Abstract
Microorganisms play important roles in the nitrogen cycles of various ecosystems. Research has revealed that a greater diversity of microorganisms is involved in the nitrogen cycle than previously understood. It is becoming clear that denitrifying fungi, nitrifying archaea, anammox bacteria, aerobic denitrifying bacteria and heterotrophic nitrifying microorganisms are key players in the nitrogen cycle. Studies have revealed a major contribution by fungi in the production of N2O and N2 in grasslands, semiarid regions and forest soils. Some fungi can grow under various O2 conditions by using three types of energy-yielding metabolism: O2 respiration, denitrification (nitrite respiration) and ammonia fermentation. The amoA-like gene copies of Crenarchaeota were shown to be more abundant in soils than in autotrophic ammonia-oxidizing bacteria, and the gene was expressed at higher levels in soil to which ammonia was added. There are some contradictory findings, however, regarding archaeal and bacterial nitrification. Anammox bacteria have been shown to be widely distributed and to play an important role in both artificial and natural environments. The contribution of heterotrophic microorganisms to nitrification has been recognized in soil, and the biochemical mechanisms of several bacteria are becoming clear. A wide variety of bacteria have been found to be able to carry out aerobic denitrification and to be distributed across diverse environments. Using molecular biological techniques for soil bacteria, Nitrosospira species of clusters 2, 3 and 4 have been shown to be the dominant group in soils. Genome analyses of autotrophic nitrifying bacteria are providing new insights into their ecology and functions in soils.
INTRODUCTION
The nitrogen cycle is one of the most important nutrient cycles in terrestrial ecosystems. Nitrogen cycling involves four microbiological processes: nitrogen fixation, mineralization (decay), nitrification and denitrification. The input of large amounts of nitrogen fertilizers to agricultural fields influences these processes, especially nitrification and denitrification, and results in increased production of N2O (CitationAkiyama et al. 2006) and pollution of ground water by nitrate leaching from fields. Thus, recent extensive research on nitrification and denitrification has focused on the detection and identification of the dominant microorganisms responsible for these processes in soil ecosystems.
In the 100 years since the nitrogen cycle was proposed, it has been believed that nitrification is carried out by two chemolithoautotrophic bacterial groups, the ammonia-oxidizing bacteria (AOB) and the nitrite-oxidizing bacteria (NOB), (, line 2) and that denitrification is mediated by denitrifying bacteria under anaerobic conditions (, line 3). Recently, however, as shown in , various microorganisms belonging to not only Bacteria but also Eukarya and Archaea have been found to be involved in the processes of denitrification and nitrification. Novel microbiological reactions, such as anammox (CitationMulder et al. 1995), fungal denitrification (CitationShoun 2006) and archaeal nitrification (CitationKonneke et al. 2005) have been discovered. Anammox bacteria belonging to the planctomycete group convert ammonium and nitrite into dinitrogen gas under anaerobic conditions (, line 5). Some of the fungi are able to produce N2O and N2 through the two pathways of fungal denitrification (, line 3) and codenitrification (, line 4). Recent studies have shown definitive evidence that Archaea mediate nitrification (, line 2). Many heterotrophic microorganisms contribute to the nitrification process, including both ammonia oxidation and nitrite oxidation (, line 2). Aerobic denitrifying ability has been shown in various bacteria genera (, line 3). Moreover molecular ecological techniques and genome analyses are providing new insights into the ecology of microorganisms responsible for conventional bacterial nitrification and denitrification. Thus, researchers are beginning to recognize that a greater diversity of microorganisms is involved in the nitrogen cycle than was previously understood ().
Figure 1 Microbiological processes in the nitrogen cycle. (1) Nitrogen fixiation, (2) bacterial nitrification, archaeal nitrification and heterotrophic nitrification, (3) aerobic and anaerobic bacterial denitrifiction, nitrifer denitrification, fungal denitrification and archaeal denitrification, (4) and (5) co-denitrification (by fungi), (5) anammox and (6) N2O production during nitrification (ammonia oxidation).
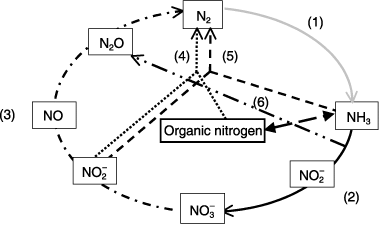
In this review we concentrate on recent developments in understanding the roles of fungi, archaea and bacteria in nitrification and denitrification.
INVOLVEMENT OF FUNGI IN THE NITROGEN CYCLE
Fungal denitrification
Denitrification is defined as the dissimilatory reduction of NO− 3 or NO− 2 to N2O and N2, respectively, by microbial reactions. For more than a century, the process has been considered to be mediated by a prokaryotic reaction, and the reaction mechanism of bacterial denitrification has been studied extensively in several bacteria, including Pseudomonas and Paracoccus (CitationZumft 1997). The bacterial denitrification process consists of four reactions catalyzed by nitrate reductase (Nar), nitrite reductase (Nir), nitric oxide reductase (Nor) and nitrous oxide reductase (Nos). However, CitationBollag and Tung (1972) reported that the soil fungi Fusarium oxysporum and Fusarium solani reduced nitrite in growing cultures and simultaneously released N2O under conditions of low O2 concentration. CitationShoun et al. (1992) found that denitrifying activity occurs in various fungi distributed across several phyla, including ascomycota such as Cylindrocarpon tonkinense and Gibberella fujikuroii and the basidiomycota Trichosporon cutaneum. Continuing research by Shoun has been clarifying the biochemical mechanisms and the physiological roles of fungal denitrification using pure cultures of fungi obtained from the type culture collection (CitationShoun 2006). In contrast, in soils, recent studies of denitrification have shown that denitrifying fungi contribute to the denitrification potential in soils and play a significant role in N2O production under both aerobic and anaerobic conditions.
The fungal denitrification system is localized in the mitochondria and functions for anaerobic respiration, as seen in the denitrification system of bacteria (CitationKobayashi et al. 1995). Dissimilatory Nar partially purified from the mitochondrial fraction of the denitrifying fungus F. oxysporum was shown to be distinct from soluble, assimilatory Nar (CitationUchimura et al. 2002). The properties of this fungal Nar were similar to those of the dissimilatory Nars of Escherichia coli and other denitrifying bacteria. Analyses of mutants defective for dissimilatory Nar or assimilatory Nar have shown that Nar is essential for fungal denitrification. Nir is one of the key enzymes in the dissimilatory denitrification process. Two structurally different Nirs are found among denitrifying bacteria: one contains copper (Cu-Nir) and is encoded by the nirK gene, and the other contains heme c and heme d1 (cd1-Nir) and is encoded by the nirS gene (CitationZumft 1997). No functionally significant differences between Cu-Nir and cd1-Nir have been reported. Fungal Nir contains copper and is an ortholog of bacterial NirK (CitationKobayashi et al. 1995). However, fungal Nor is distinct from bacterial Nor. Fungal Nor has been classified in the cytochrome P450 (P450) superfamily on the basis of its primary and tertiary structures. All bacterial Nors reported to date contain cytochromes bc in their catalytic centers. The fungal Nor, designated P450nor, reduces NO into N2O using NADH or NADPH as a direct electron donor (CitationKudo et al. 2001; CitationNakahara et al. 1993): 2NO + NAD(P)H + H+→Ν2Ο + ΝΑD(P)+ + H2O. P450nor catalyzes the reduction of two NO molecules to N2O and does not require other proteins for the reaction (CitationShiro et al. 1995). Other P450s receive an electron from NADPH via other components, such as P450 reductase. Recent gene analyses have revealed that P450nor is widely distributed among fungi.
The denitrification activities of bacteria are induced in the presence of either NO− 3 or NO− 2 under a limited O2 supply and are suppressed by excess O2, whereas fungal denitrification activity is induced in the presence of NO− 3 or NO− 2 and significant amounts of O2, but not in excess O2 (CitationZhou et al. 2001). Two types of denitrifying fungi have been categorized according to their ability to reduce NO− 3 or NO− 2 in a dissimilatory manner. A few fungi, such as F. oxysporum and Gibberella fujikuroi, reduce both NO− 3 and NO− 2 to N2O (CitationShoun and Tanimoto 1991), whereas most other reported denitrifying fungi reduce only NO− 2 to N2O (CitationShoun et al. 1992). Thus, it appears that the minimal requirements in the fungal denitrification system are Nir and P450nor.
Codenitrification
Many denitrifying fungi produce hybrid dinitrogen (N2) or N2O molecules by combining nitrogen atoms from nitrite and other nitrogen compounds (cosubstrates) under denitrifying conditions. This phenomenon was named “codenitrification” to indicate that nitrogen compounds (cosubstrates), such as azide and NH+ 4, are denitrified by the system induced by nitrite (or nitrate), but are incapable by themselves of inducing the denitrifying system (CitationShoun et al. 1992; CitationTanimoto et al. 1992). In the presence of amino acids, F. solani and C. tonkinense produced N2 by codenitrification. Fusarium oxysporum produced N2O from NO− 2 through both denitrification and codenitrification in the presence of a cosubstrate such as ammonia. Fungal P450nor was shown to catalyze the codenitrification reaction to form a hybrid N2O from NO and a cosubstrate (azide or NH+ 4 without an electron donor, such as NADH. In F. oxysporum, therefore, P450nor is a multifunctional enzyme that catalyzes denitrification or codenitrification in the presence of NADH or a cosubstrate, respectively (CitationSu et al. 2004). Codenitrification can be distinguished from denitrification by 15N-labeling experiments, which depend on the unique labeling patterns that result from the distinct biochemistry of the two processes. The codenitrification pathway combines nitrogen atoms from nitrite and other nitrogen compounds (cosubstrates), whereas the denitrifying pathway combines two molecules of nitrate or nitrite to N2 or N2O in a stepwise pathway. Thus, when 15NO− 3 is applied in anaerobic conditions, 14N15N or 14N15NO is generated by codenitrification, and 15N2 or 15N2O is generated by denitrification (CitationShoun et al. 1992; CitationTanimoto et al. 1992).
Ammonia fermentation
The third type of dissimilatory nitrate metabolism, ammonia fermentation, has also been found in the denitrifying F. oxysporum (CitationTakasaki et al. 2004; CitationZhou et al. 2002). In ammonia fermentation, nitrate is reduced to ammonia and ethanol is simultaneously oxidized to acetate to generate ATP. In the reaction, nitrate acts as the terminal electron acceptor for fermentation, but not for anaerobic respiration. Several bacteria have also been shown to have this pathway of dissimilatory nitrate reduction to ammonium (DNRA) (CitationTiedje 1988). However, in the DNRA, nitrate acts as the terminal electron acceptor for respiration, and a proton motive force is generated by the nitrate reduction (CitationSimon 2002). Ammonia fermentation supports fungal growth under conditions that are more anoxic than those of denitrification. Thus, F. oxysporum can grow under various environmental O2 conditions by using three types of energy-yielding metabolism: O2 respiration, denitrification (nitrite respiration) and ammonia fermentation. Ammonia fermentation activity has been found among many other soil fungi as well. These results suggest that fungi play an important role in the nitrogen cycle in soil ecosystems.
Role of fungi in nitrogen transformation in the soil
Fungal denitrification has been found to be dominant in forest soils, grasslands (CitationLaughlin and Stevens 2002) and semiarid regions (CitationMcLain and Martens 2006). CitationCastaldi and Smith (1998) evaluated the effect of cycloheximide on fungal denitrification activity in woodland and arable soils. Cycloheximide is one the representative antibiotics that inhibit eukaryotic protein synthesis and has been used to selectively inhibit fungal activity involved in nitrogen transformation in soils. Low cycloheximide concentrations (0.5–2.5 mg g−1 soil) drastically reduced the N2O emission induced by adding peptone to woodland soil. These results suggest a potential role of fungi in the N2O emission from this soil.
The contributions of bacteria and fungi to N2O and N2 production in a grassland soil (acid brown earth [Typic Dystrochrept]) were evaluated by combining the substrate-induced respiration inhibition method and the 15N gas-flux method (CitationLaughlin and Stevens 2002). Cycloheximide and streptomycin reduced the production of N2O by 89% and 23%, respectively. Therefore, in this grassland soil, fungi were major contributors to the production of N2O from NO− 3. In the 15N gas-flux study of the codenitrification process, a 15N atom from labeled 15NO− 2 combined with a 14N atom from a natural nitrogen source; the distribution of the 15N atoms in the N2 emitted from the soil revealed the relative contribution of fungal codenitrification and denitrification to N2 production. Approximately 92% of the labeled N2 was estimated to result from codenitrification, with 8% resulting from denitrification. In semiarid soils, fungi appear to be a vital component of nitrogen cycling and N2O production. The addition of cycloheximide to semiarid soil caused a 63% reduction in N2O production, whereas streptomycin was found to stimulate N2O production (CitationMcLain and Martens 2006).
Soil fumigation has been shown to influence overall microbial populations, diversity and activity. Recently, it has been reported that the use of Chloropicrin, which is a soil fumigant used against a wide spectrum of soil-borne diseases, increased N2O gas production in fumigated soils (CitationSpokas et al. 2006). The contribution of microbial activity to emissions of N2O following chloropicrin application was larger than that of abiotic processes in a forest soil. The relative contributions of fungi and bacteria were estimated to be 70% and 20%, respectively, of N2O production in the soil based on a selective-inhibition experiment under aerobic conditions. Experiments using 15N isotopes indicated that 12% of nitrogen from chloropicrin was incorporated into the produced N2O. Following chloropicrin fumigation, N2O production was not affected by various levels of acetylene. These results strongly suggest that under aerobic conditions fungal denitrification was the primary mechanism for chloropicrin-induced N2O production.
The ratios of fungal and bacterial biomass in soil vary depending on soil conditions, such as agricultural management, chemical properties and climate. The reported relative fungal biomass ranged from 12% to 95% in temperate soils, including arable, pasture, grassland and forest soils, and fungi often dominated the microbial biomass of these temperate soils (CitationRuzicka et al. 2000). Thus, fungi may contribute significantly to the production of N2O and N2 in various soil ecosystems.
ARCHAEAL NITRIFICATION AND DENITRIFICATION
The domain Archaea is evolutionarily distinct from the domains Eukarya and Bacteria. Archaea were previously thought to mostly inhabit extreme environments. However, surveys of 16S rRNA have provided important information about the extent of archaeal diversity in many environments. Crenarchaeota, one of the four kingdoms of Archaea, have been found in various moderate environments, including soils (CitationBintrim et al. 1997; CitationBuckley et al. 1998; CitationNicol et al. 2005; CitationOline et al. 2006).
Archaeal nitrification
Studies using radioisotopes have indicated that some archaea might be chemoautotrophs capable of light-independent carbon fixation. Until recently, however, the specific energy source for the archaea was unclear. Metagenomic studies of the Sargasso Sea have revealed the existence of an ammonia monooxygenase (AMO) gene on an archaeal- associated scaffold, although the AMO gene was thought to be unique to ammonia-oxidizing bacteria (AOB) (CitationVenter et al. 2004). CitationVenter et al. (2004) suggested that the high nitrite concentrations around the sampling site might have resulted from nitrification (ammonium oxidation) by marine Crenarchaeota, although nitrifying bacteria of the domain Bacteria were believed to be responsible for oceanic nitrification. These results indirectly indicated that nitrifying archaea play a major role in nitrification in marine ecosystems. In soil environments, the relative abundance of crenarchaeotal rDNA was estimated to be 0.5–3% using real-time polymerase chain reaction (PCR) for 16S rDNA from soil. Compared to the huge diversity of bacteria, the diversity of Crenarchaeota seems to be restricted to a few specific lineages (CitationOchsenreiter et al. 2003). Homologues of amo-like genes from soil clones were found in the Sargasso Sea samples (CitationTreusch et al. 2005). Expression of the amoA-like gene in soil samples was shown by reverse-transcription PCR (RT-PCR), and the gene was found to be expressed at higher levels in soil to which ammonia was added than in control soil (CitationLeininger et al. 2006).
An ammonia-oxidizing mesophilic Crenarchaeota (AOA), Nitrosopumilus maritimus, was isolated from a marine aquarium in Seattle, Washington (CitationKonneke et al. 2005). Nitrosopumilus maritimus is phylogenetically placed within the marine group 1.1a lineage of Crenarchaeota based on the sequences of 16S rRNA, the complete 16S–23S internally transcribed spacer region, and a small portion of the 23S rRNA. The strain was able to grow chemolithoautotrophically using ammonia as a sole source of energy, and it showed a similar growth rate and cell yield to AOB. The predicted amino acid sequences of the putative AMO-encoding genes from the strain were very similar to those of AMO-related genes reported in environmental DNA sequences of marine Crenarchaeota from the Sargasso Sea waters and from terrestrial soil. Phylogenetic analysis showed a low similarity between AOA and AOB AmoA-encoding genes; however, there were some significant similarities in the conserved amino acid residues that coordinate potential metal centers, indicating that these enzymes have a common evolutionary origin and belong to the same protein family (CitationTreusch et al. 2005). Several differences exist between the amo gene clusters of Crenarchaeota and Proteobacteria. All proteobacterial amo clusters have a conserved amoCAB operon arrangement, whereas the gene arrangement in crenarchaeal amo clusters varies. The crenarchaeal amo gene clusters contain a gene encoding a protein of unknown function between amoA and amoB, whereas the proteobacterial amo clusters do not have a gene between amoA and amoB. Further research is needed to evaluate the functional properties of the product from the Amo-encoding gene of AOA (CitationTreusch et al. 2005).
Potential amoA genes of Crenarchaeota, as well as 16S rRNA genes, have been found in environments ranging from marine to estuarine, freshwater, sediment and soil (CitationBeman and Francis 2006; CitationFrancis et al. 2005; CitationPark et al. 2006), suggesting that nitrification (ammonia oxidation) of Crenarchaeota may contribute considerably to net nitrification in a wide range of environments. In contrast, CitationOkano et al. (2004) reported that the kinetic parameters of ammonia oxidation obtained from soils treated with nitrogen fertilizer were consistent with those of pure culture of the AOB Nitrosospira. The generation time of AOB in the nitrogen-fertilized soils was similar to that determined in pure culture. The rate of ammonia oxidation per cell and the growth yields of AOB in the nitrogen-fertilized soils were in the same order of magnitude as those values obtained from pure culture. In their study, the population size of AOB in the soils was precisely estimated using a real-time quantitative PCR (RT-qPCR) assay targeting part of the amoA. These results indicated that the contribution of AOB to the nitrification activity in soil is extremely large.
Another study has shown, however, that amoA gene copies of Crenarchaeota were up to 3,000-fold more abundant than those of AOB in 12 pristine and agricultural soils from three climatic zones (CitationLeininger et al. 2006). Moreover, RT-qPCR demonstrated the expression of archaeal AMO in soil and supported the numerical dominance of archaea over AOB. These results suggested that Crenarchaeota might be the dominant ammonia-oxidizing microorganisms in soil ecosystems. In contrast, nitrite-oxidizing archaea have not been found and, thus, the contribution of archaea to the nitrite oxidation step of nitrification is unclear.
Because of these contradictory results, it remains unclear whether archaeal or bacterial nitrification is the main contributor to nitrification activity in soil ecosystems. There are no experimental methods, such as the use of specific inhibitors (acetylene, nitrapyrin), to distinguish archaeal and bacterial nitrification activity in soil. Further physiological, biochemical and genomic studies will be necessary to determine the relative contribution of nitrifying archaea to the nitrification activity in soils.
Archaeal denitrification
Several archaea, such as the hyperthermophile Pyrobaculum aerophilum and the halophile Haloferax denitrificans, are capable of denitrification (CitationCabello et al. 2004). Denitrifying archaea have been shown to reduce nitrate via nitrite, NO and N2O to N2 through the dissimilatory nitrate reduction pathway, similar to bacteria. However, recent biochemical analyses and genome sequence data have revealed differences between archaea and bacteria in the organization of the denitrifying enzyme genes, as well as in the structure and regulation of the enzymes (CitationPhilippot 2002). Very few studies have examined archaeal denitrification in natural ecosystems, including soil ecosystems. Understanding the ecology of denitrifying archaea and their contribution to the denitrification potential in soil is an open research field.
ANAMMOX (ANAEROBIC AMMONIUM OXIDATION)
Based on theoretical thermodynamic calculations, the possibility of anaerobic ammonium oxidation in biological systems has been postulated (CitationBroda 1977). The reaction was discovered in 1995 in a pilot plant treating wastewater at Gist-Brocades, Delft, in The Netherlands (CitationMulder et al. 1995). This biological process was named “anammox”, short for anaerobic ammonium oxidation. The anammox reaction combines ammonium and nitrite directly into N2 gas under anoxic conditions: NH+ 4 + NO− 2 → N2 + 2H2O. Therefore, in the experiment with labeled 15NH+ 4 and unlabeled NO− 2, 29N2 is generated by anammox (CitationVan de Graaf et al. 1995). The 15N-labeling experiment is a powerful tool for quantification of anammox.
Anammox bacteria have not been isolated in pure culture, but a dominant anammox bacterium was obtained physically from enrichment cultures using Percoll density-gradient centrifugation (CitationStrous et al. 1999). The bacterium was identified as Candidatus Brocadia anammoxidans based on phylogenetic analysis. Molecular analysis revealed that significant populations of anammox bacteria existed in wastewater treatment plants and aquatic ecosystems, and these bacteria belonged to three genera (Brocadia, Scalindua and Kuenenia) in the phylum Planctomycetes (CitationSchmid et al. 2005). Tracer experiments using 15N and a nutrient profile revealed a major contribution of anammox to the nitrogen cycle in marine environments (CitationDalsgaard et al. 2003). The anammox bacterium Candidatus Scalindua sorokinii was shown to be the direct link to the anammox reaction in the suboxic zone of the Black Sea, based on 15N tracer experiments, membrane-specific lipids and 16S rRNA analysis (CitationKuypers et al. 2003). The genome of the uncultured anammox bacterium Kuenenia stuttgartiensis was assembled and reconstructed directly from a community genome of a complex bioreactor community (CitationStrous et al. 2006). Data from the 4.3-Mb genome indicated an evolutionary relationship between anammox bacteria belonging to Planctomycetes and Chlamydiae and revealed the 200 genes involved in catabolism and respiration, including hydrazine-metabolizing enzyme and nine hydroxylamine oxidoreductase (HAO)-like proteins. It is becoming clear that anammox bacteria are more widely distributed than previously assumed, and that they play an important role in both artificial and natural environments (CitationMeyer et al. 2005; CitationPenton et al. 2006; CitationSchmid et al. 2007; CitationSchubert et al. 2006; CitationTal et al. 2005; CitationToh et al. 2002).
Members of Planctomycetes are widespread and occur in diverse environments, including oceans, lakes and soils. CitationBuckley and Schmidt (2003) found Planctomycetes in pasture, forest, tundra, arable and thermal soils using analysis of 16S rRNA libraries prepared from these soils. rRNA from Planctomycetes was estimated to account for 2–15% of the total rRNA extracted from agricultural soils, indicating that Planctomycetes may be an abundant bacterial group in soils. However, no studies have yet reported the coexistence of the anammox reaction and anammox Planctomycetes in soil environments.
AUTOTROPHIC NITRIFICATION
Nitrification is carried out in most soil ecosystems by chemolithoautotrophic AOB and NOB. Excess nitrate produced by nitrification causes the contamination of ground water, and the gaseous by-products (NO and N2O) of nitrification are two of the most potent greenhouse gases (CitationProsser 1989). Until recently, the slow growth, low yield and difficulty of isolating AOB and NOB have restricted physiological and ecological studies of these bacteria. The development of molecular biological techniques for soil bacteria has enabled a better understanding of the distribution, diversity and population dynamics of AOB and NOB (CitationKowalchuk and Stephen 2001). There are several excellent reviews on the molecular ecology and physiology of nitrifying bacteria (e.g. CitationKowalchuk and Stephen 2001; Arp and Stein 2003). In this section we focus on the recent advances in research on dominant nitrifying bacteria in agricultural soils, mechanisms of nitrifier denitrification and genome information of nitrifying bacteria.
Diversity of nitrifying bacteria in soil
The AOB are classified into three genera, Nitrosomonas (β-proteobacteria), Nitrosospira (β-proteobacteria) and Nitrosococcus (γ-proteobacteria), on the basis of the phylogenetic relationships of their 16S rRNA gene sequences (CitationHead et al. 1993). The AOB in soils have been studied mainly by targeting 16S rRNA and amoA genes. The amoA gene encodes subunit A of AMO, which catalyzes the first step of ammonia oxidation. The use of amoA is a powerful molecular tool because of its fine-scale resolution of closely related populations and its functional trait rather than a phylogenetic trait (CitationPurkhold et al. 2003). CitationIda et al. (2006) showed that pyruvate kinase was a useful molecular marker for the identification of AOB at a rank below the genus level.
The community structures of AOB have been shown to be affected by soil conditions, such as pH and nitrogen fertilizer input (CitationAvrahami et al. 2003; CitationKowalchuk and Stephen 2001). The genera Nitrosomonas and Nitrosospira were divided into at least seven clusters (Nitrosospira, clusters 1–4; Nitrosomonas, clusters 5–7) based on the 16S rRNA sequences of natural populations from a range of different environments (CitationStephen et al. 1996). Nitrosospira spp. of clusters 2, 3 and 4 were shown to be the dominant group in soils (CitationBruns et al. 1999; CitationPhillips et al. 2000). Because autotrophic nitrifying bacteria are dependent on NH+ 4 or NO− 2 as specific energy sources, the addition of NH+ 4 fertilizer to soils can increase the population size of nitrifying bacteria. Nitrosospira cluster 3 has been shown to be the dominant AOB in several neutral-pH arable fields receiving fertilizers (CitationAvrahami et al. 2003; CitationChu et al. 2006). In grassland soils treated with urea, AOB populations were dominated by Nitrosospira cluster 3 and Nitrosomonas cluster 7; the AOB communities in soil not treated with urea were more diverse than those in treated soil (CitationWebster et al. 2002). Another study reported that Nitrosomonas clusters were found in an agricultural soil that regularly received large amounts of nitrogen fertilizer (CitationHastings et al. 1997). These results indicated that the addition of ammonia fertilizer stimulates the growth of Nitrosomonas populations.
Soil pH is well known as a limiting factor for nitrification in soils (CitationDe Boer and Kowalchuk 2001). The optimum pH for nitrifying bacteria in pure culture is in the range of 7–9, and the lower limit for growth is approximately pH 6 (CitationAllison and Prosser 1993). Many investigations have shown that nitrification occurred in strongly acidic soils with pH values ranging from 3 to 5 (CitationHayatsu and Kosuge 1993; CitationWalker and Wickramasinghe 1979). The sequences representative of Nitrosospira cluster 2 were found in greater relative abundance in acidic agricultural soils, suggesting that cluster 2 may be adapted to growth at low pH (CitationLaverman et al. 2001; CitationNugroho et al. 2005). The lower pH limit for nitrification in acidic tea field soils was shown to be approximately pH 2.9. A Nitrosococcus-like acidophilic AOB was isolated from strongly acidic tea field soils (CitationHayatsu 1993).
The soil habitats in rice paddy fields are highly complex and heterogeneous. During the waterlogged period, the paddy field can be divided into five layers: floodwater, surface oxidized layer, reduced plow layer, plow pan and subsoil (CitationKimura 2000). Microbial communities examined during the waterlogged period have been shown to be different from those of other arable soils (CitationMurase et al. 2005; CitationNakayama et al. 2006; CitationSugano et al. 2005). Paddy soils, however, appear to harbor less diverse AOB communities than do grassland, meadow and forest soils (CitationAsakawa 2006; CitationBowatte et al. 2006a, Citation2006b; CitationMurase et al. 2003; CitationNicolaisen et al. 2004).
Based on cell morphology and the phylogenetic relationships of 16S rRNA gene sequences, NOB have been classified into four genera: Nitrobacter (α-proteobacteria), Nitrospina (δ-proteobacteria), Nitrococcus (γ-proteobacteria) and Nitrospira (class Nitrospira, phylum Nitrospirae) (CitationTeske et al. 1994). The genus Nitrospira contains a highly diverse group of species that are widely distributed in many natural habitats (CitationBartosch et al. 2002). Nearly all the Nitrospira and Nitrospira-like 16S rRNA sequences deposited in the public DNA database were derived from wastewater-plant microbial communities (CitationDaims et al. 2000; CitationSchramm et al. 1999) and aquatic ecosystems (CitationAltmann et al. 2003). Few studies have investigated NOB diversity and community structure in soil ecosystems. CitationFreitag et al. (2005) assessed the relationship between the diversity of NOB in agricultural grassland soils and inorganic nitrogen fertilizer management; denaturing gradient gel electrophoresis (DGGE) and clone library analyses have demonstrated that nitrogen management practices influence the diversity of functional NOB groups.
Nitrifier denitrification
Nitrous oxide is involved in the global greenhouse effect and the destruction of the ozone layer. The gas is produced in soil mainly through the microbial processes of nitrification and denitrification (CitationConrad 1996; CitationYokoyama and Ohama 2005). Denitrifiers produce N2O as an intermediate and possible end product of the reduction of NO− 3 to N2 (CitationZumft 1997). In contrast, nitrifying bacteria produce N2O in two ways: through nitrification and nitrifier denitrification (CitationArp and Stein 2003). In nitrification, N2O is produced as a by-product during ammonia oxidation by AOB. The unstable intermediate (HNO) formed during oxidation of NH2OH to NO− 2 is spontaneously decomposed to N2O (CitationHooper and Terry 1979). In nitrifier denitrification, N2O is an intermediate of the reduction of NO− 2 to N2 (CitationWrage et al. 2001). It is difficult to determine the relative importance of nitrification versus nitrifier denitrification in N2O production from soil. In contrast, the relative contributions of nitrification and denitrification to N2O production can be determined using the stable isotope technique (CitationSutka et al. 2006) or a selective inhibitor such as acetylene (CitationWrage et al. 2004). CitationSkiba and Smith (2000) reported that N2O production by nitrification increased in agricultural fields with increasing nitrogen input by fertilization.
The denitrification enzymes Nir and Nor were identified in the genome of Nitrosomonas europaea ATCC 19718 (CitationChain et al. 2003). A recent study using mutants deficient for a denitrifying enzyme (NirK or NorB) found that nitrifier denitrification was the major source of N2O produced by N. europaea ATCC 19718 (CitationSchmidt et al. 2004). The physiological role of nitrifier denitrification is not completely clear, but three main hypotheses have been proposed: (1) nitrifier denitrification may be a strategy to reduce competition for O2 from NOB by removing their substrate nitrite (CitationPoth and Focht 1985), (2) AOB use nitrite as an electron acceptor to obtain energy for their growth in low-O2 environments (CitationSchmidt and Bock 1997; CitationSchmidt et al. 2004), (3) nitrifier denitrification is used to protect AOBs’ own cells from toxic nitrite produced during ammonia oxidation (CitationBeaumont et al. 2002; CitationCho et al. 2006). In addition, the mechanism of N2 production is not fully characterized. However, the potential for nitrifier denitrification has been shown in a Nitrosospira lineage, which is considered to be the dominant AOB in soil (CitationShaw et al. 2006). These results suggested that nitrifier denitrification may be a universal trait in β-proteobacterial AOB and may contribute to N2O production in soil.
Genome analysis of nitrifying bacteria
The complete genome sequence of the β-proteobacterium N. europaea ATCC 19718 was the first reported for nitrifying bacteria (CitationChain et al. 2003). The genome sequence of a single 2.8-Mb circular chromosome was annotated and found to consist of approximately 2460 protein-encoding genes. Recently, the genome of the γ-proteobacterium Nitrosococcus oceani ATCC 19707 was analyzed (CitationKlotz et al. 2006); a single 3.48-Mb circular chromosome of N. oceani contained 3052 protein-encoding genes. The genomes of both strains contained all the genes encoding the complete central pathways of the tricarboxylic acid cycle, Embden–Meyerhof–Parnass cycle, and pentose phosphate cycle. The genome of N. europaea has two copies of the genes for AMO and HAO, whereas the genome of N. oceani contains only one copy of the genes for these enzymes. Forty genes encoding various siderophore receptors or transporters were identified in the N. europaea genome, although no genes encoding siderophore synthesize were found. The AOB need to take up large amounts of Fe from their external environment for synthesis of cytochrome, hydroxylamine oxidoreductase and other enzymes. Nitrosomonas europaea appears to depend on other bacteria that are able to produce a siderophore and take up the Fe–siderophore complex with their own siderophore receptors/transporters. However, N. oceani has genes for the synthesis of the hydroxamate-type siderophore aerobactin. The differences in their habitats might have caused the differences in the ability of N. europaea and N. oceani to synthesize a siderophore.
Genome sequence analysis of the NOB Nitrobacter winogradskyi ATCC 25391 revealed a single circular chromosome of 3.4 Mb encoding 3143 predicted proteins (CitationStarkenburg et al. 2006). Extensive similarities were found with the genes of two α-proteobacteria, Bradyrhizobium japonicum (1,300 genes) and Rhodopseudomonas palustris CG (815 genes). In addition to these two bacteria, many proteins annotated in the N. winogradskyi genome were highly similar to those of N. europaea (β-proteobacteria). The AOB and NOB generally coexist in environments, such as soils, wastewater and activated sludge. Thus, through this coexistence, genetic materials might have been exchanged between the two bacterial groups. The findings of these genome analyses provide new insights into the interaction of nitrifying bacteria and other soil microorganisms.
HETEROTROPHIC NITRIFICATION
Heterotrophic nitrification is carried out by a wide phylogenetic range of bacteria and fungi that can oxidize ammonia or reduced nitrogen from organic compounds to hydroxylamine, nitrite and nitrate (CitationFocht and Verstraete 1977). The reactions of heterotrophic nitrification appear not to be involved in energy-yielding metabolism and, thus, do not contribute to cellular growth. The physiological role of heterotrophic nitrification and the phylogenetic diversity of heterotrophic nitrifying microorganisms are unclear, although heterotrophic nitrification is considered to play a significant role in acidic soils, such as forest soils.
Recent research on the biochemical mechanisms of heterotrophic nitrification has been conducted using Paracoccus denitrificans (CitationMoir et al. 1996a), Alcaligenes faecalis (CitationJoo et al. 2005), Pseudomonas putida (CitationDaum et al. 1998) and a few other bacterial species. The bacteria possess ammonia- and hydroxylamine-oxidizing enzymes and, thus, have an ability to oxidize NH+ 4 to NO− 2. In some heterotrophic bacteria, such as P. denitrificans, heterotrophic nitrifying activity is linked to denitrifying activity. Paracoccus denitrificans AMO, which has only been purified in the active form AMO, is a quinol oxidase that consists of two subunits (CitationMoir et al. 1996a). The properties of the purified enzyme indicate that P. denitrificans AMO is similar to a family of enzymes including the AMO from N. europaea and the particulate methane monooxygenase from Methylococcus capsulatus. Physiological experiments, in particular the inhibition pattern by acetylene, have indicated that P. putida expressed a gene for AMO. Southern blot analyses, however, showed weak hybridization signals of P. denitrificans cloned DNA with N. europaea amoB (CitationCrossman et al. 1997) and the P. putida genome with N. europaea amoA (CitationDaum et al. 1998). Thus, the AMO genes of heterotrophic nitrifying bacteria seem to have only partial sequence similarities with the amoA gene of autotrophic AOB. The enzyme HAO converts NH2OH to NO− 2. The HAO of the autotrophic nitrifying bacteria N. europaea is a trimeric enzyme: each subunit contains eight hemes. In contrast, the HAOs from the heterotrophic nitrifying bacteria P. denitrificans (CitationMoir et al. 1996b) and Pseudomonas strain PB16 (CitationJetten et al. 1997) are soluble hydroxylamine oxidases that do not contain hemes.
A large number of heterotrophic microorganisms have the ability to convert NO− 2 to NO− 3. CitationSakai et al. (2000) showed that many heterotrophic bacteria were able to oxidize NO− 2 to NO− 3and that the catalase purified from Bacilllus badius I-73 had the ability to oxidize NO− 2 to NO− 3. Recently CitationMatsuzaka et al. (2003) proposed a hypothesis for the conversion of NO− 2 to NO− 3 in the heterotrophic nitrifier Burkholderia cepacia NH-17. They presumed that NO− 2 is reduced to NO by nitrite reductase and that the generated NO is oxidized to NO− 3 by NO dioxygenase, which was shown to oxidize NO to NO− 3 in aerobic conditions. Several fungal species, including Aspergillus wentii and Penicillium spp., have been shown to be able to oxidize NO− 2 to NO− 3 in pure culture (CitationFocht and Verstraete 1977). Fungal nitrification has been observed in acidic soils such as forest soils, but the biochemical mechanisms remain unclear.
The contribution of heterotrophs to nitrification in soil has long been recognized. The potentials of autotrophic and heterotrophic nitrification were distinguished by using specific inhibitors of autotrophic nitrification (CitationDe Boer and Kowalchuk 2001). The activity of autotrophic nitrifying bacteria was inhibited under extreme conditions, such as strongly acidic soil. Under such unfavorable conditions for autotrophic nitrifying bacteria, heterotrophic nitrifying microorganisms are believed to contribute to nitrification activity. Several reports have provided strong evidence for the contribution of heterotrophic microorganisms to nitrification in acidic forest soils (CitationKillham 1986). CitationBrierley et al. (2001) showed that organic nitrogen promoted nitrification in an acidic forest soil, but the addition of NH+ 4 had an inhibitory effect unless the soil was supplied with a readily available carbon source, such as acetate. The results indicated that the microorganisms responsible for nitrification in acidic soils were heterotrophs (CitationBrierley and Wood 2001). An Arthrobacter sp. isolated from the forest soil was able to survive and showed nitrification activity at pH 3 in pure culture.
AEROBIC BACTERIAL DENITRIFICATION
Bacterial denitrification usually occurs only under anaerobic conditions. Denitrifying enzyme activities are inhibited by O2 and the expressions of these genes are strictly suppressed. However, several bacteria have been shown to reduce nitrite or nitrate to gaseous nitrogen compounds, such as N2O and N2, in the presence of O2. Paracoccus denitrificans (= pantotropha; formerly Thiosphaera pantotropha), a representative aerobic denitrifying bacterium, has been characterized extensively. Paracoccus denitrificans ATCC 35512 reduced 27% of added nitrate to gaseous nitrogen in an atmosphere of 92% O2 (CitationSu et al. 2004). Aerobic denitrifying bacteria adapted to fluctuating oxic–anoxic conditions were isolated from diverse natural and managed ecosystems and characterized (CitationPatureau 2000). Aerobic denitrifying bacteria isolated from a soil in Japan were identified as Mesorhizobium sp. (CitationOkada et al. 2005) and Burkholderia cepacia (CitationMatsuzaka et al. 2003). These results implied that a wide variety of bacteria are able to carry out aerobic denitrification and that aerobic denitrifying bacteria are distributed across diverse environments. In addition, the influence of O2 concentration on the denitrifying activity differed from one denitrifying bacterium to another. Thus, aerobic denitrification is now considered as a variant represented by several denitrifying bacteria rather than a rare exception.
The results of soil incubation experiments suggested that aerobic denitrification occurs in arable soil. By using a combined stable isotope and acetylene inhibition approach, CitationBateman and Baggs (2005) were able to distinguish the relative contribution of nitrification and denitrification to N2O production. The results suggested that aerobic denitrification occurred at 20% water-filled pore space. This study has provided one of the few observations about the contribution of aerobic denitrifying bacteria to the denitrification potential in soil.
CONCLUSION
For many years, members of the domain Bacteria had been thought to be the dominant contributors to the nitrogen cycle, and autotrophic aerobic nitrification and anaerobic denitrification were considered to be the main microbial processes in the nitrogen cycle. As described in this review, however, novel microbial processes of nitrogen transformation have also been found in the domains Archaea and Eukarya (). Fungi and archaea were shown to be involved in both nitrification and denitrification. The pioneering work of Shoun and coworkers clarified the physiological roles and detailed biochemical mechanisms of denitrification by several cultured fungi. Few studies, however, have investigated the contribution of fungi to nitrification and denitrification activity in soil. Although fungal denitrification was confirmed by selective inhibition experiments, the denitrifying fungi have not been isolated from those soils. Molecular techniques, such as PCR-based methods, have revealed the contribution of archaea to the nitrification potential of soils, although only a single strain of nitrifying archaea has been isolated from a marine environment. Nitrifying archaea in soil are thought to obtain energy for their growth through ammonia oxidation, although the ammonia-oxidizing activity of the archaea was not evaluated in the soils, which were simply analyzed for amoA-like gene expression.
Molecular ecological techniques for analyzing DNA and RNA from soil are powerful tools for investigating the diversity and function of microorganisms, but these techniques are not able to determine the enzyme activity that converts substrates into different molecules. Thus, to estimate the relative contribution of the microbiological processes (e.g. bacterial nitrification and archaeal denitrification) to the nitrogen cycle, stable isotope, specific assay and selective inhibitor techniques should be developed based on the metabolic manner and properties of the enzymes revealed by studies using pure cultures of the microorganisms responsible for nitrogen transformation. Pure cultures provide useful information to determine the environmentally relevant physiological and biochemical characteristics. However, few pure cultures of these microorganisms have been obtained from soil environments. Isolation of pure cultures and cultivation-based approaches are laborious and time consuming, and many soil microorganisms remain uncultured or are considered to be unculturable. However, some isolation and culture methods have been developed in the past few years (e.g. CitationZengler et al. 2002). The true roles of microorganisms in the nitrogen cycle will likely be clarified by a combination of cultivation-based approaches and molecular ecological techniques.
ACKNOWLEDGMENT
This research was partially supported by the Ministry of Education, Science, Sports and Culture, Grant-in-Aid for Scientific Research (B), No.18380053, 2006 to 2007.
REFERENCES
- Akiyama , H , Yan , X and Yagi , K . 2006 . Estimations of emission factors for fertilizer-induced direct N2O emissions from agricultural soils in Japan: summary of available data . Soil Sci. Plant Nutr , 52 : 774 – 787 .
- Allison , SM and Prosser , JI . 1993 . Ammonia oxidation at low pH by attached populations of nitrifying bacteria . Soil Biol. Biochem , 25 : 935 – 941 .
- Altmann , D , Stief , P , Amann , R , de Beer , D and Schramm , A . 2003 . In situdistribution and activity of nitrifying bacteria in freshwater sediment . Environ. Microbiol , 5 : 798 – 803 .
- Arp , DJ and Stein , LY . 2003 . Metabolism of inorganic N compounds by ammonia-oxidizing bacteria . Crit. Rev. Biochem. Mol. Biol , 38 : 471 – 495 .
- Asakawa , S . 2006 . DNA and RNA in soil: What can we see through them? 6. Analysis of soil microbial community structure. 2. Application of DGGE to paddy field soil . Soil Sci. Plant Nutr , 52 : 137
- Avrahami , S , Liesack , W and Conrad , R . 2003 . Effects of temperature and fertilizer on activity and community structure of soil ammonia oxidizers . Environ. Microbiol , 5 : 691 – 705 .
- Bartosch , S , Hartwig , C , Spieck , E and Bock , E . 2002 . Immunological detection of Nitrospira-like bacteria in various soils . Microb. Ecol , 43 : 26 – 33 .
- Bateman , EJ and Baggs , EM . 2005 . Contributions of nitrification and denitrification to N2O emissions from soils at different water-filled pore space . Biol. Fertil. Soil , 41 : 379 – 388 .
- Beaumont , HJ , Hommes , NG Sayavedra-Soto , LA . 2002 . Nitrite reductase of Nitrosomonas europaea is not essential for production of gaseous nitrogen oxides and confers’ tolerance to nitrite . J. Bacteriol , 184 : 2557 – 2560 .
- Beman , JM and Francis , CA . 2006 . Diversity of ammonia-oxidizing archaea and bacteria in the sediments of a hypernutrified subtropical estuary: Bahia del Tobari, Mexico . Appl. Environ. Microbiol , 72 : 7767 – 7777 .
- Bintrim , SB , Donohue , TJ , Handelsman , J , Roberts , GP and Goodman , RM . 1997 . Molecular phylogeny of Archaea from soil . Proc. Natl Acad. Sci. USA , 94 : 277 – 282 .
- Bollag , JM and Tung , G . 1972 . Nitrous oxide release by soil fungi . Soil Biol. Biochem , 4 : 271 – 276 .
- Bowatte , S , Jia , Z , Ishihara , R , Asakawa , S and Kimura , M . 2006a . Characterization of ammonia-oxidizing bacteria associated with weeds in a Japanese paddy field using amoA gene fragments . Soil Sci. Plant Nutr , 52 : 593 – 600 .
- Bowatte , S , Jia , Z , Ishihara , R , Nakajima , Y , Asakawa , S and Kimura , M . 2006b . Molecular analysis of the ammonia-oxidizing bacterial community in the surface soil layer of Japanese paddy field . Soil Sci. Plant Nutr , 52 : 427 – 431 .
- Brierley , EDR and Wood , M . 2001 . Heterotrophic nitrification in an acid forest soil: isolation and characterisation of a nitrifying bacterium . Soil Biol. Biochem , 33 : 1403 – 1409 .
- Brierley , EDR , Wood , M and Shaw , PJA . 2001 . Influence of tree species and ground vegetation on nitrification in an acid forest soil . Plant and Soil , 229 : 97 – 104 .
- Broda , E . 1977 . Two kinds of lithotrophs missing in nature . Z. Allg. Mikrobiol , 17 : 491 – 493 .
- Bruns , MA , Stephen , JR , Kowalchuk , GA , Prosser , JI and Paul , EA . 1999 . Comparative diversity of ammonia oxidizer 16S rRNA gene sequences in native, tilled, and successional soils . Appl. Environ. Microbiol , 65 : 2994 – 3000 .
- Buckley , DH , Graber , JR and Schmidt , TM . 1998 . Phylogenetic analysis of nonthermophilic members of the kingdom Crenarchaeota and their diversity and abundance in soils . Appl. Environ. Microbiol , 64 : 4333 – 4339 .
- Buckley , DH and Schmidt , TM . 2003 . Diversity and dynamics of microbial communities in soils from agro-ecosystems . Environ. Microbiol , 5 : 441 – 452 .
- Cabello , P , Roldan , MD and Moreno-Vivian , C . 2004 . Nitrate reduction and the nitrogen cycle in archaea . Microbiology , 150 : 3527 – 3546 .
- Castaldi , S and Smith , KA . 1998 . Biol. Fertil Soils , 27 : 27 – 34 . Effect of cycloheximide on N2O and NO-3 production in a forest and an agricultural soil
- Chain , P , Lamerdin , J Larimer , F . 2003 . Complete genome sequence of the ammonia-oxidizing bacterium and obligate chemolithoautotroph Nitrosomonas europaea . J. Bacteriol , 185 : 2759 – 2773 .
- Cho , CM , Yan , T , Liu , X , Wu , L , Zhou , J and Stein , LY . 2006 . Transcriptome of a Nitrosomonas europaea mutant with a disrupted nitrite reductase gene (nirK) . Appl. Environ. Microbiol , 72 : 4450 – 4454 .
- Chu , H , Fujii , T Morimoto , S . 2006 . Community structure of ammonia-oxidizing bacteria under long-term application of mineral fertilizer and organic manure in a sandy loam soil . Appl. Environ. Microbiol , 73 : 485 – 491 .
- Conrad , R . 1996 . Soil microorganisms as controllers of atmospheric trace gases (H2, CO, CH4, OCS, N2O, and NO) . Microbiol. Rev , 60 : 609 – 640 .
- Crossman , LC , Moir , JW , Enticknap , JJ , Richardson , DJ and Spiro , S . 1997 . Heterologous expression of heterotrophic nitrification genes . Microbiology , 143 : 3775 – 3783 .
- Daims , H , Nielsen , PH , Nielsen , JL , Juretschko , S and Wagner , M . 2000 . Novel Nitrospira-like bacteria as dominant nitrite-oxidizers in biofilms from wastewater treatment plants: diversity and in situ physiology . Water Sci. Technol , 41 : 85 – 90 .
- Dalsgaard , T , Canfield , DE , Petersen , J , Thamdrup , B and Acuna-Gonzalez , J . 2003 . N2production by the anammox reaction in the anoxic water column of Golfo Dulce, Costa Rica . Nature , 422 : 606 – 608 .
- Daum , M , Zimmer , W , Papen , H , Kloos , K , Nawrath , K and Bothe , H . 1998 . Physiological and molecular biological characterization of ammonia oxidation of the heterotrophic nitrifier Pseudomonas putida . Curr. Microbiol , 37 : 281 – 288 .
- De Boer , W and Kowalchuk , GA . 2001 . Nitrification in acid soils: micro-organisms and mechanisms . Soil Biol. Biochem , 33 : 853 – 866 .
- Focht , DD and Verstraete , W . 1977 . “ Biochemical ecology of nitrification and denitrification ” . In Advances in Microbial Ecology , Edited by: Alexander , M . Vol. 1 , 135 – 214 . New York : Plenum Press .
- Francis , CA , Roberts , KJ , Beman , JM , Santoro , AE and Oakley , BB . 2005 . Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean . Proc. Natl Acad. Sci. USA , 102 : 14683 – 14688 .
- Freitag , TE , Chang , L , Clegg , CD and Prosser , JI . 2005 . Influence of inorganic nitrogen management regime on the diversity of nitrite-oxidizing bacteria in agricultural grassland soils . Appl. Environ. Microbiol , 71 : 8323 – 8334 .
- Hastings , RC , Ceccherini , MT , Miclaus , N , Saunders , JR , Bazzicalupo , M and McCarthy , AJ . 1997 . Direct molecular biological analysis of ammonia-oxidizing bacteria population in cultivated soil plots treated with swine manure . FEMS Microbiol. Ecol , 23 : 45 – 54 .
- Hayatsu , M . 1993 . The lowest limit of pH for nitrification in tea soil and isolation of an acidophilic ammonia-oxidizing bacterium . Soil Sci. Plant Nutr , 39 : 219 – 226 .
- Hayatsu , M and Kosuge , N . 1993 . Autotrophic nitrification in acid tea soils . Soil Sci. Plant Nutr , 39 : 209 – 217 .
- Head , IM , Hiorns , WD , Embley , TM , McCarthy , AJ and Saunders , JR . 1993 . The phylogeny of autotrophic ammonia-oxidizing bacteria as determined by analysis of 16S ribosomal RNA gene sequences . J. Gen. Microbiol , 139 : 1147 – 1153 .
- Hooper , AB and Terry , KR . 1979 . Hydroxylamine reductase of Nitrosomonas production of nitric oxide from hydroxylamine . Biochim. Biophys. Acta , 571 : 12 – 20 .
- Ida , T , Takahashi , R , Satoh , K , Kogure , M and Tokuyama , T . 2006 . Classification of chemoautotrophic ammonia-oxidizing bacteria using pyruvate kinase genes as high-resolution phylogenetic markers . Soil Sci. Plant Nutr , 52 : 284 – 290 .
- Jetten , MS , de Bruijn , P and Kuenen , JG . 1997 . Hydroxylamine metabolism in Pseudomonas PB16: involvement of a novel hydroxylamine oxidoreductase . Antonie Van Leeuwenhoek , 71 : 69 – 74 .
- Joo , HS , Hirai , M and Shoda , M . 2005 . Characteristics of ammonium removal by heterotrophic nitrification-aerobic denitrification by Alcaligenes faecalis no. 4 . J. Biosci. Bioeng , 100 : 184 – 191 .
- Killham , K . 1986 . “ Heterotrophic nitrification ” . In Nitrification: Special Publications of the Society for General Microbiology , Edited by: Prosser , JI . Vol. 20 , 117 – 126 . Oxford : IRL Press .
- Kimura , M . 2000 . “ Anaerobic microbiology in waterlogged rice fields ” . In Soil Biochemistry , Edited by: Bollag , J and Stotzky , G . 35 – 138 . New York : Marcel Dekker .
- Klotz , MG , Arp , DJ Chain , PS . 2006 . Complete genome sequence of the marine, chemolithoautotrophic, ammonia-oxidizing bacterium Nitrosococcus oceani ATCC 19707 . Appl. Environ. Microbiol , 72 : 6299 – 6315 .
- Kobayashi , M , Matsuo , Y , Takimoto , A , Suzuki , S , Maruo , F and Shoun , H . 1995 . Denitrification, a novel type of respiratory metabolism in fungal mitochondrion . J. Biol. Chem , 271 : 16263 – 16267 .
- Konneke , M , Bernhard , AE , de la Torre , JR , Walker , CB , Waterbury , JB and Stahl , DA . 2005 . Isolation of an autotrophic ammonia-oxidizing marine archaeon . Nature , 437 : 543 – 546 .
- Kowalchuk , GA and Stephen , JR . 2001 . Ammonia-oxidizing bacteria: a model for molecular microbiology ecology . Annu. Rev. Microbiol , 55 : 485 – 529 .
- Kudo , T , Takaya , N , Park , SY , Shiro , Y and Shoun , H . 2001 . A positively charged cluster formed in the heme-distal pocket of cytochrome P450nor is essential for interaction with NADH . J. Biol. Chem , 276 : 5020 – 5026 .
- Kuypers , MM , Sliekers , AO Lavik , G . 2003 . Anaerobic ammonium oxidation by anammox bacteria in the Black Sea . Nature , 422 : 608 – 611 .
- Laughlin , J and Stevens , RJ . 2002 . Evidence for fungal dominance of denitrification and codenitrification in a grassland soil . Soil Sci. Soc. Am. J , 66 : 1540 – 1548 .
- Laverman , AM , Speksnijder , AG , Braster , M , Kowalchuk , GA , Verhoef , HA and Van Verseveld , HW . 2001 . Spatiotemporal stability of an ammonia-oxidizing community in a nitrogen-saturated forest soil . Microb. Ecol , 42 : 35 – 45 .
- Leininger , S , Urich , T Schloter , M . 2006 . Archaea predominate among ammonia-oxidizing prokaryotes in soils . Nature , 422 : 806 – 809 .
- MacLain , JET and Martens , DA . 2006 . N2O production by heterotrophic N transformations in a semiarid soil . Appl. Soil Ecol , 32 : 253 – 263 .
- Matsuzaka , E , Nomura , N Maseda , H . 2003 . Participation of nitrite reductase in conversion of NO-2 to NO-3 in a heterotrophic nitrifier, Burkholderia cepacia NH-17, with denitrification activity . Microbes Environ , 18 : 203 – 209 .
- Meyer , RL , Risgaard-Petersen , N and Aller , DE . 2005 . Correlation between anammox activity and the microscale distribution of nitrite in a subtropical mangrove sediment . Appl. Environ. Microbiol , 71 : 6142 – 6149 .
- Moir , JWB , Crossman , LC , Spiro , S and Richardson , DJ . 1996a . The purification of ammonia monooxygenase from Paracoccus denitrificans . FEBS Lett , 387 : 71 – 74 .
- Moir , JWB , Wehrfritz , JM , Spiro , S and Richardson , DJ . 1996b . The biochemical characterisation of a novel non-haem iron hydroxylamine oxidase from Paracoccus denitrificans GB17 . Biochem. J , 319 : 823 – 827 .
- Mulder , A , van de Graaf , AA , Robertson , LA and Kuenen , JG . 1995 . Anaerobic ammonium oxidation discovered in a denitrifying fluidized bed reactor . FEMS Microbiol. Ecol , 16 : 177 – 184 .
- Murase , J , Itoh , K , Kano , M and Kimura , M . 2003 . Molecular analysis of β-proteobacterial ammonia oxidizer populations in surface layers of a submerged paddy soil microcosm . Soil Sci. Plant Nutr , 49 : 909 – 913 .
- Murase , J , Shimizu , M , Hayashi , M , Matsuya , K and Kimura , M . 2005 . Vertical changes in bacterial communities in percolating water of a Japanese paddy field as revealed by PCR-DGGE . Soil Sci. Plant Nutr , 51 : 83 – 90 .
- Nakahara , K , Tanimoto , T , Hatano , K , Usuda , K and Shoun , H . 1993 . Cytochrome P-450 55A1 (P-450dNIR) acts as nitric oxide reductase employing NADH as the direct electron donor . J. Biol. Chem , 268 : 8350 – 8355 .
- Nakayama , N , Okabe , A , Toyota , K , Kimura , M and Asakawa , S . 2006 . Phylogenetic distribution of bacteria isolated from the floodwater of a Japanese paddy field . Soil Sci. Plant Nutr , 52 : 305 – 312 .
- Nicol , GW , Tscherko , D , Embley , TM and Prosser , JI . 2005 . Primary succession of soil Crenarchaeota across a receding glacier foreland . Environ. Microbiol , 7 : 337 – 347 .
- Nicolaisen , MH , Risgaard-Petersen , N , Revsbech , NP , Reichardt , W and Ramsing , NB . 2004 . Nitrification-denitrification dynamics and community structure of ammonia-oxidizing bacteria in a high yield irrigated Philippine rice field . FEMS Microbiol. Ecol , 49 : 359 – 369 .
- Nugroho , RA , Roling , WF , Laverman , AM , Zoomer , HR and Verhoef , HA . 2005 . Presence of Nitrosospira cluster 2 bacteria corresponds to N transformation rates in nine acid Scots pine forest soils . FEMS Microbiol. Ecol , 53 : 473 – 481 .
- Ochsenreiter , T , Selezi , D , Quaiser , A , Bonch-Osmolovskaya , L and Schleper , C . 2003 . Diversity and abundance of Crenarchaeota in terrestrial habitats studied by 16S RNA surveys and real-time PCR . Environ. Microbiol , 5 : 787 – 797 .
- Okada , N , Nomura , N , Nakajima-Kambe , T and Uchiyama , H . 2005 . Characterization of the aerobic denitrification in Mesorhizobium sp. strain NH-14 in comparison with that in related rhizobia . Microbes Environ , 20 : 208 – 215 .
- Okano , Y , Hristova , KR Leutenegger , CM . 2004 . Application of real-time PCR to study effects of ammonium on population size of ammonia-oxidizing bacteria in soil . Appl. Environ. Microbiol , 70 : 1008 – 1016 .
- Oline , DK , Schmidt , SK and Grant , MC . 2006 . Biogeography and landscape-scale diversity of the dominant Crenarchaeota of soil . Microb. Ecol , 52 : 480 – 490 .
- Park , HD , Wells , GF , Bae , H , Criddle , CS and Francis , CA . 2006 . Occurrence of ammonia-oxidizing archaea in wastewater treatment plant bioreactors . Appl. Environ. Microbiol , 72 : 5643 – 5647 .
- Patureau , D , Zumstein , E , Delgenes , JP and Moletta , R . 2000 . Aerobic denitrifiers isolated from diverse natural and managed ecosystems . Microb. Ecol , 39 : 145 – 152 .
- Penton , CR , Devol , AH and Tiedje , JM . 2006 . Molecular evidence for the broad distribution of anaerobic ammonium-oxidizing bacteria in freshwater and marine sediments . Appl. Environ. Microbiol , 72 : 6829 – 6832 .
- Philippot , L . 2002 . Denitrifying genes in bacterial and archaeal genomes . Biochim. Biophys. Acta , 1577 : 355 – 376 .
- Phillips , CJ , Harris , D , Dollhopf , SL , Gross , KL , Prosser , JI and Paul , EA . 2000 . Effects of agronomic treatments on structure and function of ammonia-oxidizing communities . Appl. Environ. Microbiol , 66 : 5410 – 5418 .
- Poth , M and Focht , D . 1985 . 15N kinetic analysis of production by Nitrosomonas europaea: an examination of nitrifier denitrification . Appl. Environ. Microbiol , 49 : 1134 – 1141 .
- Prosser , JI . 1989 . Autotrophic nitrification in bacteria . Adv. Microb. Physiol , 30 : 125 – 181 .
- Purkhold , U , Wagner , M , Timmermann , G , Pommerening-Roser , A and Koops , HP . 2003 . 16S rRNA and amoA-based phylogeny of 12 novel beta-proteobacterial ammonia-oxidizing isolates: extension of the dataset and proposal of a new lineage within the nitrosomonads . Int. J. Syst. Evol. Microbiol , 53 : 1485 – 1494 .
- Ruzicka , S , Edgerton , D , Norman , M and Hill , T . 2000 . The utility of ergosterol as a bioindicator of fungi in temperate soils . Soil Biol. Biochem , 32 : 989 – 1005 .
- Sakai , K , Nisijima , H , Ikenaga , Y , Wakayama , M and Moriguchi , M . 2000 . Purification and characterization of nitrite-oxidizing enzyme from heterotrophic Bacillus badius 1–73, with special concern to catalase . Biosci Biotechnol Biochem , 64 : 2727 – 2730 .
- Schmid , MC , Maas , B Dapena , A . 2005 . Biomarkers for in situ detection of anaerobic ammonium-oxidizing (anammox) bacteria . Appl. Environ. Microbiol , 71 : 1677 – 1684 .
- Schmid , MC , Risgaard-Petersen , N van de Vossenberg , J . 2007 . Anaerobic ammonium-oxidizing bacteria in marine environments: widespread occurrence but low diversity . Environ. Microbiol , 9 : 1476 – 1484 .
- Schmidt , I and Bock , E . 1997 . Anaerobic ammonia oxidation with nitrogen dioxide by Nitrosomonas eutropha . Arch. Microbiol , 167 : 106 – 111 .
- Schmidt , I , van Spanning , RJ and Jetten , MS . 2004 . Denitrification and ammonia oxidation by Nitrosomonas europaea wild-type, and NirK- and NorB-deficient mutants . Microbiology , 150 : 4107 – 4114 .
- Schramm , A , de Beer , D , van den Heuvel , JC , Ottengraf , S and Amann , R . 1999 . Microscale distribution of populations and activities of Nitrosospira and Nitrospira spp. along a macroscale gradient in a nitrifying bioreactor: quantification by in situ hybridization and the use of microsensors . Appl. Environ. Microbiol , 65 : 3690 – 3696 .
- Schubert , CJ , Durisch-Kaiser , E , Wehrli , B , Thamdrup , B , Lam , P and Kuypers , MM . 2006 . Anaerobic ammonium oxidation in a tropical freshwater system (Lake Tanganyika) . Environ Microbiol , 8 : 1857 – 1863 .
- Shaw , LJ , Nicol , GW , Smith , Z , Fear , J , Prosser , JI and Baggs , EM . 2006 . Nitrosospira spp. can produce nitrous oxide via a nitrifier denitrification pathway . Environ. Microbiol , 8 : 214 – 222 .
- Shiro , Y , Fujii , M Iizuka , T . 1995 . Spectroscopic and kinetic studies on reaction of cytochrome P450nor with nitric oxide: implication for its nitric oxide reduction mechanism . J. Biol. Chem , 270 : 1617 – 1623 .
- Shoun , H . 2006 . Denitrification and anaerobic energy producing mechanisms by fungi . Tanpakushitsu Kakusan Koso , 51 : 419 – 429 . (in Japanese)
- Shoun , H , Kim , DH , Uchiyama , H and Sugiyama , J . 1992 . Denitrification by fungi . FEMS Microbiol. Lett , 73 : 277 – 281 .
- Shoun , H and Tanimoto , T . 1991 . Denitrification by the fungus Fusarium oxysporum and involvement of cytochrome P-450 in the respiratory nitrite reduction . J. Biol. Chem , 266 : 11078 – 11082 .
- Simon , J . 2002 . Enzymology and bioenergetics of respiratory nitrite ammonification . FEMS Microbiol. Rev , 26 : 285 – 309 .
- Skiba , U and Smith , KA . 2000 . The control of nitrous oxide emissions from agricultural and natural soils . Glob. Change Sci , 2 : 379 – 386 .
- Spokas , K , Wang , D , Venterea , R and Sadowsky , M . 2006 . Mechanisms of N2O production following chloropicrin fumigation . Appl. Soil Ecol , 31 : 101 – 109 .
- Starkenburg , SR , Chain , PS Sayavedra-Soto , LA . 2006 . Genome sequence of the chemolithoautotrophic nitrite-oxidizing bacterium Nitrobacter winogradskyi Nb-255 . Appl. Environ. Microbiol , 72 : 2050 – 2063 .
- Stephen , JR , McCaig , AE Smith , Z . 1996 . Molecular diversity of soil and marine 16S rRNA gene sequences related to beta-subgroup ammonia-oxidizing bacteria . Appl. Environ. Microbiol , 62 : 4147 – 4154 .
- Strous , M , Fuerst , JA Kramer , EH . 1999 . Missing lithotroph identified as new planctomycete . Nature , 400 : 446 – 449 .
- Strous , M , Pelletier , E Mangenot , S . 2006 . Deciphering the evolution and metabolism of an anammox bacterium from a community genome . Nature , 440 : 790 – 794 .
- Su , F , Takaya , N and Shoun , H . 2004 . Nitrous oxide-forming codenitrification catalyzed by cytochrome P450nor . Biosci. Biotechnol. Biochem , 68 : 473 – 475 .
- Sugano , A , Tsuchimoto , H , Tun , CC , Asakawa , S and Kimura , M . 2005 . Succession and phylogenetic profile of eubacterial communities in rice straw incorporated into a rice field: estimation by PCR-DGGE analysis . Soil Sci. Plant Nutr , 51 : 51 – 60 .
- Sutka , RL , Ostrom , NE Ostrom , PH . 2006 . Distinguishing nitrous oxide production from nitrification and denitrification on the basis of isotopomer abundances . Appl. Environ. Microbiol , 72 : 638 – 644 .
- Takasaki , K , Shoun , H Yamaguchi , M . 2004 . Fungal ammonia fermentation, a novel metabolic mechanism that couples the dissimilatory and assimilatory pathways of both nitrate and ethanol: role of acetyl CoA synthetase in anaerobic ATP synthesis . J. Biol. Chem , 279 : 12414 – 12420 .
- Tal , Y , Watts , JE and Schreier , HJ . 2005 . Anaerobic ammonia-oxidizing bacteria and related activity in Baltimore inner harbor sediment . Appl. Environ. Microbiol , 71 : 1816 – 1821 .
- Tanimoto , T , Hatano , K , Kim , D , Uchiyama , H and Shoun , H . 1992 . Co-denitrification by the denitrifying system of the fungus Fusarium oxysporum . FEMS Microbiol. Lett , 93 : 177 – 180 .
- Teske , A , Alm , E , Regan , JM , Toze , S , Rittmann , BE and Stahl , DA . 1994 . Evolutionary relationships among ammonia- and nitrite-oxidizing bacteria . J. Bacteriol , 176 : 6623 – 6630 .
- Tiedje , JM . 1988 . “ Ecology of denitrification and dissimilatory nitrate reduction to ammonium ” . In Biology of Anaerobic Microorganisms , Edited by: Zehnder , AJB . 179 – 244 . New York : John Wiley & Sons .
- Toh , SK , Webb , RI and Ashbolt , NJ . 2002 . Enrichment of autotrophic anaerobic ammonium-oxidizing consortia from various wastewaters . Microb. Ecol , 43 : 154 – 167 .
- Treusch , AH , Leininger , S , Kletzin , A , Schuster , SC , Klenk , HP and Schleper , C . 2005 . Novel genes for nitrite reductase and Amo-related proteins indicate a role of uncultivated mesophilic crenarchaeota in nitrogen cycling . Environ. Microbiol , 7 : 1985 – 1995 .
- Uchimura , H , Enjoji , H , Seki , T , Taguchi , A , Takaya , N and Shoun , H . 2002 . Nitrate reductase-formate dehydrogenase couple involved in the fungal denitrification by Fusarium oxysporum . J. Biochem. (Tokyo) , 131 : 579 – 586 .
- Van de Graaf , AA , Mulder , A , de Bruijn , P , Jetten , MS , Robertson , LA and Kuenen , JG . 1995 . Anaerobic oxidation of ammonium is a biologically mediated process . Appl. Environ. Microbiol , 61 : 1246 – 1251 .
- Venter , JC , Remington , K Heidelberg , JF . 2004 . Environmental genome shotgun sequencing of the Sargasso Sea . Science , 304 : 66 – 74 .
- Walker , N and Wickramasinghe , KN . 1979 . Nitrification and autotrophic nitrifying bacteria in acid tea soils . Soil Biol. Biochem , 11 : 231 – 236 .
- Webster , G , Embley , TM and Prosser , JI . 2002 . Grassland management regimens reduce small-scale heterogeneity and species diversity of β-proteobacterial ammonia oxidizer populations . Appl. Environ. Microbiol , 68 : 20 – 30 .
- Wrage , N , Velthof , GL , Laanbroek , HJ and Oenema , O . 2004 . Nitrous oxide production in grassland soils: assessing the contribution of nitrifier denitrification . Soil Biol. Biochem , 36 : 229 – 236 .
- Wrage , N , Velthof , GL , van Beusichem , ML and Oenema , O . 2001 . The role of nitrifier denitrification in the production of nitrous oxide . Soil Biol. Biochem , 33 : 1723 – 1732 .
- Yokoyama , K and Ohama , T . 2005 . Effect of inorganic N composition of fertilizers on nitrous oxide emission associated with nitrification and denitrification . Soil Sci. Plant Nutr , 51 : 967 – 972 .
- Zengler , K , Toledo , G Rappe , M . 2002 . Cultivating the uncultured . Proc. Natl Acad. Sci. USA , 99 : 15681 – 15686 .
- Zhou , Z , Takaya , N , Nakamura , A , Yamaguchi , M , Takeo , K and Shoun , H . 2002 . Ammonia fermentation, a novel anoxic metabolism of nitrate by fungi . J. Biol. Chem , 277 : 1892 – 1896 .
- Zhou , Z , Takaya , N , Sakairi , MA and Shoun , H . 2001 . Oxygen requirement for denitrification by the fungus Fusarium oxysporum . Arch. Microbiol , 175 : 19 – 25 .
- Zumft , WG . 1997 . Cell biology and molecular basis of denitrification . Micobiol. Mol. Biol. Rev , 61 : 533 – 616 .