extend data linkages. On the other hand, the authenticated type contains data that NIMS has capitalized and can be used under a certain contract. Currently, PoLyInfoRDF data are available for access only from these endpoints, and not for GUI services based on fixed stories. However, users are free to conduct customized searches and inferences for their own purposes using any SPARQL.
The next challenge is to establish a way to present the entire RDF and generalize semantic technologies, such as an integrated search. To ensure generality, it is clear that all RDF schemas must be described in a well-known format, rather than in local grammar or in a graph representation using typical examples. To this end, we have chosen to describe the schema in the Shape Expressions (ShEx) language [Citation34]. ShEx can flexibly represent RDF node and edge structures; it can specify the data formats of nodes such as IRIs, blank nodes, and literals, and for graph structures composed of these nodes and predicates (edges), it can specify the cardinality. Although a similar language, the Shapes Constraint Language (SHACL), can be used to express RDF schemas and is a W3C Recommendation [Citation35], ShEx is superior in terms of extensibility and descriptiveness and has been adopted by Wikidata [Citation36] and Gene Ontology [Citation37], among others. Thus, in terms of performance and popularity, it is better than SHACL [Citation38].
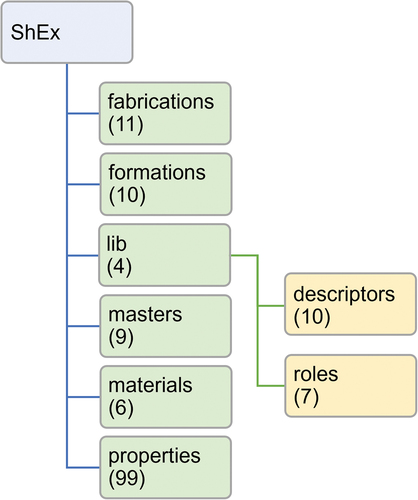
The schemas of PoLyInfo data can be modularized using ShEx; for example, common parts can be modularized among more than 100 properties and then imported to each property, extended, or restricted as desired. These schemas can be used for validation to create a new RDF that is conceptually consistent with PoLyInfo. All ShEx schemas that can reproduce PoLyInfoRDF are published in an open data repository (Materials Data Repository, MDR) [Citation39]; and the folder tree structure of the schema is shown in . The ShEx folders correspond to the radial namespace groups in , whereas the common modules are stored in the ‘lib’ folder. Additionally, related modules are grouped in the subfolders ‘descriptors’ and ‘roles’. Here, the numbers in parentheses indicate the number of files in each folder; many schemas are prepared mainly in the properties folder. Examples of specific knowledge expressions that use schemas are presented in the following section.
Figure 4. Folder tree structure of published ShEx files at https://doi.org/10.48505/nims.4415.
5. Contents of PoLyInfo schema
5.1. Master information
In Section 5, we introduce ShEx as a means to normalize various instances in PoLyInfo and render the polymer knowledge expression machine-readable. Although we have compactly normalized PoLyInfoRDF, it is still exceedingly large to be included in this paper; therefore, we will highlight only a representative example of each namespace group and describe its main points. Nonetheless, this provides an introduction to trace back to the original ShEx. With the help of international standard specifications, no further supplementation will be necessary for a complete understanding of the schemas.
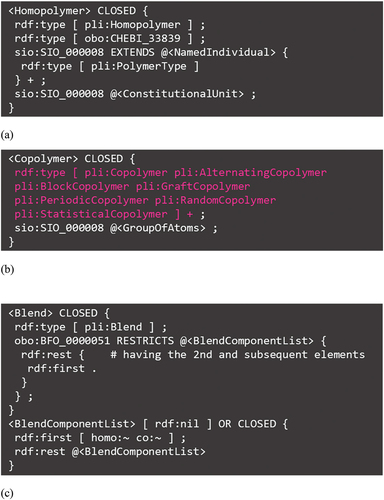
The most important files in the ‘masters’ namespace group would be ShEx for homopolymers, copolymers, and polymer blends. shows a portion of Homopolymer.shex. As shown in this figure, Homopolymer is a type of homopolymer (pli:Homopolymer, where ‘pli’ indicates that it is defined in the PoLyInfo ontology; see end of Section 2) and also a type of macromolecule (obo:CHEBI_33839) defined in an external ontology, Chemical Entities of Biological Interest (ChEBI) of the OBO Foundry. The homopolymer has one or more (+) polymer types (named individual of pli:PolymerType), such as polyolefins, as attributes (sio:SIO_000008, has attribute). Herein, the characters ‘+’, ‘*’, and ‘?’ characters in ShEx are cardinalities according to the notation of the XML specification, indicating one or more, zero or more, and zero or once, respectively. Another attribute, <ConstitutionalUnit>, is CU modularized in structureDescriptors.shex in the lib folder, which ensures master management at the molecular level specific to PoLyInfo.
As mentioned in PoLyInfo (I), PoLyInfo has the following two data management points with regard to copolymers:
The constitutional repeating unit (CRU) of copolymers is represented by a combination of the CU and junction unit (JU)
Copolymers with the same CRU but different middle-range orders (Statistical, Random, Alternating, etc.) are managed using different copolymer IDs (COID)
These features are reproduced in Copolymer.shex in the ‘masters’ namespace group. As shown in , there is a list of classes for identifying the middle-range order (shown in pink), and a CRU is given as an attribute <GroupOfAtoms> with CU and JU modularized in structureDescriptors.shex.
In the case of polymer blends, we found that the homopolymers and copolymers included as components can be described using a nested structure that draws out recursively, as summarized in Blend.shex in . Because their decomposition into CUs can be handled by the schemas in , molecular-level polymer management, which is a feature of PoLyInfo, can be expressed using the schemas in .
The advantage of this expression is its robustness to changes in the number of component polymers in the polymer blend; the recursive drawing of polymer components into a first component and other components reproduces the multiple components in any polymer blend. shows an example for polyethene//polyethene//poly[ethylene-co-(hex-1-ene)] (BDID BD000003). This figure shows two types of copolymers, polyethene (P010001) and poly[ethylene-co-(hex-1-ene)] (P900354), being drawn out sequentially.
Figure 6. Recursive management of polymer components in polymer blend. An example for polyethene//polyethene//poly[ethylene-co-(hex-1-ene)] (BDID BD000003).
![Figure 6. Recursive management of polymer components in polymer blend. An example for polyethene//polyethene//poly[ethylene-co-(hex-1-ene)] (BDID BD000003).](/cms/asset/98667da8-fc3d-42f4-9423-d58ceea14e50/tstm_a_2354651_f0006_oc.jpg)
Finally, end-groups are managed in PoLyInfo, and the corresponding ShEx file is EndGroup.shex in the folder of this category. It is worth noting here that molecular-level data management in PoLyInfo extends to the terminal structure of the polymer chain.
5.2. Material information
The ‘materials’ namespace group in organizes the relationship between the sample ID and its normalized ID, the PID, in terms of the constituent materials; the Identification.shex in the namespace group shows this in a straightforward manner. As discussed in PoLyInfo (I), all the samples with higher-order structures in PoLyInfo can be categorized as primary structures. Although the primary structure is often used in machine learning as a structural descriptor, it is necessary to understand that this is not a hash value for higher-order structures but merely a taxonomic superclass name for polymer components. The exact relationship between the polymer sample (pli:PolymerSample) and polymer (pli:Polymer) in PoLyInfo can be understood from the ShEx file, as shown in .
Figure 7. ShEx for relationship between polymer sample (pli:PolymerSample) and polymer (pli:Polymer) in PoLyInfo.
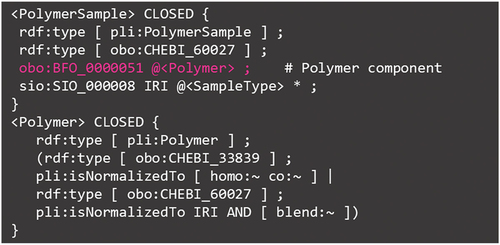
The important points of this ShEx are as follows. (1) The polymer sample is explicitly indicated to contain polymers (shown in pink), and the predicate obo:BFO_0000051 (has part) reproduces the radial connection in (c.f., item #2 in . On the other hand, the polymer sample also includes fillers and additives that are not polymers, and these substances are expressed with Filler.shex and Additive.shex in the materials namespace group. The actual situation is expressed in this ShEx citing ChEBI; polymer sample (pli:PolymerSample) is a type of obo:CHEBI_60027, a sub-concept of ‘mixture’, while homopolymer (pli:Homopolymer) and copolymer (pli:Copolymer) in polymer (pli:Polymer) are a type of obo:CHEBI_33839, a material with ‘multiple repetition of units’. (2) Although composites and compounds have the same PID as neat resins when normalized, in many cases, the effects and impacts of the additives are more important to the industry than the properties of neat resins. ShEx clarifies that the distinction between neat resins, composites, and compounds is summarized in Named Individual of sample type (pli:SampleType). Herein, the PoLyInfo ontology shows that this sample type also includes chelate and inorganic polymers. However, this is due to a temporal idea of metal complexes and semimetals as special additives in the long history of PoLyInfo curation, and the number is not large: only 12 chelate polymers and 2884 inorganic polymers. More importantly, even this infrequent information can be reliably recognized using ShEx and ontology, and immediately approached using SPARQL, if necessary.
5.3. Fabrication information
In Section 5.2, we examined a radial connection between a polymer sample and neat polymer and rendered it machine-readable. The next thing to consider in Sections 5.3 and 5.4 would be the machine readability of ‘fabrications’ and ‘formations’ for polymers, and the establishment of the radial connections in .
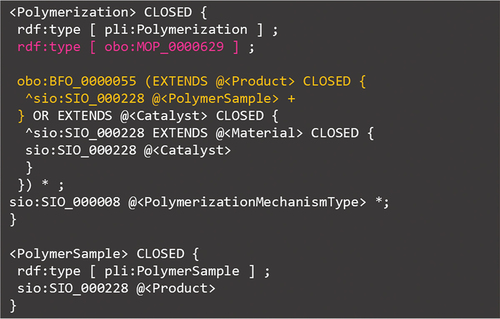
The main focus of this subsection with regard to fabrications is on polymerization information. In PoLyInfo (I), it is reported that this information is processed and catalogued on the GUI. Although PoLyCun originally contains sufficient items to have a search function for polymerization, it is not easy to formulate the relationship between them and organize it as a GUI service. As a solution to this problem, the machine readability and semantic expressions of PoLyInfo introduced herein are extremely useful. The static data management of PoLyInfo organizes processes by defining roles, and ShEx generalizes these roles and stores them as modules under the lib folder, as shown in . The ShEx file PolymerizationInformation.shex first imports these modules and compactly summarizes the polymerization information. shows a brief ShEx description of the polymerization information using the product and catalyst roles as examples. Other roles are omitted for the sake of clarity; the exact schema can be obtained from the original ShEx file.
The important points of this ShEx are as follows: (1) As shown in pink, Polymerization refers to the Molecular Process Ontology (MOP) [Citation40] definition obo:MOP_0000629 (polymerisation) published by OBO Foundry: ‘The process of converting a monomer mixture of monomers into a polymer. [database_cross_reference: https://doi.org/10.1351/goldbook.P04740]’. From here, it can be observed that the human-readable IUPAC Gold book definitions are cited via MOP. The integration of knowledge through data linkage is one of the advantages of this initiative. (2) As highlighted in orange, the polymerization is connected to each role through the predicate obo:BFO_0000055 (realizes), whereas the corresponding materials are connected to the roles via the predicate sio:SIO_000228 (has role). These connections satisfy the predicate constraints defined in the BFO and SIO, and it can be concluded that even the domain knowledge, i.e. polymer chemistry, is able to inherit the international description rules.
As shown in and , each namespace group is radially arranged around the polymer sample, and there are several ways to connect them. Considering (2) in detail, the polymer sample is connected to the polymerization process through a product role (pli:Product). The predicate here is sio:SIO_000228 (has role), which corresponds to item #3 in ). Through that polymerization, the polymerization mechanism type (pli:PolymerizationMechanismType) is connected as an attribute. On the other hand, the stereoregularity strongly related to the polymerization process is connected as an attribute of the polymer sample in the file StereoregularityInformation.shex in this ‘fabrications’ namespace group. Consequently, hybrid SPARQL queries can clarify these two relationships, as discussed in PoLyInfo (I).
Because the average molecular weight is important information, PoLyCun records it in detail using the measurement method and conditions. The degree of polymerization, solution viscosity, and melt flow rate are also similarly curated. Although these properties are categorized into fabrication information, the detailed schema involving measurements will be explained in the properties namespace group in Section 5.5, owing to their similarity. Herein, we emphasized that the normalized ShEx for the average molecular weight explicitly indicates its type, which is also defined in the PoLyInfo ontology. It is possible in SPARQL to sort them and display only the corresponding polymer samples; therefore, the types and numbers of cases are as follows:
Average molecular weight (pli:AverageMolecularWeight): 2,246
gel permeation chromatography/size exclusion chromatography (GPC/SEC) peak molecular weight (pli:GpcSecPeakMolecularWeight): 190
Number-average molecular weight (pli:Number-AverageMolecularWeight): 1,970
Viscosity average molecular weight (pli:ViscosityAverageMolecularWeight): 1,970
Weight average molecular weight (pli:WeightAverageMolecularWeight): 18,629
Z average molecular weight (pli:ZAverageMolecularWeight): 76
This result is not made available by the storytelling-oriented PoLyInfo GUI and thus demonstrates the flexibility of PoLyInfoRDF in retrieval.
5.4. Formation information
In this category for ‘formations’, information on the higher-order structure of polymers is compiled. For industrial applications, it is important that the synthesized polymers are mixed with other materials and molded. The ShEx values of higher-order structures can be divided into the following three categories:
Microscopic attributes: summarizes the results of measurements of the crystal structure, crystallinity, orientation, etc.
Macroscopic attributes: morphology, moldingSampleShape, and other macroscopic shapes
Forming process: summarizes the macroscopic processes used to complete a polymer sample, such as molding and molding treatments
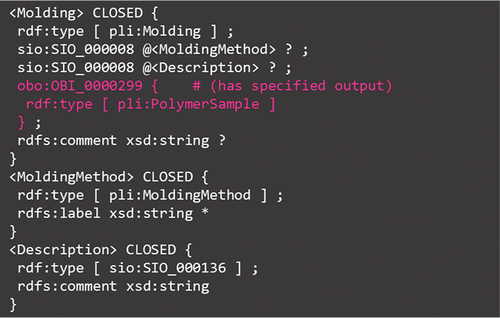
This subsection focuses on molding in the ‘forming process’ category. This is because the molding is connected to the polymer sample in a unique manner via the predicate shown in , whereas the other micro- and macroscopic attributes are connected to the polymer sample in a similar manner as in the other subsections. summarizes Molding.shex.
The main points of this ShEx are as follows: (1) Molding is defined in the PoLyInfo ontology and has two attributes: molding method and description. It seems that the molding methods (pli:MoldingMethod) could be defined as named individuals in the ontology and given URIs but are actually given in literal form because there are 538 items, owing to the fluctuation of the terminology in papers. On the other hand, for the description, the type sio:SIO_000136 is used to semantically clarify that it is a concrete specification rather than an accompanying information for the molding method. (2) It is conceptually important to note that the molding ‘has specified output (obo:OBI_0000299)’ of polymer sample (highlighted in pink). The predicate used here corresponds to item #1 in ). This notation is applicable to molding treatment (pli:MoldingTreating) and uniformization (pli:Uniformization). Molding changes only the shape of the polymer sample, which is different from the polymerization process, in which the polymer sample plays the role of a product, as discussed in Section 5.3.
5.5. Properties information
Details on approximately 100 properties recorded in PoLyInfo are omitted here and can instead be obtained from the NIMS website [Citation41].
Because most of the properties have commonality when expressed in ShEx, and because all properties can be reproduced by replacing minor parts, we have modularized the bases of the ShEx. We have divided ShEx into three main bases:
<Property_base>
<Measurement_base>
<IAO_0000109_base>
With these bases, a ShEx for general properties is formulated, as shown in ; a polymer sample has properties as attributes (in this case, glass transition temperature; pli:GlassTransitionTemperature), and the predicate for the radiative connection that realizes the namespace group structure shown in is sio:SIO_000008 (with attribute) (highlighted in pink). The specific property values and units are aggregated in obo:IAO_0000109 (datum), which is related to the property with obo:IAO_0000221 (is quality measurement of). The property value of the glass transition temperature is numerical, and its representation is detailed in values.shex in lib folder shown in . On the other hand, as shown in , the datum is formalized with obo:OBI_0000299 (has_specified_output) as the output of the measurement process (pli:Measurement). Furthermore, as shown in , the measurement method, measurement standard, and measurement conditions are given as attributes of the measurement process; herein, the dependence of the properties on the measurement conditions, as discussed in PoLyInfo (I), is machine-readable.
Figure 10. ShEx for polymer properties. RDF linkage of (a) property-datum and (b) measurement-datum. (c) Content details of datum.
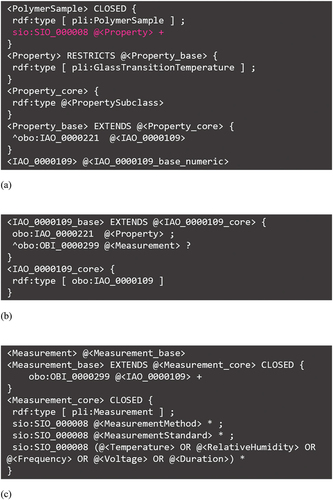
The actual properties recorded in PoLyInfo are not all those with relatively simple data structures, such as glass-transition temperatures. In this initiative, properties that include various external factors are also successively normalized in ShEx. Here, we list the three representative categories and provide an overview of the original file names for each typical example.
(1) Dynamic properties
Dielectric dispersion (pli:DielectricDispersion) is one of the electrical property values obtained by dynamic measurement and is generally expressed as complex numbers, i.e. comprising a real part (ε'), imaginary part (ε''), and phase (tan δ) of dielectric constant ε. ShEx extends <Property_base> to consider ε', ε'', and tan δ as component parts of pli:DielectricDispersion and formulates them using BFO_0000051 (has part) (see DielectricDispersion.shex).
These component parts are defined as pli:DielectricConstantAc, pli:DielectricLossFactor, and pli:DielectricLossTangent in the PoLyInfo ontology.
(2) Properties dependent on other substances
Intrinsic viscosity (pli:IntrinsicViscosityEta) clearly depends on the solvent used. In this case, <Measurement_base> is extended to indicate that the measurement process realizes a solvent role (pli:Solvent). This is the same relationship between process and role described in Section 5.3. It can be seen in IntrinsicViscosityEta.shex that the actual solvent can be related to this role using sio:SIO_000228 (has role).
(3) Model-dependent properties
Gas diffusion coefficient (pli:GasDiffusionCoefficientD) can be generalized using the Arrhenius formula. In this case, <IAO_0000109_base> is extended and formulated to be estimated using the Arrhenius formula by the predicate pli:isEstimatedBy defined in the PoLyInfo ontology. Ultimately, it can be understood that the Arrhenius formula can be modeled by identifying the activation energy (pli:ActivationEnergy) and the prefactor (pli:ArrheniusEquationPrefactor) in GasDiffusionCoefficientD.shex.
6. Challenges and prospects
As polymer chemistry is becoming more advanced, and molecular structures and properties are becoming more diverse, we have been developing a PoLyInfo knowledgebase with a history of several decades. This section addresses challenges from the standpoint of polymer knowledge and from the standpoint of machine readability of data structures.
From the polymer knowledge standpoint, there is an expectation that the limitations of the SMILES described in PoLyInfo(I) will be improved by concept detailing. For example, the fact that the same polymer can be defined by different SMILES indicates a principled lack of uniqueness in SMILES. However, considering that the structural identification of complex polymers in the current data curation is generally achieved by additional algorithms, it would be possible to address this issue by writing the equivalence of the polymers in an ontological manner. Specifically, if the concept description of the coordinates of the constituent elements and symmetry operations such as translation is sufficient, it should be possible to represent the structure of any polymer. Although such concept descriptions are not included in the current PoLyInfo ontology, but this mechanism could be implemented in a form similar to the rules for reasoning using PoLyInfoRDF. This is also similar for the representation and retrieval of various topologically complex polymer structures. Ultimately, machine-readable detailing of polymer geometry through mathematical knowledge, rather than SMILES-like hashing, will be the accurate structural descriptor. This ongoing work to make knowledge machine-readable will determine the goal of this initiative.
On the other hand, challenges from the standpoint of machine readability of data structures are as follows. Beyond the structuring of the database, the machine readability of expert knowledge, i.e. the characteristics, constraints, and intuitions of properties, as described in PoLyInfo help, has not yet been realized. In other words, we believe that the domain ontology has been created, whereas the application ontology is not. It would be difficult to make all the knowledge of polymers machine-readable for the purpose of application ontology. A realistic example in which ontologies are exploited could be as follows:
To extract canonical knowledge more accurately than conventional RDBs using semantic relationships, specializing in domain ontologies
To integrate with external ontologies and databases through direct entity (ID) linking in line with ontological realism
To create on-demand conceptual linkages for purpose-specific issues and to integrate PoLyInfo with external knowledge
To combine several conceptual linkages and reasoning about the unknowns surrounding polymer chemistry
In such utilization, a taxonomy with concept inheritance, which is well aligned with ontology, will be useful. Taxonomies that make it relatively easy to uniquely determine concept inheritance, such as crystallographic taxonomies and taxonomies based on the chemical groups contained in polymers, should be the subject of further studies.
Furthermore, the verification of whether such a large knowledge integration will be a breakthrough in the limits of conventional polymer chemistry requires engineering preparation that takes into account the usability of the knowledge system, as well as a return to the fundamental debate on whether deterministic knowledge description can generate extrapolative ideas or not. However, considering that the same argument is being made for probabilistic knowledge description in machine learning such as artificial intelligence, it is thought that materials for verification are insufficient to draw a conclusion at this point. We believe that the main purpose of this paper, to preserve knowledge and make it available at any time, will result in the creation of verification materials for discussions on the academic exploration of this unknown area.
From a broader perspective, collaboration may occur beyond the top ontologies. The integration of material ontologies that conform to each top ontology would be a more realistic plan, and the Elementary Multiperspective Material Ontology (EMMO) is one such target for collaboration [Citation42,Citation43]. Ultimately, we hope that knowledge that solves societal problems will become available worldwide.
7. Conclusion
To collaboratively form a large machine-readable knowledgebase covering science, starting with polymers, we introduced an ontology for polymer chemistry. This ontology is based on the well-known top ontology Basic Formal Ontology (BFO), which guarantees linkages with extensive external knowledge in the framework. The created ontology covers all entities in the NIMS polymer database PoLyInfo and ultimately produces the resource description framework (RDF) of PoLyInfo (PoLyInfoRDF). This semantically integrated database format was accessible from the NIMS endpoints and established data linkages that complemented the missing parts of PoLyInfo, such as small molecules.
Acknowledgements
This study was partially supported by the
MEXT Program: Data Creation and Utilization-Type Material Research and Development Project Grant Number JPMXP1122714694
Council for Science, Technology and Innovation (CSTI), Cross-ministerial Strategic Innovation Promotion Program (SIP), the 3rd period of SIP ‘Materials Informatics Infrastructure Linkage and Human Resource Development for Fostering Material Unicorns’ (Funding agency: NIMS)
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- PoLyInfo [Internet]. Tsukuba, Japan: PoLyInfo; [cited 2024 Feb 28]. Available from: https://polymer.nims.go.jp/
- Gracheva E, Lambard G, Samitsu S, et al. Prediction of the coefficient of linear thermal expansion for the amorphous homopolymers based on chemical structure using machine learning. Sci Technol Adv Mater Methods. 2021;1(1):213–16. doi: 10.1080/27660400.2021.1993729
- Wu S, Kondo Y, Kakimoto MA, et al. Machine-learning-assisted discovery of polymers with high thermal conductivity using a molecular design algorithm. Npj Comput Mater. 2019;5(66):1–11. doi: 10.1038/s41524-019-0203-2
- Ishii M, Ito T, Sado H, et al. NIMS polymer database PoLyInfo (I): an overarching view of half a million data. Sci Technol Adv Mater. 2024 forthcoming. doi: 10.1080/27660400.2024.2354651
- The World Wide Web Consortium [Internet]. Wakefield (MA): World Wide Web Consortium; [cited 2024 Feb 28]. Available from: https://www.w3.org/
- Web ontology language (OWL) [internet]. Wakefield (MA): World Wide Web Consortium; [cited 2024 Feb 28]. Available from: https://www.w3.org/OWL/
- Resource description framework (RDF) [internet]. Wakefield (MA): World Wide Web Consortium; [cited 2024 Feb 28]. Available from: https://www.w3.org/2001/sw/wiki/RDF
- SPARQL Query Language for RDF [Internet]. Wakefield (MA): World Wide Web Consortium; [cited 2024 Feb 28]. Available from: https://www.w3.org/2001/sw/wiki/SPARQL
- OWL 2 web ontology language, structural specification and functional-style syntax (second edition) [internet]. Wakefield (MA): World Wide Web Consortium; [cited 2024 Feb 28]. Available from: https://www.w3.org/TR/owl2-syntax/
- RDF 1.1 Concepts and Abstract Syntax [Internet]. Wakefield (MA): World Wide Web Consortium; [cited 2024 Mar 1]. Available from: https://www.w3.org/TR/rdf11-concepts/
- SPARQL 1.1 query language [internet]. Wakefield (MA): World Wide Web Consortium; [cited 2024 Feb 28]. Available from: https://www.w3.org/TR/sparql11-query/
- Bioinformatics embraces Semantic Web technologies [internet]. Cambridgeshire (UK): EMBL-EBI; [cited 2024 Feb 28]. Available from: https://www.ebi.ac.uk/about/news/technology-and-innovation/RDF-platform/
- Fu G, Batchelor C, Dumontier M, et al. PubChemRDF: towards the semantic annotation of PubChem compound and substance databases. J Cheminform. 2015;7(34):1–15. doi: 10.1186/s13321-015-0084-4
- Kimura T, Kushida T. Openness of Nikkaji RDF data and integration of chemical information by Nikkaji acting as a hub. J Inf Process Manag. 2015;58(3):204–212. doi: 10.1241/johokanri.58.204
- PubChemRDF version 1.8.0 beta, released in February 2023 [internet]. Bethesda (MD): PubChem; [cited 2024 Apr 26]. Available from: https://pubchem.ncbi.nlm.nih.gov/docs/rdf-version#section=Version-1-8-0-beta
- PubChemRDF is launched [internet]. Bethesda (MD): PubChem; [cited 2024 Feb 28]. Available from: https://pubchem.ncbi.nlm.nih.gov/docs/pubchemrdf-1-0beta
- PubChemRDF official documentation [Internet]. Bethesda (MD): PubChem; [cited 2024 Apr 26]. Available from: https://pubchem.ncbi.nlm.nih.gov/docs/rdf
- Hastings J, Chepelev L, Willighagen E, et al. The chemical information ontology: provenance and disambiguation for chemical data on the biological semantic web. PLoS ONE. 2011;6(10):e25513. doi: 10.1371/JOURNAL.PONE.0025513
- Smith B, Ashburner M, Rosse C, et al. The OBO foundry: coordinated evolution of ontologies to support biomedical data integration. Nat Biotechnol. 2007;25(11):1251–1255. doi: 10.1038/nbt1346
- The International Chemical Identifier [Internet]. Cambridge (UK): InChI Trust; [cited 2024 Feb 28]. Available from: https://www.inchi-trust.org/
- Weininger D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J Chem Inf Comput Sci. 1988;28(1):31–36. doi: 10.1021/ci00057a005
- NBDC NikkajiRDF [Internet]. Saitama Japan: Japan Science and Technology Agency; [cited 2024 Feb 28]. Available from: https://rdfportal.org/dataset/nikkaji
- Unichem [Internet]. Cambridgeshire (UK): EMBL-EBI; [cited 2024 Apr 26]. Available from: https://www.ebi.ac.uk/unichem/
- Arp R, Smith B, Spear A. Building ontologies with basic formal ontology. Cambridge (MA): MIT Press; 2015.
- information-artifact-ontology/IAO [internet]. San Francisco (CA): GitHub, Inc.; [cited 2024 Feb 28]. Available from: https://github.com/information-artifact-ontology/IAO
- RDF 1.2 Schema [Internet]. Wakefield (MA): World Wide Web Consortium; [cited 2024 Feb 28]. Available from: https://www.w3.org/TR/rdf12-schema/
- Dumontier M, Baker CJ, Baran J, et al. The Semanticscience Integrated Ontology (SIO) for biomedical research and knowledge discovery. J Biomed Semant. 2014;5(14):1–11. doi: 10.1186/2041-1480-5-14
- PoLyInfo Help [Internet]. Tsukuba, Japan: PoLyInfo; [cited 2024 Apr 26]. Available from: https://polymer.nims.go.jp/PoLyInfo/guide/en/help_index.html
- (IUPAC) Gold book [Internet]. Research Triangle Park (NC): International Union of Pure and Applied Chemistry; [cited 2024 Apr 26]. Available from: https://goldbook.iupac.org/
- OntoBee [Internet]. Ann Arbor (MI): He Group; [cited 2024 Apr 26]. Available from: https://ontobee.org/
- PoLyInfo ontology registered in the Materials Data Repository of NIMS. doi: 10.48505/nims.4414
- Chung DDL. Processing-structure-property relationships of continuous carbon fiber polymer-matrix composites. Mater Sci Eng. 2017;113:1–29. doi: 10.1016/j.mser.2017.01.002
- D2RQ [Internet]. San Francisco (CA): GitHub, Inc.; [cited 2024 Feb 28]. Available from: http://d2rq.org/
- Shape Expressions Language 2.1, Final Community Group Report 8 October 2019 [Internet]. Wakefield (MA): World Wide Web Consortium; [cited 2024 Feb 28]. Available from: http://shex.io/shex-semantics/
- Shapes Constraint Language (SHACL) W3C Recommendation 20 July 2017 [Internet]. Wakefield (MA): World Wide Web Consortium; [cited 2024 Feb 28]. Available from: https://www.w3.org/TR/shacl/
- Wikidata:Database reports/EntitySchema directory [Internet]. San Francisco (CA): Wikimedia Foundation, Inc.; [cited 2024 Feb 28]. Available from: https://www.wikidata.org/wiki/Wikidata:Database_reports/EntitySchema_directory
- GO_Shapes (Gene Ontology) [Internet]. San Francisco (CA): GitHub, Inc.; [cited 2024 Feb 28]. Available from: https://github.com/geneontology/go-shapes
- ShEx & SHACL compared [Internet]. Iasi, Romania: Figshare; [cited 2024 Feb 27]. Available from: https://figshare.com/articles/presentation/ShEx_and_SHACL_compared/13174583
- PoLyInfo Schema registered in the Materials Data Repository of NIMS. doi: 10.48505/nims.4415
- RXNO: reaction ontologies [Internet]. San Francisco (CA): GitHub, Inc.; [cited 2024 Feb 27]. Available from: https://github.com/rsc-ontologies/rxno
- PoLyInfo Property List [Internet]. Tsukuba, Japan: PoLyInfo; [cited 2024 Apr 26]. Available from: https://polymer.nims.go.jp/PoLyInfo/guide/en/term_polymer.html#chap21
- Temal L, Rosier A, Dameron O, et al. Mapping BFO and DOLCE. Stud Health Technol Inform. 2010;160(2):1065–1069. doi: 10.3233/978-1-60750-588-4-1065
- The Elementary Multiperspective Material Ontology (EMMO) [Internet]. San Francisco (CA): GitHub, Inc.; [cited 2024 Mar 1]. Available from: https://emmo-repo.github.io/
Appendix A
List of prefix/URI correspondences in ShEx
PREFIX compound: <http://rdf.ncbi.nlm.nih.gov/pubchem/compound/>
PREFIX mono: <http://dice.nims.go.jp/ontology/PoLyInfo-ont/polyinfo-rdf/Monomer#>
PREFIX obo: <http://purl.obolibrary.org/obo/>
PREFIX owl: <http://www.w3.org/2002/07/owl#>
PREFIX pli: <http://dice.nims.go.jp/ontology/PoLyInfo-ont/Schema#>
PREFIX rdf: <http://www.w3.org/1999/02/22-rdf-syntax-ns#>
PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>
PREFIX sio: <http://semanticscience.org/resource/>
PREFIX skos: <http://www.w3.org/2004/02/skos/core#>
PREFIX xsd: <http://www.w3.org/2001/XMLSchema#>