Abstract
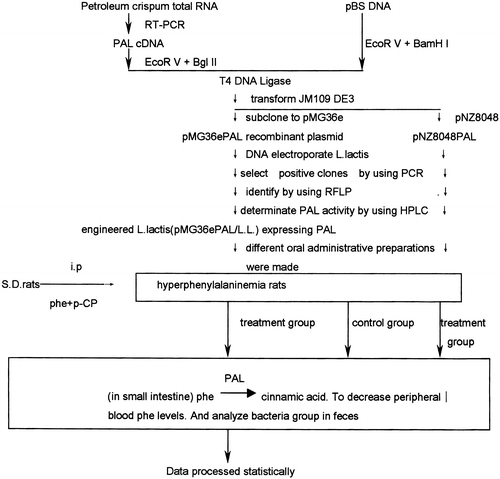
To replace the low phenylalanine (phe) diet treatment and to improve the quality of life of the phenylketonuria (PKU) patients, A phenylalanine ammonia-lyase (PAL) cDNA from petroselinum crispum was subcloned into expression vectors, pMG36e and pNZ8048, and transformed L. lactis by electroporation. The pMG36e PAL/L. lactis and pNZ8048 PAL/L.L. were screened and characterized by using PCR and HPLC, and prepared as a liquid type preparation that were given orally to treat hyperphenylalaninemia-rats. The phe levels of the rat plasma sample were determined by HPLC. The data showed that the plasma phe levels in hyperphenylalanemia (HPA) rats receiving preparations made from the engineered L.lactis were significantly reduced compared with non-treated HPA rats. The effect of the pNZ8048PAL/L.L. showed a higher expression of PAL and better cure results than pMG36ePAL/L.L. These results point a potential way for PKU treatment.
INTRODUCTION
Phenylketonuria (PKU) is an autosomal, recessive genetic disorder which occurs in approximately 1/16,000 births. Classical PKU is caused by mutations of the gene in chromosome 12 that codes for phenylalanine hydroxylase(PAH). PAH is essential for the conversion of phenylalanine (phe) to tyrosine. The deficient PAH activity in phe metabolism results in an increase of the level of phe and its abnormal metabolites that cause widespread brain damage and severe mental retardation. A low phe diet is now the only available treatment. But it costs a lot and it is difficult for patients to accept and sustain this diet over long periods (from neobirth to brain mature).Citation[[1]]
An enzyme, phenylalanine ammonia lyase (PAL) was used to treat PKU as early as 1986.Citation[2-3] But the studies were not continued because PAL was not available in sufficient amounts at reasonable cost. Orum et al. cloned and expressed a PAL gene from Rhodosporidium toruloides in Escherichia coli.Citation[[4]] Sarkissian and Chang et al. used a mutant N-ethyl-N′-nitrosourea (ENU)-treated mouse as orthologous models of human PKU and HPA to study enzyme substitution therapy with PAL by i.p. injection and oral gavage with PAL enzyme protected by the E. coli cells that lowed plasma phe levels clearly.Citation[[5]]
As the first stage of a novel strategy to treatment of PKU (), we cloned and expressed a PAL cDNA from petroselinum crispum in Eschezichia Coli.Citation[[6]] Then the PAL cDNA 2.2 Kb fragment was subcloned and expressed into pMG36e/L. lactis. The host is a small intestine-beneficial bacteria.Citation[[7]] The engineered pMG36e/L. lactis was used to teat the HPA rats by oral gavage administration that showed the plasma phe level in treatment group were lowed clearly.Citation[[8]] Further we subcloned the PAL cDNA fragment into a new vector pNZ8048 and expressed in a stain of L. lactis.Citation[[9]] A Higher expression level was obtained and used to cure the HPA rats that give positive results. These results point out an very hopefully altenative approach to cure the PKU.
MATERIALS AND METHODS
Materials
The plasmid pBS PAL containing Petroselinum crispum PAL cDNA was constructed in our laboratory.Citation[[6]] L. lactis MG1363, in which endogenous plasmids were eliminated and proteinase were inactivity, was donated by Dr. J. Kok in Groningen University. Expressive plasmid vector pMG36e was obtained from Professor G. Venema. Plasmid pNZ8048 and L.L.NZ9000 were from NIZO Food Research, The Netherland. Restriction enzymes, AMV reverse transcriptase, T4 DNA ligase were purchased from Promega. Taq DNA polymerase, RNasin, DEPC, IPTG, antibiotics were from Huamei Co. Trans-cinnamic acid, trans-p-coumaric acid, para-chlorophenylalanine methyl ester were from Sigma. L-phenylalanine was from Gibco. Tryptone, Yeast extract were from Oxoid. M17 broth was from Difco. Trypticase soy broth were from Becton Dickison and Company. Eggerth–Gagnon broth, Glucose blood liver broth were from Japan.
Two pairs of primers were designed to identify instantly pMG36ePAL and pNZ8048 PAL positive clone containing PAL cDNA insert by PCR:
Pvec-TCACAAAATGCTATACTAGGTAGG, PAL-TTAACAGATTGGAAGAGGAGC. And Pnis 5′ GATCACATGATTACGTTCGAAG 3′, P63 5′ CTAAGATCTAGAGCATGTCAG TTAAC 3′. They were synthesized in the Institute of Microbiology of the Chinese Academy of Sciences.
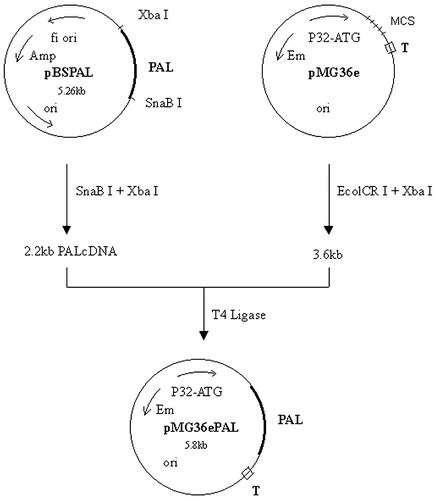
Construction of Constitutive Expression Recombinant Plasmid pMG36ePAL and Inducible Recombination Expression Plasmid pNZ8048PAL
2.2 kb PAL cDNA was prepared from digests of pBS PAL cDNACitation[[6]] with restriction enzymes SnaB I and Xba I, and then purified. pMG36e was digested by enzymes Ecol CR I and Xba I. The target cDNA and vector segments were mixed in ratio 3:1. They were ligated according to routine method (). The procedure of pNZ8048PAL construction was omitted.
Transformation of L. Lactis by Electroporation
The sensitive (competent) L. lactis cells were prepared according to reference.Citation[[10]] Bio-Rad Gene Pulser was used for electroporation, 2.5 kV/2 mm, 25 uf, 200 Ω. The recovery medium was SGM17MC (adding 50 ng/ml Em and 250 ug/ml phe). The solid medium for plate was SGM17+agar (adding 5 ug/ml Em and 250 ug/ml phe). Incubation was anaerobic at 30°C for 2–3 days.
PCR to Screen the Positive Clones Containing PAL cDNA Insert
20 ul PCR system contains 1xPCR buffer,1.5 mM MgCl2, 0.2 mMdNTP, one pair of primers each 50 ng, 1 U Taq. After 95°C 3 min, twenty five cycles were carried out at 94°C 1 min→52°C 1 min→72°C 2 min 30 seconds, then 72°C 10 min following the last cycle.
SDS-PAGE
The SDS-PAGE of cell lysis of the pNZ8048PAL/NZ9000 were conducted with routine method.Citation[[11]] The negative control was L. lactis containing original vector.
Determination of Expressed PAL Activity by HPLC
One milliliter of the cultured mixture of the engineered L. lactis were obtained at designed times. After centrifugation, supernatant was acidified by HCI, diluted 10 times by dH2O, filtered through a 0.45 um membrane filter. Cinnamic acid concentration was measured by using HPLC (Backman, UV Detecter,166NM).Citation[[12]] The mobile phase was a mixture of methanol and water(50:50) with 0.1% acetic acid. Elution rate was 1 ml/min, detector wavelength was 313 nm. The column was a uBondpak C18, 0.46×25 cm. For phe determination,Citation[[13]] an aliquot of 0.2 ml supernatant of the culture liquid, 0.2 ml ethanol and 0.4 ml chloroform were mixed and shaken vigorously, and then centrifuged at 12,000 rpm for 15 min. 10 ul of the upper layer was injected into the column. The mobile phase: methanol–water (10–90) with 0.01 M KH2PO4 and 1×10−4 M H3PO4, final pH 4.3, elution rate 1 ml/min, detector wavelength 210 nm.
Freeze-Drying of Engineering L. Lactis and Preparation of Its Enteric-Coated Microcapsules
PMG36ePAL/L.L, pMG36e/L.L, was suspended in suitable protectant () in ratio 1:3 (v/v), shell-frozen, and then freeze-dried till water content less then 4%. The resulting powder was then pressed through a No. 50 stainless steel sieve. This powder was then shared in amples, sealed, and stored. The percentage of alive cells in the powder stored different periods was tested by cultivating in anoxic on M17(+phe+Em) at 30°C for 72 h and counting the bacteria clones.
Table 1. The Blood Phe Concentration of All Rats on Different Days During Treatment
Enteric-coated microcapsules were made from the powder of freeze-dried L. lactis with mixed lipids. The protective efficiency of the microcapsules were tested by passing a simulated gastric juice or/and then passing a simulated intestine juice.
Preparations of the Engineered L. Lactis
Anti-acid buffer: pMG36ePAL/L.L cultivated anaerobically in MRS (containing 0.1 M PBS, pH8.0) to 109 CFU/ml.
Liquid Preparation of pNZ8048PAL/NZ9000
The cultured bacterium was suspended in volume of fresh culture medium to a concentration of 109 CFU/ml. It will be used to cure HPA rats.
Treatment of HPA Rats by the Preparations of the Engineered L.L
Hyperphenylalaninemia model rats were produced by using the combined administration of phe and para-chlorophenylalanine methyl ester for 3 days.Citation[[14]]
First batch of therapeutic test. Treatment group I (11 rats) were received the PAL engineering L. lactis (109 CFU/day) in enteric-coated microcapsules; Treatment group II (10 rats) were receiving the PAL engineered L. lactis(109 CFU/ml) in culture containing anti-acid buffer 2 ml per day; Control group (10 rats) were receiving L. lactis (109 CFU/day) carrying empty-vector in enteric-coated microcapsules.
Second batch of therapeutic test. Treatment group (15 rats) were receiving pNZ8048 PAL/L. lactis(109 CFU/day) suspended in anti-acid buffer; Model rats of control group 1(15 rats) were received L. lactis (109 CFU/day) carrying empty-vector in anti-acid buffer.
Blood samples were obtained from rat tails on different days after treatment. The concentration of phe were measured by using HPLC.
Determination of Total Bacteria and L. Lactis in Rats' Faces
Fresh feces of rats were collected on 1st, 10th day of test. 1 g of the feces was suspended in 9.0 ml N.S., diluted 102, 104, 106 times. 50 ul of each was speared on BL, EG, TS and M17 (+phe+Em) culture medium in 9 cm dish in diameter. TS medium was cultured oxically at 37°C for 24 hours. The others were cultured anaerobically at 37°C for 72 hours. Then they were stained by using Gram's method, analyzed, statistically counted total bacteria and L. lactis, and calculated the percentage of L. lactis in total bacteria.
RESULTS AND DISCUSSION
Construction of pMG36e PAL was proceeded according to . PAL cDNA segments and vector segments were mixed in ratio 3:1.
Electroporation transformation efficiency was about 1.0–1.3×103 of recombinant/ug of plasmid DNA.
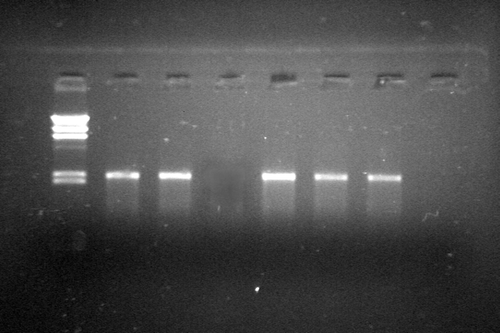
Selection and Identification of pMG36e PAL/L. Lactis by PCR
showed the electrophoretic result of rapid selection of positive clone using PCR technique. Clones with 2.2 Kb band were the right pMG36ePAL/L.L. clones with an insertion of PAL cDNA and correct direction.
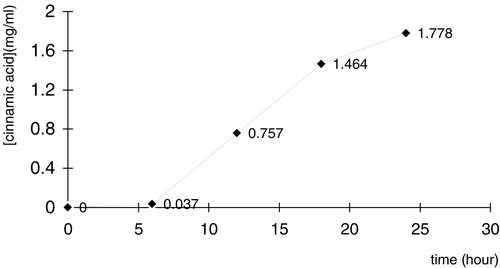
The Activity and Specificity of the Expressed PAL
Samples from a cultural mixture of a positive clone pMG36e PAL/L.L were obtained at 6, 12, 18, 24 hour, in which cinnamic acid were measured by using HPLC (see ). With a continue culturation, cinnamic acid concentration was increased gradually from 0 to 1.778 mg/ml in 24 hours. It showed that PAL expressed by engineered L. lastis has the activity to convert phe to cinnamic acid. The concentrations of phe were also measured by using HPLC. The result showed that phe decreased gradually during the culturation.
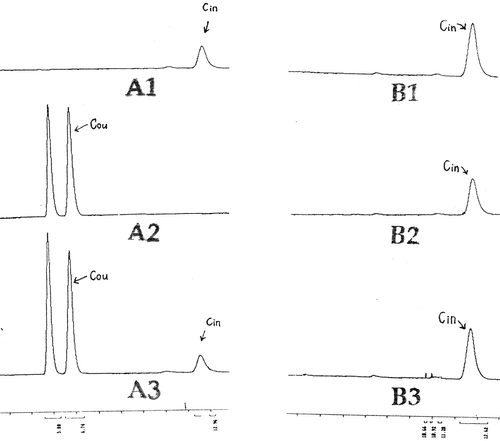
But the expressed PAL could not convert tyrosine to coumaric acid that indicates a very good specificity ().
Figure 5. Analysis of PAL specificity in pET23bPAL/JM109DE3 by using HPLC. B1, B2 and B3 were from same clone pET23bPAL/JM109DE3. The only different between them was in culture medium: B1: NZCYM+0.1 mM Phe; B2: NZCYM+0.1 mM tyrosine; B3 NZCYM+0.1 mM Phe+0.1 mM tyrosine. Coumaric acid, the TAL deamination product from tyrosine, was not found in the three culture media above. Only Cinammic acid, the PAL product from Phe, was detected. The result shows: there is no TAL activity in the PAL product of pET23bPAL/JM109DE3, i.e. the specificity of the PAL is ideal.
Selection of Protectants for Freeze-Dry of the Engineered L. Lactis
Skimmed-milk adding 10% sucrose was the best in the 5 kinds of freeze-dry protectants tested.
Result of Effect of the Oral Administration Preparations of the pMG36ePAL/L.L. on HPA Rats
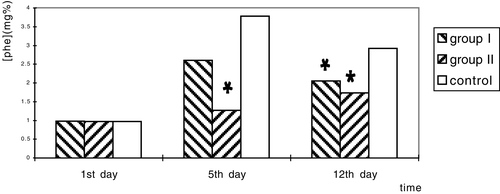
The result of as showed in . each group had 11 rats. And was treated with oral administration of enteric-coated microcapsules, anti-acid buffer liquid or original vector L. lactis respectively. Phe levels in peripheral blood of hyperphenylalaninemia rats' decreased obviously in treatment group I and group II. Compared with the control group. The differences of the phe levels between group II and control group (day 5), and group I and control, II and control (day 12) have statistically significance. The anti-acid liquid preparation showed a better effect (group II).
Figure 6. Phe concentration in blood of the hyperphenylalanemia rats treated with two different preparations. *There is a significant difference between treatment group and control group (P<0.05).
After determination of bacteria amount in test rats' faces on 1st and 10th day, we found that L. lactis amount on 10th day of the treatment group II was higher than that of group II and the difference is significant (P<0.01).
SDS-PAGE Result of the PNZ8048PAL/L.L
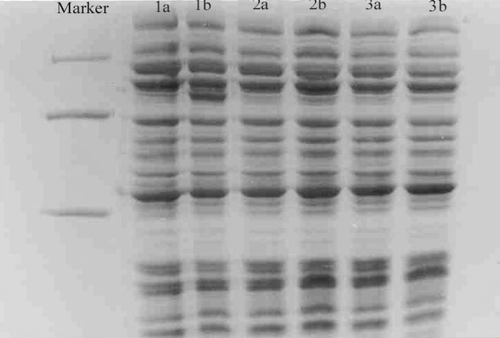
A 78 KD protein band occurred in the lane 3,5 that indicats the expressed PAL. () in the clone of lane 3 is better than another clone of lane 5.
Figure 7. Coomassie blue-stained gels after SDS-PAGE of extracts L.L. containing pNZ(8048-PAL)1 or pNZ(8048-PAL)2 producing PAL protein. Lane 1, molecular weight marker (in kilodaltons); Lane 2, uninduced pNZ(8048-PAL)1/L.L. cells; Lane 3, induced pNZ(8048-PAL)1 cells; Lane 4, uninduced pNZ(8048-PAL)2/L.L. cells; Lane 5, induced pNZ(8048-PAL)2/L.L. cells; Lane 6, uninduced pNZ(8048-PAL)2/L.L. cells; Lane 7, induced pNZ(8048-PAL)2/L.L. cells.
Results of Effect of the Preparation of PNZ8048PAL/L.L. on the HPA Rats
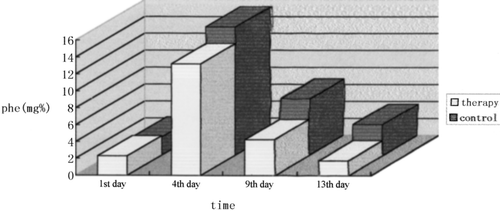
and show that on the 4th day after treatment, phe level in rats' blood of treatment group was lower than that of control group, but there was no significant difference. On the 9th and 13th day treatment group was lower than control group, and there were significant differences between treatment group and control group (P<0.05).
Figure 8. The blood Phe Concentration of each group on different days during treatment. Notes: (1) On the 1st day, blood Phe concentration was similar between two groups. (2) On the 4th day the Phe concentration in the therapy group was slightly lower than control group, but there was no significant difference between two groups. (3) On the 9th day the Phe concentration in the therapy group was distinctly lower than control group. There is significant difference between two groups (P=0.008<0.05). (4) On the 13th day the Phe concentration in the therapy group was 2 times lower than control group. There was extremely significant difference between two groups (P=0.000<0.01). The result shows that feeding engineering L.L. containing PAL gene can reduce the blood Phe concentration of PKU model rats.
This experiment was repeated, and a similar result was observed.
The low phe diet significantly reduces serum phe levels and can largely reduce or prevent the mental impairment characterized of PKU if initiated early in the neonatal period. It requires rigorous restriction of natural protein intake. Besides being unpalatable, this kind of treatment is ineffective in controlling systemic phe levels during mild fever, infection or pregnancy. Furthermore it is also a high risk for fetus in PKU woman. The UK MRC study group on PKU has published their recommendation that emphasized the above problems and concluded that there is a need for an alternative to the low phenylalanine diet.Citation[[15]] We have addressed a novel strategy to treatment of PKU as described above. The expressed PAL together with the host bacterium was delivered into small intestine of the HPA rats or PKU patients in the future. The PAL converts phe to harmless cinnamic acid. Thus phe level in blood was decreased. Because the recombinant plasmid only passed through intestines and didn't enter somatic cell, and L.L. is a food-grade bacteria, the safety should be good. The L. lactis cells may protect the PAL against inactivation by gastric acidity and intestinal digestive enzymes. The patient's diet would not be extremely restricted as the low phe diet requested. So the life quality of the PKU patients would be greatly improved. On the other hand because the post-expression procedures are very simple, the cost of production of the oral preparations will be low that is beneficial for both of the PKU family and pharmacy.
ACKNOWLEDGMENTS
A grant support for this research from Beijing Natural Science Foundation. Some good suggestion and discussion from Dr. Xuetuo Li, the Institute of Chemistry and Physics, Chinese Academy of Sciences, and from Dr. Shengdong Lu, the Chinese Academy of Medical Sciences; Dr. Venema provided pMG36e, University of Groningen, the Netherlands, are gratefully acknowledged.
REFERENCES
- Scriver C. R. The Hyperphenylalanine. The Metabolic Basis of Inherited Disease6th Ed., C. R. Scriver, et al. Mc Craw-Hill Co., New York 1989; Vol. 1: 495–546
- Bourget L., Chang T.M. S. Phenylalanine ammonia-lyase immobilized inmicrocatsules for the depletion of pherylalanine in plasma in phenylaketonurea rat model. BBA 1986; 883: 432–438
- Ambrus C. M., Anthone S., Horvath C., et al. Extracorporeal enzyme reactors for depletion of phenylalanine in phenylketonuria. Ann. Intern. Med. 1987; 106: 531–537
- Orum H., Rasmussen F. Appl. Microbiol. Biotechnol. 1992; 36: 745–748
- Sarkission C. N., Shao Z. Q., Blain F., et al. A different approach to treatment of phenylketonurea: phenylalanine degradation with recombinant phenylalanine ammonia lyase. P.N.A.S. USA 1999; 96: 2339–2344
- Liu J. Z., Xiang H., Hu W., et al. Cloning and expression of phenylalanine ammonia lyase cDNA in Escherichia coli. Chin. J. Biotechnol. 1998; 14(4)384–388
- Xiang H., Liu J. Z., Hu W., et al. Expression in lactococcus lactis of catalytically acive phenylalanine ammonia-lyase from parsley. Acta Microbiol. Sin. 1999; 39(3)196–204
- Jia X. Y., Liu J. Z., Xiang H., et al. A new strategy of gene theraputics for hyperphenylalaninemia rats. Natl. Med. J. China 2000; 80(6)464–467
- Kuipers O. P., Beerthuizen H. M., Siezen R. J., et al. Characterization of the nisin gene cluster nis ABTCIPR of Lactococcus lactis requirement of oxprcssion of the nis A and nis I genes for development of immcrity. Eur. J. Biochem. 1993; 216: 281–291
- Well J. M., Willson P. W., Lepage R.W. F. Improved cloning vectors and transformation procedure for Lactococcus lactis. J. Appl. Bacteriol. 1993; 74: 629–636
- Sambrook J., Fritsch F. F., Maniatis T. Molecular Cloning—A Laboratory Manual2nd Ed. Cold Spring Harbor Laboratory Press, New York 1989
- Scott D. A. Identification by high performance liquid chromatography of tyrosine ammonia-lyase activity in purified fractions of phaseolus valgaris phenylalanine ammonie-lyase. J. Chromatogr., Biomed. Appl. 1992; 573: 309–312
- Zhang L. F., Yu Y. L., Yang R. Y. Direct determination of phe in serum extracts of phenylketonuria patients by reversed-phase high performance liquid chromatography. J. Chromatogr. 1983; 282: 333–339
- Greengard O.D. G., Delvalle J. Models for the Study of Inborn Errors Metabolism, F. A. Hommes. Elseviery North/Holland Bimedical Press, Amsterdam 1979; 125–130
- Medical Research Council Working Party on Phenylketonuria. Brit. Med. J. 1993; 396: 115