Abstract
This study was designed to evaluate the seroprevalence of hepatitis G virus (HGV) infection, its impact, and its relationship with other hepatotropic viruses among chronic renal failure patients undergoing hemodialysis at the Lok Nayak Hospital, New Delhi. The study group consisted of 100 consecutive cases of patients with chronic renal failure undergoing hemodialysis and equal healthy controls matched for age and sex. The patients were included on the basis of detailed history, clinical examination, and liver function profile. HGV RNA was detected in serum samples of all patients as well as of healthy controls using nested reverse transcription polymerase chain reaction (RT-PCR). The primers used were derived from the NS3 helicase region of the viral genome. Serological assay was used for screening the viral markers for hepatitis B and C (HbsAg and Anti HCV). A history of blood transfusion was recorded in 65% of the cases. HGV RNA was detected in only six out of 100 (6%) cases of chronic renal failure. The seroprevalence of HCV infection was detected in 27 (27%), while HBV infection was seen in 10 (10%) out of 100 cases. The mixed infection of HGV and HCV was seen in 33.3% (two out of six) of the chronic renal failure cases, while the coinfection between HGV and HBV was not observed. In the 100 cases of healthy controls, HGV RNA was detected in only three (3%) subjects. Serological markers for Anti HCV antibody and HbsAg were positive in only one (1%) and two (2%) of the subjects, respectively. The seroprevalence of HGV infection in chronic renal failure was found to be statistically nonsignificant when compared to that of healthy controls. Also, there was no difference in clinical course and liver function profile of HGV-positive and HGV-negative cases. However, alanine aminotransferase (ALT) was significantly out of range in HCV-positive patients compared with HCV-negative patients. The presence of HGV infection reflected a postparental exposure to blood and blood-contaminated products in hemodialysis patients. It is suggested that HGV infection in cases of chronic renal failure is unlikely to influence the course of the disease and may be considered an innocent bystander.
Introduction
Hemodialysis patients comprise a high-risk population, whose members may develop liver disease due to bloodborne viral agents.Citation[1] A new RNA virus hepatitis G virus (HGV) family has been identified recently and is known to be highly prevalent in such high-risk groups. The HGV shares about 25% homology with hepatitis C virus (HCV) genome,Citation[2] and the genome consists of approximately 9,400 base pairs that encode a single large polyprotein.Citation[3] This virus is readily transmitted through transfusion of blood and blood products. Three to 57% of HGV infection has been reported in serum samples of hemodialysis patients using RT-PCR technique.Citation[3-6] HGV has been found in 1–2% of blood donors and in 10–20% intravenous drug abusers.Citation[7&8] HGV accounts for 1–4% of the community-acquired non-A, non-B hepatitis patients.Citation[9] As regards its course, acute HGV hepatitis is very mild and is not associated with jaundice. HGV has also been found in sera of chronic hepatitis patients, who have frequent parenteral exposure, including intravenous drug users, bone marrow transplant patients, and organand blood recipients. There are several studies that have attempted to establish the role of HGV in the major high risk groups, and they are based on serology and RT-PCR that reported 15–38% in intravenous drug abusers, 3–57% in hemodialysis patients, and 14–38% in hemophiliacs.Citation[10] No study has been carried out so far in North India on the role of HGV infection in this high-risk population. However, one study has been recently reported from south India in a high-risk population.Citation[11] In view of the paucity of information on the prevalence of HGV from North India, the present study was designed to evaluate the role of HGV infection in patients with chronic renal failure who were attending the dialysis clinic of Lok Nayak Hospital in Urban Delhi.
Materials and Methods
The study included 100 such consecutive subjects of chronic renal failure, who visited for hemodialysis at the dialysis unit of Lok Nayak Hospital during the period of 1 year. All of these patients were on dialysis at the end stage of kidney failure. The study group also included equal healthy controls who were matched to the subjects (i.e., same geographic and socioeconomic status). Serum samples were collected from all patients, and controls were stored at − 20°C deep freezer until analysis. Healthy controls included the voluntary blood donor from the blood bank of the hospital. The patients were evaluated on the basis of a standard Performa with relevant information with respect to case history, risk factors, liver function test, and serological tests, which included HbsAg, anti-HCV. The written informed consent was obtained from all patients and control subjects. The study had Institutional Review Board (IRB) approval.
Serological Assay
Sera samples of hemodialysis patients and healthy controls were screened for HbsAg using third generation ELISA kit (General Biological Corporation, Taiwan). Anti-HCV antibody was tested by third generation ELISA kit (Biokit; S.A., Spain). The microtiter plates of the anti-HCV kit were coated with recombinant antigens representing epitopes, core, NS3, NS4, and NS5 of HCV.
RNA Extraction
RNA was extracted from 100 uL serum by the acid-guanidinium-phenol-chloroform method of Chomezynski and SachiCitation[4] with slight modifications.Citation[12] In brief, 100 uL of patient's serum was used, to which 600 uL of lysis buffer [4 M guanidinium thiocyanate, 0.7 M sodium citrate (pH 7.0), 2% sarcosyl] and 0.1 M (final concentration) β-mercaptoethanol were added. To this, 50 uL of 2 M sodium acetate (pH 4.0) was added, followed by 600 µL of (diethoxy pyrocarbonate) DEPC saturated phenol and 131 µL chloroform isoamyl alcohol (49: 1), and the mixture was left in ice. After centrifugation at 12,000 rpm for 20 min at 4°C, the pallet was resuspended in 150 uL lysis buffer (without β-mercaptoethanol), precipitated in ice cold isopropanol, kept at − 70°C for 90 min, centrifuged, and washed once with 75% ethanol. Finally, the pallet was dissolved in 25 uL of DEPC water and stored at − 70°C until use.
Reverse Transcription-PCR
Reverse transcription for preparation of complementary DNA (cDNA) with the MMuLV reverse transcriptase (New England Biolab) and the PCR were performed in a single reaction tube. The outer and inner primer sequences for nested PCR were selected from the NS3 helicase region of the HGV genome, and the primer sequences are given as follows:
Outer primers:
Sense S1 : 5′-GGC ACC TCG TGT TCT GCC A-3′
Antisense A1 : 5′-AGG TCT CCG TCC TTG ATG AT-3′
Inner primers:
Sense S2 : 5′-CAT TC (AC) AAG GCG GAG TGC GAG-3′
Antisense A2 : 5′-(AG) TC (CT) TT GAT GAT GGA ACT GTC-3′
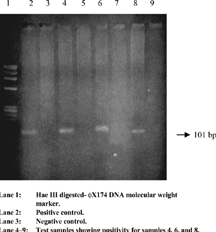
For reverse transcription and the first round of PCR, 5 uL of extracted template RNA was added to 25 uL of PCR master mix containing reaction buffer, outer primers A1 and S1, and both reverse transcriptase (40 units) and Taq DNA polymerase (one unit), incubated at 42°C for 1 hour for cDNA synthesis. Subsequently, PCR amplification was carried out after inactivating reverse transcriptase and denaturing cDNA at 94°C for 5 min and 35 cycles with 30 sec each of denaturation at 94°C, annealing at 57°C, and extension at 72°C in a DNA thermal cycler (M.J. Research, USA). Finally, in the last cycle, the extension was prolonged for 8 min at 72°C. The second round of PCR was performed by using 5 uL of the first PCR product as the template and amplifying with the second PCR mix containing the inner primers (A2 and S2). All reagent compositions and temperature profiles were the same as those of the first PCR. Both positive and negative controls were run parallel along with the test samples. An expected final PCR product of 101 bp () was detected in an ethidium bromide-stained 3% Nusieve agarose gel.
Statistical Analysis
Statistical analysis was performed by Fisher's exact test, Student t-test, and Mann–Whitney U test using software EPISTAT and SPSS wherever applicable. Theresults were expressed as mean ± SD. Statistical significance was set at p = 0.05. The p value < 0.05 was considered significant.
Results
Epidemiology
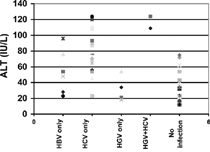
One hundred patients (63 males/37 females) with chronic renal failure on hemodialysis were in the age group of 19–64 years, with the mean age of 42.40 ± 15.8 years. All the patients were from Delhi and nearby metro cities. The duration of dialysis was 24 ± 09 months (mean), and 65% of the cases gave histories of blood transfusion. The 100 healthy controls were matched to the subjects (i.e., same geographic and socioeconomic status) and had the mean age ± SD of 33.00 ± 7.6 years. There were no statistically significant differences with respect to age, ALT level, number of transfusions, and hemodialysis sessions in between HGV-positive and HGV-negative cases (). All these factors were also statistically nonsignificant in the comparison of HBV-positive and HBV-negative cases. However, ALT level serum was found to be elevated in a significant number of HCV-positive samples in comparison to HCV-negative cases, as per the statistical analysis, and all other factors were found to be nonsignificant in the HCV group ().
Table 1. Analysis of Factor Associated with Hepatitis G Virus and Other Hepatotrophic Viruses in Hemodialysis Patients.
Clinical Relevance
In this context (), liver function profiles of HGV-positive and HGV-negative cases were comparable and found to be statistically nonsignificant. With respect to ALT levels believed to be correlated with disease activity, the mean alanine amino transferase levels in HGV-positive and HGV-negative cases were found to be comparable, with no statistical difference (p = 0.52). All patients with exclusive HGV infection exhibited liver function profiles within the normal range, while in those cases where patients had mixed infection of HGV and HCV, elevation in the ALT was shown (). Among the HCV-positive and HCV-negative cases, significant statistical difference (p = 0.04) was observed with respect to ALT levels. However, ALT level was not significantly de-ranged in the HBV-positive cases in comparison with HBV-negative cases as per statistical analysis. Those patients who showed evidence of exclusive HGV infection were also clinically asymptomatic.
Table 2. Liver Function Profiles of Hemodialysis Patients.
Seroprevalence of HGV, HCV, and HBV Infection
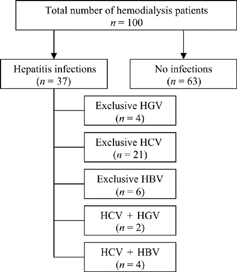
In this series (), we included 100 patients of chronic renal failure on hemodialysis and 100 healthy controls. When we screened the serum samples of these groups for HBV and HCV markers and HGV RNA, HGV infection was found in 6% of hemodialysis patients and 3% of healthy controls. The seroprevalence of HBV and HCV infection in hemodialysis patients was found to be 10% and 27%, respectively. In the group of healthy controls, the seroprevalence of HBV and HCV was detected in 2% and 1%, respectively. Out of 100 cases of hemodialysis patients and healthy controls, the seroprevalence HGV infection (6%) in hemodialysis patients was found to be nonsignificant (p = 0.24) compared to 3% seroprevalence of healthy controls (). However, serological evidence of HBV (p = 0.02) and HCV (0.001) infection in hemodialysis patients was found to be statistically significant comparedto healthy controls. The HCV infection in hemodialysis patients was found to be highly significant (p = 0.001).
Table 3. Prevalence of HGV, HBV, and HCV in Hemodialysis Patients and Healthy Controls.
Coinfection
In the group of six HGV positive hemodialysis patients, four (66.6%) patients had exclusive HGV infection, while two (33.3%) showed coinfection with HCV (). However, coinfection between HGV and HBV was not observed in any of the hemodialysis patients (). Of 10 hemodialysis patients who had HBV infection, coinfection with HCV was shown in four cases, while the remaining six cases had exclusive HBV infection (). Out of 27 HCV-positive hemodialysis cases (), 21 cases showed exclusive HCV infection, while multiple infections were observed in 6% (4% HBV, 2% HGV) of the cases. Out of 100 hemodialysis cases, 63% of the patients were negative for HBV, HCV, and HGV.
Risk Factors
The seroprevalence of anti-HCV was found to be higher and showed coinfection with HGV. Of 27 HCV-infected patients, 19 recorded past histories of blood transfusion. A past history of blood transfusion was also recorded in four out of six HGV-positive hemodialysis patients. However, no major difference was found in the number of transfusions between HGV-positive and HGV-negative cases.
Discussion
The clinical significance of HGV has been assessed in different countries worldwide. Controversies regarding the mode of transmission, clinical implication, and seroprevalence among different disease groups i.e., chronic liver disease, acute viral hepatitis, fulminant hepatic failure, and chronic renal failure, were observed from earlier studies. Our earlier study reported seroprevalence of HGV infection in acute viral hepatitis (14.3%), fulminant hepatitis (11.4%), commercial blood donors (46.6%), and the general population (4%).Citation[5], Citation[13] Another report from India stated that the seroprevalence was 16.3% (7/43) HGV infection among the paid plasma donors from a commercial plasmapheresis unit and with hemophiliacs was 11.4% (5/44). HGV infection was significantly higher compared to voluntary blood donors (0/51).Citation[14] The study was designed to evaluate the seroprevalence of HGV infection in patients with chronic renal failure who had undergone hemodialysis from urban Delhi in order to assess hemodialysis as a risk factor for HGV infection. In the present study, overall prevalence of HGV RNA was 6% (n = 100) in hemodialysis patients, which was comparable to the reports from Japan (4.9%). However, a higher seroprevalence of HGV (7.9–26%) infection was reported from European countries,Citation[15-19] with a much higher seroprevalence of HGV (55%) reported in French patients.Citation[20] The 58.6% seroprevalence of HGV infection has recently been reported in India in high-risk renal transplant patients.Citation[11] However, exclusive HGV infection was only shown in 10 cases.Citation[11] The high rate of seroprevalence of HGV may be because of coinfections with other hepatotropic viruses, like HBV, HCV, HDV, and TTV.Citation[11] Out of six HGV-positive hemodialysis cases, two showed coinfection with HCV and both showed de-ranged liver function tests. However, coinfection with HGV and HBV was not seen in any of the hemodialysis cases. In our dialysis unit, the blood was screened for HbsAg and anti-HCV antibody before transfusion. Even though seroprevalence of HCV in dialysis patients was found to be 27%, which was surprisingly high, this may be due to the presence of a mutant virus strain, or nosocomial transmission may be the possible route of HCV transmission, associated with inadequate universal precautions.
The long-term evolution of HGV infection may be different in various risk groups. However, a lower prevalence of HGV infection in hemophiliacs has been tentatively attributed to shorter duration of HGV viremia than HCV.Citation[16] This might be the reason for potential clearance of HGV viremia in immunocompetent patients.Citation[10] However, HCV showed genetic heterogeneity in its genome that leads to evolution of quasispecies with immune pressure to allow persistent HCV infection.Citation[21] It has been demonstrated that humoral immune response to the HGV envelop antigen developed in a substantial proportion of posttransfusion cases and was associated with the loss of detectable HGV viremia.Citation[22] However, there are reports on the influence of primer combination on detection of HGV RNA or various PCR-based detection. The 5′ NC region primers appeared to be most effective in detecting the HGV RNA as compared to primers derived from the NS3 region.Citation[23] We used the NS3-region-derived primers, as the protocol was already standardized in our laboratory for the detection of HGV RNA. That may be one of the possible reasons for the low prevalence of HGV in this study.
The HGV infection may not be due only to blood transfusion, there may be other factors. No difference was observed in the transfusion history between HGV-positive and HGV-negative cases. Among the four cases that showed exclusive HGV infection, there was one that did not have any past history of transfusion. This suggests that besides parenteral mode, there may be some other mode of transmission that deserves further investigation. However, one study suggested patient-to-patient transmission of HGV in hemodialysis.Citation[9] The liver function profiles of HGV-positive subjects were found to be comparable with HGV-negative hemodialysis subjects. The mean ALT level did not differ between HGV-positive and HGV-negative cases but was higher in HCV-positive patients. Our finding with detailed evaluation showed that HGV may be just an “innocent bystander.” However, the ALT level in HCV-positive cases was significantly high (p = 0.04) compared to that in HCV-negative cases. In view of these observations, the study suggests that HBV and HCV have clinical relevance with abnormal biochemical parameters and HGV and have no clinical relevance, as supported by other studies.Citation[5], Citation[24]
It has been concluded that hemodialysis may not be the only risk factor influencing the seroprevalence of HG0V infection. There may be some other mode of transmission, i.e., postparenteral exposure to blood and blood contaminated products, tattooing, and IV drug abusers. However, the information generated from this study suggests that the clinical course of HGV infection is benign, and the liver functions are not disturbed, and routine screening of HGV infection in blood for blood banks is not necessary.
The HGV infection was found to be clinically irrelevant, and it seems possible that the disease groups do not link with this virus infection.
Acknowledgments
This study was supported by a grant from the Department of Science and Technology, Ministry of Science and Technology, Govt. of India, New Delhi. We thank Rambabu Yadav and Nivesh Bhardwaj for their kind assistance in ELISA of patients included in this study.
References
- Campo, N.; Sinelli, N.; Brizzolara, R.; Torre, F.; Gurreu, G.; Russo, R.; Saffoli, S.; Celle, G.; Picciotto, A. Hepatitis G virus infection in hemodialysis and in peritoneal dialysis patients. Nephron 1999, 82, 17–21. [PUBMED], [INFOTRIEVE], [CSA]
- Karayiannis, P.; Thomas, H C. Current status of hepatitis G virus/GBV-C in transfusion: is it relevant? Vok Sang 1997, 73, 63–69. [CROSSREF], [CSA]
- Leary, T P.; Muerhoff, A S.; Simmons, J N.; Pilot-Malias, T J.; Erker, J C.; Chlumus, L.; Schlauder, G G.; Dawson, G J.; Desai, S M.; Moshahwar, J K. Sequence and genomic organization of HGV/GBV-C: a novel member of the Flaviviridae associated with human non A–F hepatitis. J. Med. Virol. 1996, 48, 60–67. [PUBMED], [INFOTRIEVE], [CSA]
- Chomezynski, P.; Sachi, N. Single step method of RNA isolation by acid guanidium thiocyanate phenol chloroform extraction. Anal. Biochem. 1987, 162, 156–159.
- Kar, P.; Bedi, P.; Berry, N.; Chakravorty, A.; Gupta, R K.; Saha, R.; Das, B C. Hepatitis G virus (HGV) infection in voluntary and commercial blood donors in India. Diagn. Microbiol. Infect. Dis. 2000, 38 (1), 7–10. [PUBMED], [INFOTRIEVE], [CROSSREF], [CSA]
- Panigrahi, A K.; Saxena, A.; Acharya, S K.; Panda, S K. Hepatitis G virus in multitransfused thalassemics from India. J. Gastroenterol. Hepatol. 1998, 13 (9), 902–906. [PUBMED], [INFOTRIEVE], [CSA]
- Aikawa, T.; Sugai, Y.; Okamoto, H. Hepatitis G infection in drug abusers and chronic hepatitis C. N. Engl. J. Med. 1996, 334, 195–196. [PUBMED], [INFOTRIEVE], [CROSSREF], [CSA]
- Alter, H J. The cloning and clinical implications ofHGV/GBV-C. N. Engl. J. Med. 1996, 334, 1536–1537. [PUBMED], [INFOTRIEVE], [CROSSREF], [CSA]
- Masuko, K.; Mitsui, T.; Iwano, K.; Yamazaki, C.; Okuda, K.; Meguro, T.; Murayama, N.; Inoue, T.; Tsudo, F.; Okamoto, H.; Miyakawa, Y.; Mayumi, M. Infection with hepatitis G virus/GBV-C in patients on maintenance dialysis. N. Engl. J. Med. 1996, 34, 1485–1490. [CROSSREF]
- Mphahele, M J.; Lou, G K.K. HGV: the identification, biology and prevalence of an orphan virus. Liv 1998, 18, 143–155.
- Abraham, P.; John, G T.; Raghuraman, S.; Radhakrishnan, S.; Thomas, P P.; Jacob, C K.; Sridharan, G. GB virus C/hepatitis G virus and TT virus infections among high risk renal transplant recipients in India. J. Clin. Virol. 2003, 28, 59–69. [PUBMED], [INFOTRIEVE], [CROSSREF], [CSA]
- Madan, K.; Gopalkrishna, V.; Kar, P.; Sharma, J K.; Das, U P.; Das, B C. Detection of hepatitis C and E virus genomes in sera of patients with acute viral hepatitis and fulminant hepatitis by their simultaneous amplification in PCR. J. Gastroenterol. Hepatol. 1998, 13, 125–130. [PUBMED], [INFOTRIEVE], [CSA]
- Kapoor, S.; Gupta, R K.; Das, B C.; Kar, P. Clinical implications of hepatitis G virus (HGV) infection in patients of acute viral hepatitis and fulminant hepatic failure. Ind. J. Med. Res. 2000, 112, 121–127.
- Arankalle, V A.; Deshmukh, T M.; Chobe, L P.; Chada, M S.; Walimbe, A M. Hepatitis G virus infection in india: prevalence and phylogenetic analysis based on 5′ noncoding region. Ind. J. Gastroenterol. 2001, 20, 13–17. [CSA]
- Forns, X.; Fernandez-lama, L.; Costa, J. Hepatitis G virus infection in a hemodialysis unit: prevalence and clinical implications. Nephrol. Dial. Transplant. 1997, 12, 956–960. [PUBMED], [INFOTRIEVE], [CROSSREF], [CSA]
- Jarvis, L M.; Davison, F.; Hanley, J P.; Yap, P L.; Ludlan, C A.; Simmon, P. Infection with hepatitis G virus among recipients of plasma products. Lancet 1996, 8 (1), 1352–1355. [CROSSREF]
- Schulte-Frohlinde, E.; Schmolke, S.; Reindl, W. Significance of antibodies to recombinant E2 protein of hepatitis G virus to hemodialysis patients. J. Viral Hepatitis 1998, 5, 341–344. [CROSSREF], [CSA]
- Szabo, A.; Viazov, S.; Heeman, U. GBVC/HGV infection in renal dialysis and transplant patients. Nephrol. Dial. Transplant. 1997, 12, 2380–2384. [PUBMED], [INFOTRIEVE], [CROSSREF], [CSA]
- Tribl, B.; Oesterreicher, C.; Pohanka, E. GBV-C/HGV in hemodialysis patients: anti-E2 antibodies and GBV-C/RNA in serum and peripheral blood mononuclear cells. Kidney Int. 1998, 53, 212–216. [PUBMED], [INFOTRIEVE], [CROSSREF]
- De Lamballerie, X.; Charrel, R N.; Dussol, B. Hepatitis GB virus C in-patients on hemodialysis. N. Engl. J. Med. 1996, 334, 1334–1349. [CROSSREF], [CSA]
- Bukh, J.; Miller, R H.; Purcell, R H. Genetic heterogeneity of hepatitis C virus Quasispecies and genotypes. Semin. Liver Dis. 1995, 15, 19–23.
- Tacke, M.; Kiyosawa, K.; Stark, K. Detection of antibodies to a putative hepatitis G virus envelop protein. Lancet 1997, 349, 318–320. [PUBMED], [INFOTRIEVE], [CROSSREF]
- Zhang, X H.; Shinzama, H.; Shao, L.; Ishibashi, M.; Saito, K.; Ohno, S.; Yamada, N.; Miasawa, H.; Togashi, H.; Takahashi, T. Detection of HGV RNA in patients with hepatitis B, hepatitis C and non A–E hepatitis by RT-PCR using multiple primer sets. J. Med. Virol. 1997, 52 (4), 385–390. [PUBMED], [INFOTRIEVE], [CSA]
- Linnen, J.; Wages, J R.J.; Zhang-Keckzy, Z Y. Molecular cloning and disease association of hepatitis G virus: a transfusion transmitted agent. Science 1996, 271, 505–508. [PUBMED], [INFOTRIEVE]