Abstract
To clarify the degrading mechanisms of a newly synthesized herbicide, Beflubutamid ([RS]-N-benzyl-2-[4-fluoro-3-trifluoromethylphenoxy]butanamide; BFB), in soil, we started a series of studies. The results on the characteristics of degradation in soil, and on the isolation and identification of BFB-degrading microbes are summarized as follows. BFB was immediately degraded in non-sterilized soil, but degraded little in sterilized soil, indicating that soil microbes are involved in BFB degradation in soil. Results suggested that the application of sawdust and cattle manure (CM) increased the population and activities of microbes because the degrading activity was greater in CM-applied soil than in soil supplied with chemical fertilizer. According to the morphological characteristics and 18S rDNA sequences, the isolated strain MF1 was categorized as the class Ascomycota, belonging to the order Hypocleales, and closely related to Gibberella fujikuroi, Fusarium oxysporum f. sp. Vasinfeatum and Gibberella puricaris. The results suggested that BFB degrades fastest under acidic conditions because the growth of strain MF1 was at an optimum at pH 6.5 and was depressed at neutral and alkaline pHs.
INTRODUCTION
After World War II, many synthetic organic pesticides developed in Europe were introduced in Japan, and contributed to an increase in food production (CitationKanazawa 1992). Today, approximately 0.5 Mg of pesticides are produced in our country (CitationUmezu and Ozawa 1994a). Among these pesticides, herbicides have contributed not only to an increase in food production by controlling weeds, but also to a reduction in labor (CitationKanazawa 1992). Although the use of pesticides and improvements in agricultural techniques have reduced labor, these factors have also led to a decrease in the number of farmers in recent years, with the number of new graduates who take up agriculture decreasing dramatically from more than 60,000 in 1960 to just 1,700 in 1998 (CitationUmezu and Ozawa 1994a). This indicates how serious the labor shortage in farming communities is, and under such conditions pesticides have become more necessary than ever before.
Owing to a lack of understanding about the necessity of pesticides and misinformation from the mass media, pesticides have been one of the targets for criticism since the 1960s when environmental pollution first became a serious concern (CitationKuwatsuka 1979a; CitationUmezu and Ozawa 1994b). In fact, many kinds of chlorine-based and high residue-prone pesticides, such as 1,1,1-trichloro-2,2-bis(4-chlorophenyl)ethane (DDT) and pentachloronitrobenzene (PCNB), were found to have high toxicity to the human body, and their use has been forbidden. However, these days, the toxicity of pesticides to human health has decreased because newly synthesized pesticides must undergo strict examination with regard to safety and metabolism before being put into use (CitationUmezu and Ozawa 1994b). Furthermore, high residue-prone pesticides with a long half-life, more than 1 year, cannot be registered for application (CitationYamamoto 1994). Accordingly, most pesticides have become relatively decomposable. In contrast, it has been reported that successive applications of these pesticides reduce their effectiveness and that the accumulation of decomposition products damages crops (CitationYamamoto 1994). As the degradation of pesticides in soils mainly relies on soil microorganisms (CitationYamamoto 1994), to solve the problems caused by pesticides, it is necessary to identify the pesticide-degrading microbes and clarify their degrading mechanisms, especially in the case of newly synthesized pesticides.
Beflubutamid ([RS]-N-benzyl-2-[4-fluoro-3-trifluoromethylphenoxy]butanamide; BFB) is an amide herbicide newly synthesized by Ube Ind. Ltd that is used for regulating the growth of broad-leaved weeds in wheat and barley fields (CitationTakamura et al. 1999). This herbicide is degraded relatively quickly (DT50 = 5.4 days) and is believed to be degraded by soil microorganisms. However, as yet the degraders of this herbicide have not been isolated and little is known about the degrading mechanisms.
In this study, with the aim of clarifying the microbial BFB-degrading mechanisms, we isolated and identified BFB-degrading microbes and then characterized their BFB degradation. Here, we report the characteristics of BFB degradation in upland soils and the isolation and identification of BFB-degrading microbes.
MATERIALS AND METHODS
Soils
Two kinds of soils, categorized as medium textured yellow soil, were sampled from the plowed layer of upland fields of the Fukuoka Agricultural Research Center, Chikushino City, Fukuoka Prefecture in February 2003: One soil received chemical fertilizer (NPK soil), and the other soil received successive applications of sawdust and cattle manure (CM soil). In these fields, soybeans and wheat had been planted in rotation from 1983 to 1994, and squash and cabbage have been cropped in rotation since 1995.
To exclude bulk plant residues and coarse fragments and to make them uniform, the soils were passed through a 2-mm mesh sieve and stored at 4°C. Before using, the soils were moistened to 40% of their maximum water-holding capacity and pre-incubated for 1 week at 28°C.
Degradation of Beflubutamid in soils
Beflubutamid was added to the soils at 2.55 mg kg−1, which is equivalent to the conventional amount of application of BFB in upland fields. Then, 20 g of each soil was placed into a 100-mL glass bottle, and incubated at 28°C for 12 days in triplicate. After 0, 3, 6 and 12 days of incubation, the soils were sampled and the residual BFB in the soil was measured. The BFB was extracted and purified as follows. Soils (20 g) in the 100-mL glass bottles were placed in a 200-mL Erlenmeyer flask and shaken (120 rpm) with 0.8 m3 m−3 acetone (60 mL) for 20 min. The extract was filtered, collected, and then concentrated in vacuo at a temperature lower than 40°C. Moreover, 0.8 m3 m−3 acetone (40 mL) was added to the residue in a 200-mL Erlenmeyer flask, shaken well and filtered. Each extract was combined, concentrated to approximately 5 mL in vacuo, and then shaken vigorously with ethyl acetate (30 mL) and 50 g L−1 sodium chloride solution (30 mL) in a separating funnel for 1 min. This procedure was repeated once again. Non-polar lipid was collected into a 50-mL plastic bottle, and disodium sulfate anhydrous (3 g) was added to dehydrate, after which the solution was filtered through Toyo No.1 filter paper (Toyo Roshi, Tokyo, Japan), and then concentrated in vacuo at a temperature lower than 40°C. Finally, acetonitrile (5 mL) was added to dissolve BFB and analyzed using reverse-phase high-performance liquid chromatography (HPLC).
To clarify the characteristics of BFB degradation in the soils, the time-course of BFB degradation in sterilized and non-sterilized soil was measured. Soil that was autoclaved (121°C, 30 min) three times every other day was used as sterilized soil. BFB (2.55 mg kg−1) was added to the sterilized and non-sterilized CM-soil (20 g) in 100-mL glass bottles, and then they were incubated at 28°C for 1 week in triplicate. Before and after incubation, the soils (20 g) were sampled and the residual BFB was measured using HPLC.
Analysis of Beflubutamid
Reverse-phase HPLC equipped with a photodiode array detector (Waters Co., Milford, MA, USA) was used to measure the concentrations of BFB. The analytical system used was as follows: instrument, Waters 625 LC System (Waters Co.); column, Inertsil ODS-3 (4.6 mm internal diameter × 250 mm, GL Sciences, Tokyo, Japan); detector, Waters 996 Photodiode Array Detector (Waters Co.); mobile phase, acetonitrile + methanol + 20 mmol L−1 phosphate dihydrate (59 + 11 + 30 by volume); flow rate, 1 mL min−1; column temperature, room temperature; data analysis, Waters Millennium32 program (Waters Co.).
Isolation of BFB-degrading microbes
To enrich the BFB-degrading microbes, BFB (1 mg m−3) was applied to the CM soil and incubated at 28°C for 1 week. After incubation, the soils (5 g) were collected and homogenized (10,000 g) with 45 mL of sterilized pure water using a wahling blender for 3 min. This mixture was serially diluted 102-fold with sterilized pure water. Each diluted solution (100 µL) was inoculated and spread on a soil extract agar medium containing 100 mg L−1 of BFB. The medium was composed of 0.1 g BFB, 0.2 g K2HPO4 and 15 g agar in 1,000 mL of soil extract solution (pH 6.8). The BFB is so insoluble in aquatic solutions that it (0.1 g) was dissolved in dimethylsulfoxide (DMSO) (700 µL) and mixed well with surfactant, Solpol 3004XL (0.2 g, TOHO Chemical Ind. Co., Tokyo, Japan), and then added to the medium. Colonies that established clear zones on the plate during the incubation were isolated.
Preparation of a spore mixture
Strain MF1 was cultured to sporulate on a nutrient broth (Difco) agar slant medium (pH 6.8) at 28°C for 2 weeks. Sodium dodecylsulfate (SDS; 0.1 g L−1) solution (5 mL) was added and the surface of the medium was scratched with a glass stick to release the spores from the fungal body. This solution was moved into another test tube and mixed with sterile glass beads (ϕ2.5 mm) for 1 min, after which the mixture was filtered through a funnel plugged with sterile cotton to exclude the hyphae. The spores were enumerated under a microscope, and the mixture was diluted to the density of 105 spores mL−1.
Microscopic observation
For the extension of the hyphae, strain MF1 was cultured in a nutrient broth agar medium (pH 6.8) at 28°C for 3 days. Part of the medium was removed, put on a slide glass, and observed under a Nikon ECLIPSE E600 microscope (Nikon Co., Tokyo, Japan) equipped with a Nikon DS-5M digital camera (Nikon Co.).
Quinone content
A spore mixture (300 µL, 105 spores mL−1) was inoculated into a nutrient broth liquid medium (30 mL, pH 6.8), and cultured by shaking (100 rpm) at 28°C for 3 days. Quinones were extracted from the isolates as previously described (CitationYamada and Kondo 1973). The medium was percolated with methanol (10 mL), sodium hydrate (4 g) and pyrogallol (1 g) at 100°C for 40 min. After cooling down, the percolated solution was removed into a 100-mL centrifugation tube and shaken three times with n-hexane (40 mL) for 1 min to dissolve the quinones into the n-hexane. To separate the n-hexane layer from an aquatic one, the tube was centrifuged at 5,000 g for 10 min. The n-hexane layer was collected and concentrated in vacuo at a temperature lower than 40°C. Finally, the quinones were dissolved in acetone (200 µL).
The quinones extracted from strain MF1 were analyzed using reverse-phase HPLC (CitationFujie et al. 1998). The analytical system used was as follows: instrument, Waters 625 LC System (Waters Co.); column, Zorbax ODS (4.6 mm internal diameter × 250 mm, GL Sciences Inc.); detector, Waters 996 Photodiode Array Detector (Waters Co.); mobile phase, methanol + diisopropyl ether (9 + 2 by volume); flow rate, 1 mL min−1; column temperature, room temperature; data analysis, Waters Millennium32 program (Waters Co.). To identify the species of quinone extracted from strain MF1, the following microorganisms were used as standards: Q-8, Burkholderia solanacearum MAFF730101; Q-9, Pseudomonas putida IFO14164; Q-10 (H2), Fusarium oxysporum IFO9966.
Optimal pH value
A spore mixture (50 µL, 105 spores mL−1) was inoculated into a nutrient broth liquid medium (50 mL, pH 4.5, 5.0, 5.5, 6.0, 6.5, 7.0 or 8.0), and cultured by shaking (100 rpm) at 28°C for 3 days in triplicate. After incubation, the medium was filtered through a glass filter (ADVANTEC, GC-50, Toyo Roshi) to collect the fungal bodies. The weight of the dried filter (105°C, 12 h) was measured before and after filtration, and the difference was taken to be the fungal biomass.
Extraction of DNA
A spore mixture (100 µL, 105 spores mL−1) was inoculated into a nutrient broth liquid medium (10 mL, pH 6.8), and cultured by shaking (100 rpm) at 28°C for 3 days. An aliquot of the culture (1 mL) was removed into a micro tube and centrifuged at 10,000 g for 10 min to harvest a fungal body. The DNA was extracted as follows. An SDS lysis mixture (100 mmol L−1 NaCl; 500 mmol L−1 Tris [pH 8.0]; 100 g L−1 SDS, 400 µL), a 120 mmol L−1 phosphate buffer (pH 8.0, 400 µL) and a phenol + chloroform + isoamylalcohol mixture (25 + 24 + 1 by volume, 400 µL) were added to the pellet, and then homogenized with glass beads (ϕ2 mm [0.06 g]; ϕ1 mm [0.30 g]; ϕ0.1 mm [0.80 g]) at 5,000 rpm for 30 sec with a MINI BEADBEATER (Biospec Products, Bartlesville, OK, USA). The supernatant (600 µL) was transferred into a new tube, the same amount of a chloroform + isoamylalcohol mixture (24 + 1 by volume) was added, and the mixture was voltexed for 20 sec. After centrifugation (15,000 g, 5 min) at room temperature, the supernatant (500 µL) was transferred into another new tube, and 3 mol L−1 CH3COONa (50 µL) was added and mixed well. Then, ethanol (≈1.5 mL) was added and mixed well, and incubated at room temperature for 10 min. After centrifugation (15,000 g, 5 min) at 4°C, the supernatant was removed. The pellet was washed with 0.7 m3 m−3 ethanol (1 mL), and centrifuged at 15,000 g for 2 min at 4°C, after which the supernatant was removed completely, and the pellet was dried at room temperature for 5 min. Distilled water (50 µL) was added to dissolve the DNA.
Polymerase chain reaction amplification and sequencing of 18S rDNA
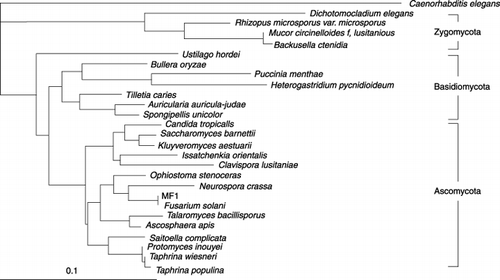
To clarify the phylogenetic position of strain MF1, we sequenced 18S rDNA region, which is often used in phylogenetic studies (CitationLeeflang et al. 2002; CitationBorneman and Hartin 2000). A schematic diagram of 18S rDNA is
Figure 1 Diagrams of 18S rDNA and the annealing position of the primers. White and black regions indicate the conserved and variable regions, respectively.
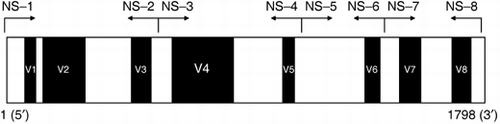
Table 1 Sequence and annealing position of the primers used in this study
A polymerase chain reaction (PCR) was carried out with the DNA extracted from strain MF1 using Takara Premix Taq (Premix Taq, Takara Bio, Shiga, Japan). The program Temp Control System PC707 (Astec Co., Fukuoka, Japan) was used for the reaction. The DNA (10 ng) and eukaryote specific primers (20 pmol), NS1 and NS8 (CitationWhite et al. 1990), were added to Premix Taq (25 µL), and filled with distilled water up to 50 µL. The temperature program for PCR was as follows: 94°C 2 min; 43°C 30 sec; 70°C 2 min; 94°C 1 min (30 cycles); 72°C 10 min.
Direct sequencing was carried out on the PCR amplification products using the dye-primer method and a Thermo sequenase fluorescently labelled primer cycling kit with 7-deaza-dGTP (Amersham Biosciences, Picataway, NJ, USA). Approximately 100 ng of PCR product was used as a template, and primers labeled with Cy5, NS1-8, were used as sequence primers. The temperature program for the reaction was as follows: 95°C 2 min; 95°C 30 sec; 55°C (annealing temperature[AT]) 2 min; 70°C 1 min (30 cycle) ; 72°C 5 min (except for NS1 and NS4). The AT for NS1 and NS4 was 43°C and 50°C, respectively. Electrophoresis was carried out with an automatic sequencer (ALF express II DNA sequencer, Amersham Biosciences). Sequences were determined using an ALFwin Sequence analyzer (ALFwin software, Amersham Biosciences).
Phylogenetic analysis
Phylogenetic analysis of strain MF1 was based on the sequences of the V4 region of 18S rDNA, the most variable region of an 18S rDNA sequence, and screened against the sequences in the DNA Data Bank of Japan using NCBI-BLAST program ver. 2.2.6. (CitationAltschul et al. 1997). Moreover, to clarify the phylogenetic position of the isolate, 18S rDNA sequences of several kinds of fungi submitted to the database were obtained. These data were aligned using CLUSTALW program ver. 1.8.3 (CitationThompson et al. 1994) to quantify the difference among the sequences. The quantified data were converted into genetic distances, and a phylogenetic tree was developed using the Tree View program (CitationPage 1997).
RESULTS AND DISCUSSION
Degradation of BFB in soils
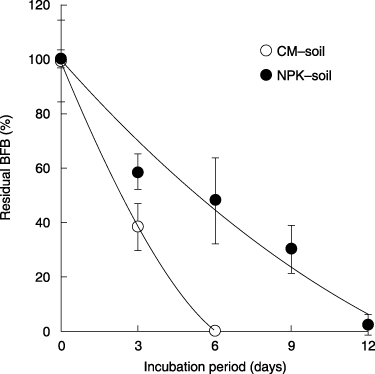
The time-course of BFB degradation in soils is shown in . In the CM soil, half of the BFB was degraded in the first 3 days of incubation, and BFB was completely degraded in 6 days. However, in the NPK soil, it took 6 days to degrade half of the BFB, and most of the BFB applied to the soil was degraded in approximately 12 days. It is known that the application of plant residues (such as straw, alfalfa and compost) and a source of nutrients, such as glucose, increase pesticide-degrading activity (CitationKuwatsuka 1979b). In the case of degradation with co-metabolism, the application of organic
Figure 2 Time-course of the changes in Beflubutamid (BFB) in sawdust and cow manure soil (CM) and chemical fertilizer (NPK) soil. The error bars indicate the standard deviations for three independent determinations.
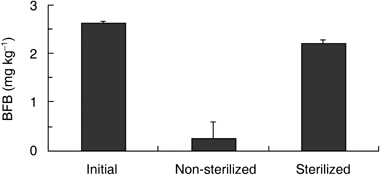
The degradation of BFB in sterilized and non-sterilized CM soil is shown in . As the amount of BFB (2.63 mg kg−1) measured before incubation bordered on the theoretical value (2.55 mg kg−1), it was confirmed that most of the BFB in the soil could be recovered. Thus, the adsorption of BFB by the soil did not affect the extraction of BFB from the soil. In sterilized soil, approximately 90% of BFB (2.22 mg kg−1) remained in the soil, while in non-sterilized soil the BFB was degraded to approximately 10% of the initial concentration (0.27 mg kg−1). It has been reported that, in many kinds of pesticides, the degradation rate in soils that have been sterilized is decreased in several ways (CitationKuwatsuka 1979b). Therefore, soil microbes are believed to be one of the most important factors in the degradation of pesticides (CitationYamamoto 1994). Consistent
with these findings, the soil microbes in the soils used in this study degraded BFB.Isolation of BFB-degrading microbes
The metabolism of pesticides is classified broadly into assimilation and co-metabolism (CitationKatayama and Yamamoto 1999). It is thought that because recent pesticides have been designed to be used in small amounts and that the concentrations in the soil are too low to be used by the microbes as a substrate, assimilating microbes cannot increase in pesticide-amended soils (CitationKatayama and Yamamoto 1999). In addition, it is assumed that most of the microbial degradation of artificial organic compounds, such as pesticides, occurs via co-metabolism (CitationKatayama and Yamamoto 1999). These reports indicate that assimilating microbes are not involved in degrading pesticides in actual soil. Traditionally, a mineral-basal medium with pesticides as the exclusive carbon source was used to isolate pesticide-degrading microbes. However, only assimilating microbes can be isolated using this method, indicating that it is not suitable for isolating microbes that mainly degrade pesticides in actual soils.
To isolate BFB-degrading microbes under conditions similar to those in an actual soil environment, a soil extract solution was used as the main component of the medium.
In addition, we attempted to prepare a cloudy medium with BFB emulsification and we succeeded. According to the clear zones established as a result of the degradation of BFB, BFB-degrading microbes were determined. BFB is so insoluble in aquatic solutions that it is difficult to mix in a medium. Using several types of water-insoluble materials, mediums for isolating degrading microbes have been prepared in several ways. CitationKiyohara et al. (1982) used the following method to isolate solid hydrocarbons, phenanthrene-, anthracene- and biphenyl-degrading microbes. First, these compounds were dissolved in an ethereal solution and then sprayed
Figure 4 Process of forming the halo-established medium after autoclaving and dispensing to the Petri dish.
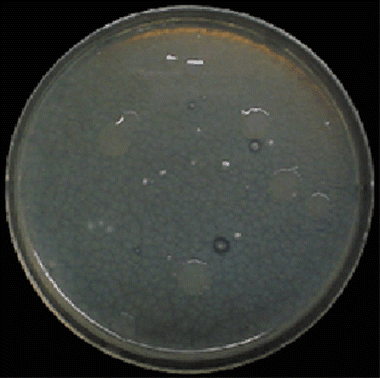
When the 10−1-diluted soil mixture was inoculated, several haloes were established on the medium (). Colonies with a halo were removed and re-inoculated on the nutrient broth (pH 6.8) agar plates. Subcultures on the same medium were inoculated two more times. Consequently, ten and three strains were isolated from the CM and NPK soils, respectively. However, as a result of denaturing gradient gel electrophoresis (DGGE) analysis, all isolates were identified as the same species. We named them “strain MF1”.
Morphological properties
A micrograph of strain MF1 is shown in . Strain MF1 formed a septate mycelium and established elliptical ascospores composed of two or more cells with a horizontal septate. From these morphological characteristics, strain MF1 was judged to be a fungus belonging to the class Ascomycota, related to the genus Fusarium.
Respiratory quinone
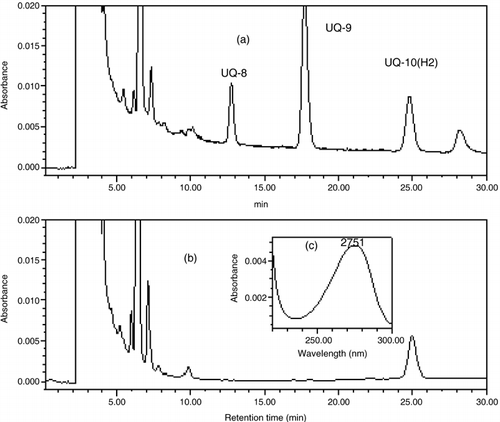
Respiratory quinones are thought to be one of the most important factors in the chemotaxonomy of microorganisms because each microbial community has its own predominant quinone species (CitationKatayama 2000). Fungus is known to have ubiquinones with nine or ten isoprenoid units (UQ-9 and UQ-10), and with ten isoprenoid units di- and tetra-hydrated (UQ-10[H2] and UQ-10[H4], respectively) (CitationWorld Data Centre for Microorganisms) 1995; CitationMinistry of Economy, Trade and Industry 2002). The HPLC chromatographs of standard quinones and those extracted from strain MF1 are shown in . A
single peak was obtained from strain MF1 extract and was identified as UQ-10(H2) according to its retention time.Effect of pH on the growth of strain MF1
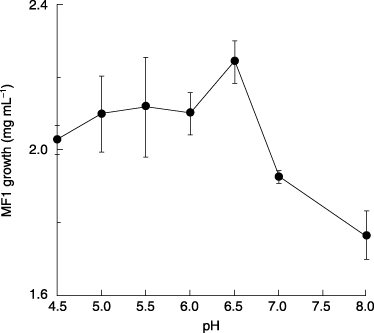
The effect of pH on the growth of strain MF1 is shown in . For 3 days of incubation, strain MF1 exhibited significant growth at pH 6.5, acescence conditions, and decreased with a rise in the pH value from 7.0 to 8.0. The growth of strain MF1 also decreased under acidic conditions, pH 6.0 to 4.5. In general, a fungus is supposed to grow vividly under acescence conditions (CitationWilkinson 1986). From these results, strain MF1 grows actively under acescence conditions around pH 6.5 and degrades BFB faster at this level than at any other level of pH.
Phylogenetic analysis of strain MF1 based on the 18S rDNA sequences
As all organisms have rRNA that is a biological molecule with common functions, by analyzing the rRNA genes, it is possible to examine the genetic relationship among a wide range of organisms (CitationNishiyama 1998). In recent studies, microbes have been identified from their oligonucleic acid sequences of rDNA, for example, 16S/23S rDNA in bacteria and 18S/28S rDNA (CitationHennequin et al. 1999) in fungi. A phylogenetic analysis on the basis of 18S rDNA can be applied to a wide range of taxa and is able to reveal the order or family, sometimes genus or species, level of classification.
We obtained approximately 1.7-kbp sequences of 18S rDNA of strain MF1 and conducted a homology search based on the V4 region, a variable region in the 18S rDNA sequence. From a BLAST search, we found that strain MF1 shares high homology with fungi belonging to the class Ascomycota, such as Gibberella fujikuroi (Accession No. AB237662, 100%), Fusarium oxysporum f. sp. Vasinfeatum (Accession No. AF219124, 100%) and Gibberella puricaris (Accession No. AF149875, 100%). All these fungi belong to the same order, Hypocleales. Furthermore, phylogenetic analysis also showed that MF1 belongs to the class Ascomycota ().
Microbial degradation of amide-pesticides
Many researchers have isolated and characterized several kinds of amide pesticide-degrading microbes, and there are many reports on amide-bond hydrolysis by their enzymes, such as aryl acylamine amidohydrolase (EC 3.5.1.4) (CitationLanzilotta and Pramer 1970a; CitationSharabi and Bordeleau 1969). Penicillium sp. and its cell-free extract degrade Karsil (N-[3,4-dichlorophenyl]-2-methylpentanamide) to 2-methyl-valeric acid and 3,4-dichloroaniline (CitationSharabi and Bordeleau 1969). Fusarium solani degrades propanil (3′,4′-dichloropropionanilide) to 3,4-dichloroaniline (CitationLanzilotta and Pramer 1970b). Bacillus sphaericus ATCC 12123 and its cell-free extract hydrolyze the amide-bond of linuron (3-[3,4-dichlorophenyl]-1-methoxy-1-methylurea), acylanilides and methoxy-substituted phenylureas (CitationEngelhardt et al. 1971). Successively, characteristics of this enzyme, an aryl acylamidase (EC 3.5.1.13) from Bacillus sphaericus ATCC 12123, were investigated, and it was shown that it is induced by the phenylamides of acylanilide-, phenylcarbamate- and methoxysubstituted phenylurea-type compounds (CitationEngelhardt et al. 1973). Recently, CitationYe et al. (2004) isolated mefenacet-degrading bacterium, Sphingomonas sp., which also have an amide-bond hydrolyzing ability.
In addition, there are some reports on the other metabolic mechanisms of amide-pesticides. Cunninghamella elegans and Mucor ramannianus R-56 degrade DEET (N,N-diethyl-m-toluamide) to N-ethyl-m-toluamide by deethylation (CitationSeo et al. 2005). Additionally, C. elegans is also able to produce two other metabolites, N,N-diethyl-m-toluamide-N-oxide and N-ethyl-m-toluamide-N-oxide, by N-oxidation (CitationSeo et al. 2005). Streptomyces strains are able to transform suspected endocrine-disrupting chemicals such as alachlor (2-chloro-2′,6′-diethyl-N-[methoxymethyl] acetanilide), mainly into 1-chloroacetyl-2,3-dihydro-7-ethylindole, 7-ethylindole, 7-ethyl-3-metyl-2-methoxy-2,3-dihydroindole, 8-ethylquinoline and 7-ethyl-N-methylindole.
In the BFB soil degradation experiments conducted before registration, 2-(4-fluoro-3-trifluoromethylphenoxy) butanoic acid (UR-50604) is detected as one of the major metabolites of BFB, and it is considered that BFB is mainly degraded by amide-hydrolases of soil
microbes. The soil used in this study is different from that used in previous experiments, but it may be that strain MF1 produces amide hydrolases and degrades BFB into UR-50604 because Penicillium sp. and Fusarium solani, related species of strain MF1, also produce amide hydrolases. To confirm this assumption, we should identify the BFB metabolites mediated by strain MF1 in future experiments.Although many soil microbes are known to have amide pesticide degrading abilities, we have captured only one kind of BFB-degrading microbe at this time. We believe that the reason for this result is as follows. One, BFB-degrading microbes cannot sufficiently be picked out using the naked eye. Two, although we investigated the effect of surfactant on the number of established colonies and decided its appropriate concentration at the inception of isolation, surfactant may inhibit the BFB-degrading activity of the other microbes except for strain MF1. To find more BFB degraders, we should reconsider the isolating methods used in the culture conditions and detection of BFB degradation.
REFERENCES
- Kanazawa , J . 1992 . “ Pesticides in the modernized agriculture ” . In Environmental Science of Pesticide , Edited by: Kanazawa , J . 11 – 22 . Tokyo : Godo-shuppan . (in Japanese)
- Umezu , K and Okawa , H . 1994a . “ [Pesticides-related industry] ” . In Aspects of Pesticide in Environment and Agriculture , Edited by: Umezu , K and Okawa , H . 1 – 21 . Tokyo : Soft Science . (in Japanese)
- Kuwatsuka , S . 1979a . “ Degradation and metabolism of pesticides ” . In Pesticide design – strategy and tactic , Edited by: Yamamoto , I and Hukami , J . 539 – 575 . Tokyo : Soft Science . (in Japanese)
- Umezu , K and Okawa , H . 1994b . “ [Pesticides, human health and environmental conservation type agriculture] ” . In Aspects of Pesticide in Environment and Agriculture , Edited by: Umezu , K and Okawa , H . 23 – 56 . Tokyo : Soft Science . (in Japanese)
- Yamamoto , H . 1994 . “ [Environmental problems and soil biochemistry ” . In Soil Biochemistry , Edited by: Kimura , M . 188 – 204 . Tokyo : Asakurashoten Publishing Company . (in Japanese)
- Takamura , S , Okada , T Fukuda , S . 1999 . UBH-820 – a new selective herbicide for weed control in cereals . BCPC-Weeds , 1 : 41 – 46 .
- Yamada , Y and Kondo , K . 1973 . Coenzyme Q system in the classification of the yeast genera Rhodotorula and Cryptococcus andyeast-like genera Sporobolomyces and Rhodosporidium . JGenApplMicrobiol , 19 : 59 – 77 .
- Fujie , K , Hu , HY , Tanaka , H , Urano , K , Katsuaki , S and Katayama , A . 1998 . Analysis of respiratory quinones in soil for characterization of microbiota . Soil SciPlant Nutr , 44 : 393 – 404 .
- Leeflang , P , Smit , E , Glandorf , DCM , van Hannen , FJ and Wernars , K . 2002 . Effect of Pseudomonas putidaWCS358r and its genetically modified phenazine producing derivative on the Fusariumpopulation in a field experiment, as determined by 18S rDNA analysis . Soil BiolBiochem , 34 : 1021 – 1025 .
- Borneman , J and Hartin , RJ . 2000 . PCR primers that amplify fungal rRNA genes from environmental samples . ApplEnvironMicrobiol , 66 : 4356 – 4360 .
- White , TJ , Bruns , T , Lee , S and Taylor , J . 1990 . “ Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics ” . In PCR Protocols , Edited by: Innis , MA , Gelfand , DH , Sninsky , JJ and White , TJ . 315 – 324 . San Diego : Academic Press .
- Altschul , SF , Madden , TL Schaffer , AA . 1997 . Gapped BLAST and PSI-BLAST: a new generation of protein database search programs . Nucleic Acids Res , 25 : 3389 – 3402 .
- Thompson , JD , Higgins , DG and Gibson , TJ . 1994 . CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice . Nucleic Acids Res , 22 : 4673 – 4680 .
- Page , RDM . 1997 . Tree View , Glasgow : University of Glasgow . version 1.5
- Kiyohara , H , Nagao , K and Yana , K . 1982 . Rapid screen for bacteria degrading water-insoluble, solid hydrocarbons on agar plates . ApplEnvironMicrobiol , 43 : 454 – 457 .
- Alexander , M . 1994 . Biodegradation and Bioremediation , San Diego : Academic Press .
- Katayama , A and Yamamoto , H . 1999 . “ [Pesticides and microbes] ” . In Environmental Issues and Soil Microbiology , Edited by: Japanese Society of Soil Microbiology . 29 – 69 . Tokyo : Hakuyuusha . (in Japanese)
- Katayama , A . 2000 . [Behavior and structure of microbial degrading agro-chemicals in soil . JPesticide Sci , 25 : 300 – 309 . (in Japanese)
- World Data Centre for Microorganisms1995 Quinone Database. [Updated 3 October 1995.] Available from URL: http://wdcm.nig.ac.jp/wdcm/Quinone.html (http://wdcm.nig.ac.jp/wdcm/Quinone.html)
- Ministry of Economy, Trade and Industry . 2002 . “ Data about the classification and identification of microbes ” . [Updated 29 May 2002.] Available from URL: http://www.meti.go.jp/policy/bio/dna/topview.htm
- Wilkinson , JF . 1986 . Introduction to Microbiology , 3rd edn , Oxford : Blackwell Scientific Publications .
- Nishiyama , M . 1998 . “ [A classification and a molecule system of a soil microbe] ” . In Molecular Biology of Soil Microorganisms , Edited by: Japanese Society of Soil Microbiology . 5 – 28 . Tokyo : Hakuyuusha . (in Japanese)
- Hennequin , C , Abachin , E Symoens , F . 1999 . Identification of Fusariumspecies involved in human infections by 28S rRNA gene sequencing . JClinMicrobiol , 37 : 3586 – 3589 .
- Lanzilotta , RP and Pramer , D . 1970a . Herbicide transformation. II. Studies with an acylamidase of Fusarium solani . ApplMicrobiol , 19 : 307 – 313 .
- Sharabi , NE and Bordeleau , LM . 1969 . Biochemical decomposition of the herbicide N-(3,4-dichlorophenyl)-2-methylpentanamide and related compounds . ApplMicrobiol , 18 : 369 – 375 .
- Lanzilotta , RP and Pramer , D . 1970b . Herbicide transformation. I. Studies with whole cells of Fusarium solani . ApplMicrobiol , 19 : 301 – 306 .
- Engelhardt , G , Wallnofer , PR and Plapp , R . 1971 . Degradation of linuron and some other herbicides and fungicides by a linuron-inducible enzyme obtained from Bacillus sphaericus . ApplMicrobiol , 22 : 284 – 288 .
- Engelhardt , G , Wallnofer , PR and Plapp , R . 1973 . Purification and properties of an aryl acylamidase of Bacillus sphaericus, catalyzing the hydrolysis of various phenylamide herbicides and fungicides . ApplMicrobiol , 26 : 709 – 718 .
- Ye , YF , Min , H and Du , YF . 2004 . Characterization of a strain of Sphingobacterium sp. and its degradation to herbicide mefenacet . JEnvironSci(China) , 16 : 343 – 347 .
- Seo , J , Lee , YG , Kim , SD , Cha , CJ , Ahn , JH and Hur , HG . 2005 . Biodegradation of the insecticide N, N-diethyl-m-toluamide by fungi: identification and toxicity of metabolites . ArchEnvironContamToxicol , 48 : 323 – 328 .
- Kuwatsuka , S . 1979b . “ Ecotoxicology of pesticides ” . In Pesticide design – strategy and tactic , Edited by: Yamamoto , I and Hukami , J . 1082 – 1105 . Tokyo : Soft Science . (in Japanese)