Abstract
The present study examined T4-type phage communities in rice straw (RS) under the composting process by analyzing the composition of the major capsid gene (g23) of T4-type bacteriophages. The g23 clones were obtained from RS throughout the composting process from RS materials to composting RS in the curing stage (for 124 days). Most of the g23 clones were phylogenetically closely related to those in rice field soils and rice field floodwaters, and Paddy Groups II and III appeared to characterize the g23 genes in the composting RS. The diversity of g23 genes in the composting RS was highest in the RS material (day 0 after the onset of composting) and in the early thermophilic stage (day 7), and decreased markedly in the middle and curing stages. This change was in contrast to that of the bacterial community, which showed higher diversity in the middle and curing stages. There was no specific clone that characterized any stage during the composting process. These findings indicate that the phage community is not the major controlling agent in determining eubacterial succession and that the thermophilic stage in the composting process efficiently annihilated T4-type phages in the composting pile.
Introduction
The world produces 550 million tons of rice annually from an area of approximately 140 million hectares; 91% of the total production comes from Asia (CitationKyuma 2004). Rice straw (RS), a by-product of rice production, is often returned to the rice field in the form of compost to improve soil fertility (CitationInoko 1984; CitationTakahashi et al. 1978). Composting is a microbial degradation process that involves aerobic decomposition and a thermophilic stage (CitationFinstein and Morris 1975).
Microbiota in RS have shown temporal succession in the composting process stages: the thermophilic, middle and curing stages. Molecular analyses of microbial communities targeting phospholipid fatty acid (PLFA) and ssu rRNA genes have demonstrated that Gram-positive bacteria including thermophilic Bacillus spp. and actinomycetes dominate the thermophilic stage, and Clostridia and Gram-negative bacteria such as Cytophaga proliferate in the middle and curing stages (CitationCahyani et al. 2002, Citation2003). Methanogens and microeukaryotes also show stage-dependent succession in the composting process (CitationCahyani et al. 2004a,Citationb).
Viruses are the most abundant biological entities on earth and many studies have shown the ecological importance of viruses in controlling microbial communities in the environment (CitationWeinbauer 2004). As the host specificity of viruses is generally very strict (CitationKimura et al. 2008; CitationMoebus 1992), virus communities may change as microbial communities change during the composting process of RS. Recent developments in viral genomics have revealed that comparable genomic information is preserved among viral subsets and this information can be used as a tool for the phylogenetic evaluation of viral communities in the environment (CitationWeinbauer and Rassoulzadegan 2004). The most popular candidate gene for the evaluation of subset viral communities is the capsid gene, g23, of T4-type bacteriophages (phages), Myoviridae (CitationBreitbart et al. 2004). From a sequence comparison of tail and capsid genes, g18, g19 and g23, CitationTétart et al. (2001) and CitationDesplats and Krisch (2003) grouped the T4-type bacteriophage family, mainly enterophages, based on increasing divergence from the bacteriophage T4, into T-evens, PseudoT-evens, SchizoT-evens and ExoT-evens. Then, by applying primers targeting the g23 gene of the bacteriophage T4 to T4-type bacteriophage communities in diverse marine environments, CitationFilée et al. (2005) observed that most of the sequences of the polymerase chain reaction (PCR) products belonged to five previously uncharacterized groups (Groups I–V). Furthermore, it was recently elucidated that the g23 gene assembly in rice soils is different from, and more divergent than, those in marine and freshwater environments (CitationFujii et al. 2008; CitationWang et al. 2009a,Citationb), and CitationFujii et al. (2008) and CitationWang et al. (2009a,Citationb) grouped soil-specific g23 amino acid sequences of soil clones into Paddy Groups I–IX.
In the present study, we examined T4-type phage communities in RS along with the composting process by analyzing g23 sequences to evaluate the effect of eubacterial succession on phage communities in the composting process. This is the first study to examine phage communities during the composting process of RS.
Materials and methods
Rice straw samples
All RS samples used in the present study were the same as those used in our previous studies to elucidate the succession of biota (eubacteria, methanogens and eukaryotes) during the composting process of RS by PLFA analysis (CitationCahyani et al. 2002) and by polymerase chain reaction-denaturing gradient gel electrophoresis (PCR-DGGE) analysis followed by sequencing (CitationCahyani et al. 2003, Citation2004a,Citationb).
Setup of the compost pile and the sampling method
Composting of RS was conducted in a storehouse at Aichi-ken Anjo Research and Extension Center, central Japan (34°48′N, 137°30′E). Composting was carried out based on a conventional method used in Japan with RS and ammonium sulfate ([NH4]2SO4) as the materials (CitationCahyani et al. 2002). The size of the compost pile was approximately 210 cm long, 180 cm wide and 80 cm high. At each sampling time, five subsamples were collected randomly from the pile at a depth of approximately 30 cm, where the measured temperature was not different from that of the central part of the compost. The samples were then stored at _20°C. Samples used in the present study were taken on days 0 (RS material), 7, 14, 21, 28, 61, 89 and 124 over the composting period.
DNA extraction and purification
Rice straw material and mixtures of the five subsamples of RS compost taken on the above seven sampling dates were subjected to DNA extraction. The DNA extraction was carried out in duplicate as described previously (CitationCahyani et al. 2003). In brief, the frozen samples were lyophilized overnight and homogenized using a mortar and a pestle that were cooled with liquid nitrogen. The samples were subjected three times to freezing at –196°C and thawing at 65°C, with the addition of DNA extraction buffer and cetyltrimethylammonium bromide (CTAB). Afterwards, the samples were treated with lysozyme and proteinase K. The extracted DNA was purified using Sephadex G-200 (Pharmacia, Uppsala, Sweden).
Polymerase chain reaction amplification
The g23 genes in the T4-type phages were amplified with the primers MZIA1bis (5′-GAT ATT TGI GGI GTT CAG CCI ATG A-3′) and MZIA6 (5′-CGC GGT TGA TTT CCA GCA TGA TTT C-3′) (CitationFilée et al. 2005). The PCR reaction mixture and the cycle conditions were the same as those used in our previous study for elucidating T4-type phage communities in Mn nodules in Japanese paddy fields (CitationCahyani et al. 2009). The PCR was carried out with the TaKaRa PCR Thermal Cycles Model TP 240 (TaKaRa, Tokyo, Japan). The total volume of the reaction mixture was 50 μL, containing 0.4 μL of each primer (50 μmol L−1 each), 5 μL of 2.5 mmol L−1 dNTP mixture, 5 μL of 10× Ex Taq buffer (20 mmol L−1 Mg2+; TaKaRa), 1 μL of 0.1% bovine serum albumin (TaKaRa), 0.5 μL of Ex Taq DNA polymerase (TaKaRa), 1 μL of extracted DNA template and 36.7 μL of milli-Q water. The cycle conditions for the PCR amplification were as follows: one cycle of initial denaturation at 94°C for 5 min, followed by 35 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 1 min and extension at 72°C for 1 min, and one cycle of final extension at 72°C for 5 min.
Denaturing gradient gel electrophoresis analysis, cloning and sequencing
The DGGE was carried out by loading the PCR products onto an 80 g L−1 polyacrylamide gel with a denaturing gradient ranging from 20 to 60% in an electrophoresis cell D-code System (Bio-Rad Laboratories, Hercules, CA, USA) with 1× TAE buffer (40 mmol L−1 Tris, 20 mmol L−1 acetic acid and 1 mmol L−1 ethylenediaminetetraacetic acid [EDTA] at pH 8.0). The electrophoresis conditions for the DGGE analysis were similar to those of CitationFujii et al. (2008) and CitationCahyani et al. (2009); the analysis was run at 60°C and 150 V for 15 h. Visualization of the bands was achieved by staining with SYBR Green I nucleic acid gel stain (BMA, Rockland, ME, USA).
All recognizable DGGE bands were excised and amplified again with the same primers. The PCR products of the excised DGGE bands were purified with the QIAquick Gel Extraction kit (Qiagen, Tokyo, Japan). Cloning was carried out before sequencing. The cloning procedures were the same as those described previously (CitationCahyani et al. 2003). The PCR products were cloned into pT7 Blue T-vector (Novagen, Darmstadt, Germany). One positive clone from each transformation was chosen for sequencing. The isolated vector plasmids were used as templates for cycle sequencing with the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA), according to the manufacturer’s instructions. After ethanol/EDTA precipitation, sequencing was carried out with an ABI 3130 Genetic Analyzer (Applied Biosystems).
Sequence analysis
Although substitution, deletion or addition of nucleotides in the sequence is small at the nucleotide level, it often brings about significant changes in the amino acid sequence, and in the conformation and function of the g23 protein. Therefore, the obtained nucleotide sequences were translated to deduced amino acid sequences. The translated amino acid sequences were compared to other sequences in the database of the National Center for Biotechnology Information (NCBI) using a BLAST search. Sequences that had conserved regions in the fragments (between amino acid sequences from 111 to 127 and from 261 to 304 in coliphage T4 sequence; CitationParker et al. 1984) were judged to be g23 genes.
To examine whether the clones obtained in the present study were RS-compost specific, the alignments of the g23 fragments in the present study were compared first with those of T-evens, PseudoT-evens, SchizoT-evens, ExoT-evens and marine clones (CitationFilée et al. 2005) and then with environmental DNA samples that had been obtained from soils and RS in paddy fields (CitationCahyani et al. 2009; CitationFujii et al. 2008; CitationJia et al. 2007; CitationWang et al. 2009a,Citationb,Citationc). The sequences were aligned and a neighbor-joining tree was constructed using Clustal X (CitationThompson et al. 1997). The bootstrap analysis was based on 1,000 replications. The consensus tree was drawn with Tree View software (version 1.6.6; CitationPage 1996). DNA sequences of partial g23 genes were deposited in the DNA Data Bank of Japan (DDBJ) under accession numbers AB375934 to AB375983.
Results
Denaturing gradient gel electrophoresis band patterns of g23 fragments in composting rice straw
The number of DGGE bands was 18, 18, 6, 14, 6, 6, 2 and 2 on days 0, 7, 14, 21, 28, 61, 89 and 124, respectively, and in total, 50 g23 clones were obtained from those bands (). A succession of g23 fragments was observed during the composting process. Many g23 fragments with different mobilities appeared in the RS material (day 0), and most of these fragments remained on day 7, although some new bands appeared and some bands disappeared on day 7. In contrast, the number of DGGE bands was small on day 14. Then, some bands appeared or reappeared on day 21 (). However, the new bands on day 21 disappeared on day 28 and onwards, and the number of DGGE bands decreased along with composting time with only two bands remaining on day 124.
Figure 1 Denaturing gradient gel electrophoresis patterns of g23 gene fragments recovered from composting rice straw (RS). C0 to C124 indicate the period (0–124 days) of composting. Stages I to IV: (I) pre-composting stage (the initial RS materials), (II) thermophilic stage, (III) middle stage, (IV) curing stage.
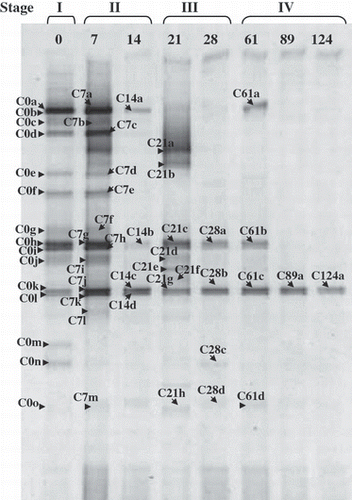
The DGGE patterns of the g23 fragments were very simple during the composting period. There was no conspicuous succession after day 28, and a broad band (positions of C0k and C0l) was present continuously from the RS material to the RS samples in the curing stage.
Sequence of the g23 clones
Fifty clones (15, 13, 4, 8, 4, 4, 1 and 1 clones from RS samples on days 0, 7, 14, 21, 28, 61, 89 and 124, respectively) were successfully sequenced as g23 fragments and aligned according to deduced amino acid sequences. The number of deduced amino acids between primers ranged from 124 to 180, in which 26 clones had 138 amino acid residues. Band C61a had the largest number of amino acid residues (180).
There were three groups of clones that showed the same sequence at the amino acid level, the first group consisted of C0b, C0c, C0j and C7a, the second group consisted of C0f, C7d and C7e, and the third group consisted of clones C0i, C14b, C14d, C21e, C21f, C21g, C61b, C61c, C89a and C124a. Within these three groups, clones C0j and C7a, clones C0f and C7e and clones C14b, C14d, C21f, C21g, C61b, C61c, C89a and C124a had the same sequence with each other at the nucleotide level.
Closest relatives of the g23 clones
Except for clones C21a (identity = 71%) and C7b (identity = 88%), all clones in the present study showed high identity, ranging from 97 to 100%, with clones in the NCBI database (). Nineteen out of 50 clones were closely related to clones retrieved from Japanese rice soils, with identities ranging from 71 (Band C21a) to 100% (Bands C0e, C0f, C0g and C7c) (CitationCahyani et al. 2009; CitationWang et al. 2009a, Citationc), and 31 clones were closely related to clones obtained from the floodwaters of a Japanese rice field, with identities ranging from 97 (Band C7g) to 100% (Bands C0i, C0o, C14b and C14d) (CitationNakayama et al. 2009).
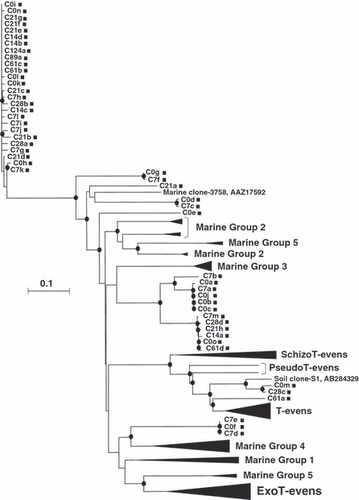
Phylogenetic analysis of the g23 clones in rice straw under the composting process
The phylogenetic tree in shows the clustering of the g23 clones in RS under the composting process, and g23 clones in marine environments and enterophage g23 genes. Almost all of the clones from composting RS formed clusters distinct from the clusters of marine clones and enterophages. Exceptions were clones C0m, C28c and C61a, and these clones formed a cluster with g23 genes in T-evens, PseudoT-evens and SchizoT-evens. Clone C21a, with low identity to its closest relative (71%), was remote from any marine and T-even cluster.
shows the relationships of the g23 genes obtained in the present study with those from soils and RS in paddy field environments, including the closest relatives shown in (CitationCahyani et al. 2009; CitationJia et al. 2007; CitationFujii et al. 2008; CitationWang et al., 2009a,Citationb,Citationc). Most clones from composting RS were distributed into Paddy Groups I, II, III, V, VIII and IX, and no clones belonged to Paddy Groups IV, VI and VII. Clone C21a was left ungrouped with a clone from rice field soil, and clones C0m, C28c and C61a were grouped with T-evens and PseudoT-evens. Most of the clones (26 clones) belonged to Paddy Group III, followed by Paddy Group II (with 12 clones). After day 7, no clones belonged to Paddy Groups I, V, VIII and IX.
Discussion
Denaturing gradient gel electrophoresis band patterns of g23 fragments in composting rice straw
Four stages have been recognized in the composting process of RS based on the change in temperature of the composting pile, the pattern of PLFA compositions and the DGGE band patterns of eubacterial and eukaryotic communities. These stages are: (I) the pre-composting stage (the initial RS materials), (II) the thermophilic stage, (III) the middle stage, (IV) the curing stage (CitationCahyani et al. 2002, Citation2003, Citation2004b). The RS material stage was represented by the sample taken on day 0, and the thermophilic, middle and curing stages were represented by the samples collected on days 7 and 14, on days 21 and 28, and on days 61, 89 and 124, respectively, in the present study.
The number of g23 fragments in was greatest on days 0 and 7, and lowest in the curing stage (two different fragments only on days 89 and 124). In contrast, the number of DGGE bands ranged from 21 to 42 for bacterial communities and from 15 to 38 for eukaryote communities. The number of bands was small on day 0, increased on days 7 and 14, and was largest in the middle stage, with a gradual decrease in the curing stage for bacterial and eukaryote communities (CitationCahyani et al. 2003, Citation2004b). Thus, DGGE bands were more numerous in the middle and curing stages than in the RS material and thermophilic stages. This finding indicates that the diversity of T4-type phage communities does not reflect the diversity of the bacterial communities in the composting process of RS, although phages are strictly host specific in general and phage communities are expected to be closely related to bacterial communities (CitationKimura et al. 2008).
As this was the first study to examine phage communities in the composting process, there is no information on phage communities in the composting process with which we could discuss the diversity of g23 genes in composting RS. However, several studies have examined DGGE of g23 genes retrieved from soils and floodwaters of rice fields (CitationCahyani et al. 2009; CitationFujii et al. 2008; CitationNakayama et al. 2009; CitationWang et al. 2009a,Citationb,Citationc), and all studies detected many DGGE bands in every sample. Therefore, the diversity of g23 genes in composting RS was estimated to be very low, particularly in the middle and curing stages compared with the diversities in rice field environments. CitationYu et al. (2006) isolated many Thermus phages belonging to Myoviridae from hot springs under isolation conditions at 70°C. However, there was no new DGGE band that appeared during the thermophilic stage on days 7 and 14 in the present study (). The present result indicates that most phages of thermophilic bacteria were not T4-type phages in composting RS, even if the phages of thermophilic bacteria existed in the thermophilic stage.
Table 1 Closest relatives in the National Center for Biotechnology Information database to the denaturing gradient gel electrophoresis bands obtained from rice straw under the composting process
The low diversity of the g23 genes could be ascribed to the composting process in which the temperature of the composting pile reached 66°C on day 7 (CitationCahyani et al. 2002). The high temperature during the thermophilic stage might have annihilated T4-type phages in the subsequent composting stages. It is well known that pasteurization (sterilization at 60°C) is the broadest and most currently available method for the removal of potential viral contaminants from plasma-derived products (CitationChandra et al. 2002).
Different mobilities among clones with identical nucleotide sequences
As described previously, clones C0j and C7a were identical in sequence at the nucleotide level. However, they had different mobilities (). There were also three different mobilities for the clones with the same nucleotide sequence (C14b, C14d and C21f). The differences in mobility among the clones with the same nucleotide sequence were estimated to result from the use of the degenerate primer MZIA1bis. This finding warns against a careless comparison of DGGE banding patterns, which misleads the false change in T4-type phage communities among the samples. However, we sequenced nearly all of the DGGE bands and found a conspicuous decrease in band number (). Therefore, the general conclusion that T4-type phage communities become very simple in the curing stage might be reasonable.
Phylogenetic analysis of the g23 clones in rice straw under the composting process
Almost all g23 clones in RS under the composting process formed clusters distinct from the clusters of enterophages and marine clones (). In the composting process, only RS and ammonium sulfate were used as materials without the addition of animal feces, which might have resulted in the distribution of only three clones (C0m, C28c, C61a) to the T-even cluster, the enterophage cluster. Most clones were closely related to the clones obtained from the soils and floodwaters of Japanese paddy fields and had high identity values (97–100%), except for clones C21a (71%) and C7b (88%). This finding indicates that T4-type phages in composting RS are not compost specific, but rather similar to those in rice fields. This inference was supported by the distribution of the clones from the composting RS in the phylogenetic tree in , in which most composting RS clones belonged to the Paddy Groups, particularly Paddy Groups II and III. Paddy Groups II and III may be the groups that characterize T4-type phages in composting RS. CitationCahyani et al. (2004a) observed many methanogenic archaea in RS materials and attributed their origins to contaminant soil because proliferation of methanogenic archaea on dry RS material is unlikely. Similarly, the T4-type phage communities in RS on days 0 and 7 appear to have been derived from contaminated soil (soil dust).
Figure 2 Neighbor-joining phylogenetic tree comparing g23 amino acid sequences of clones obtained in the present study with those of enterophages and marine clones (Filée et al. 2005). The filled circles indicate internal nodes with at least 50% bootstrap support. The scale bar represents the abundance of amino acid substitutions per residue. The filled squares indicate clones obtained from composting rice straw (RS) in the present study. The filled triangles indicate the clusters of enterophages and marine clones (Filée et al. 2005).
The DGGE band number was highest on days 0 and 7 and decreased markedly from day 28 onwards; no bands with different mobility appeared after day 14 (). In addition, almost all clones obtained on day 14 and onwards were distributed in Paddy Groups II and III, and any difference in the distribution of clones was rarely observed on days 0 and 7 (), which was in contrast to the succession of eubacterial communities over four succession stages (CitationCahyani et al. 2003). This finding indicated that there is no relationship between T4-type phage succession and bacterial succession in the composting process of RS.
Figure 3 Neighbor-joining phylogenetic tree comparing g23 amino acid sequences of clones obtained in the present study with those of soil and rice straw clones in paddy fields (Cahyani et al. 2009; Fujii et al. 2008; Jia et al. 2007; Wang et al. 2009a,b,c). The filled circles indicate internal nodes with at least 50% bootstrap support. The scale bar represents the abundance of amino acid substitutions per residue. The filled triangles indicate the clusters of soil and rice straw clones in paddy fields (Cahyani et al. 2009; Fujii et al. 2008; Jia et al. 2007; Wang et al. 2009a,b,c).
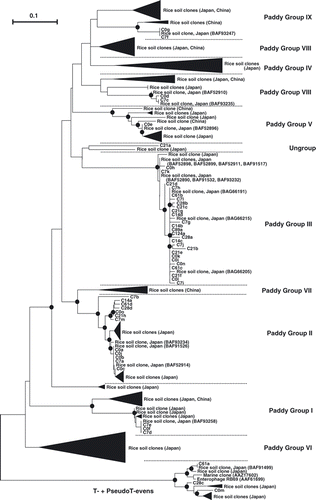
CitationJia et al. (2007) obtained four g23 clones from RS placed in flooded paddy soil. The clones were R-1, R-2, R-5 and R-15, and these clones were distributed in Paddy Groups IV and VI in , which contained no clone from composting RS in the present study. The T4-type phages in RS in the paddy fields and under composting may be different from each other.
In conclusion, the present study examined T4-type phage communities in RS under composting by analyzing g23 sequences. Most of the g23 sequences were closely related to those in paddy soils and paddy field floodwaters, and Paddy Groups II and III appeared to be characteristic of the g23 genes in composting RS. The diversity of g23 genes in composting RS was markedly low in composition compared with those in the soils and floodwaters. It was highest in the pre-composting stage (day 0) and early thermophilic stage (day 7) and decreased markedly in the middle and curing stages. This change was different from the changes observed in eubacterial and eukaryote communities; diversities were higher in the middle and curing stages than in the RS material and thermophilic stages. There was no specific clone that characterized any stage during the composting process. These findings indicate that the phage community is not the major controlling agent in determining eubacterial succession and that the thermophilic stage in the composting process efficiently annihilated T4-type phages in the composting pile.
References
- Breitbart , M , Miyake , JH and Rohwer , F . 2004 . Global distribution of nearly identical phage-encoded DNA sequences . FEMS Microbiol. Lett. , 236 : 249 – 256 .
- Cahyani , VR , Watanabe , A , Matsuya , K , Asakawa , S and Kimura , M . 2002 . Succession of microbiota estimated by phospholipids fatty acid analysis and changes in organic constituents during the composting process of rice straw . Soil Sci. Plant Nutr. , 48 : 735 – 743 .
- Cahyani , VR , Matsuya , K , Asakawa , S and Kimura , M . 2003 . Succession and phylogenetic composition of bacterial communities responsible for the composting process of rice straw estimated by PCR-DGGE analysis . Soil Sci. Plant Nutr. , 49 : 619 – 630 .
- Cahyani , VR , Matsuya , K , Asakawa , A and Kimura , M . 2004a . Succession and phylogenetic profile of methanogenic archaeal communities during the composting process of rice straw estimated by PCR-DGGE analysis . Soil Sci. Plant Nutr. , 50 : 555 – 563 .
- Cahyani , VR , Matsuya , K , Asakawa , A and Kimura , M . 2004b . Succession and phylogenetic profile of eukaryotic communities in the composting process of rice straw estimated by PCR-DGGE analysis . Biol. Fertil. Soils , 40 : 334 – 344 .
- Cahyani , VR , Murase , J , Ishibasi , E , Asakawa , S and Kimura , M . 2009 . T4-type bacteriophage communities estimated from the major capsid genes (g23) in manganese nodules in Japanese paddy fields . Soil Sci. Plant Nutr. , 55 : 264 – 270 . DOI
- Chandra , S , Groener , A and Feldman , F . 2002 . Effectiveness of alternative treatments for reducing potential viral contaminants from plasma-derived products . Thromb. Res. , 105 : 391 – 400 .
- Desplats , C and Krisch , HM . 2003 . The diversity and evolution of the T4-type bacteriophages . Res. Microbiol. , 154 : 259 – 267 .
- Filée , J , Tétart , F , Suttle , CA and Krisch , HM . 2005 . Marine T4-type bacteriophages, a ubiquitous component of the dark matter of the biosphere . P. Natl Acad. Sci-Biol. , 102 : 12471 – 12476 .
- Finstein , MS and Morris , ML . 1975 . “ Microbiology of municipal solid waste composting ” . In Advances in Applied Microbiology , Edited by: Perlman , D . Vol. 19 , 113 – 153 . New York : Academic Press .
- Fujii , T , Nakayama , N , Nishida , M , Sekiya , H , Kato , N , Asakawa , S and Kimura , M . 2008 . Novel capsid genes (g23) of T4-type bacteriophages in a Japanese paddy field . Soil Biol. Biochem. , 40 : 1049 – 1059 .
- Inoko , A . 1984 . “ Compost as a source of plant nutrients ” . In Organic Matter and Rice , 137 – 145 . Los Banos : International Rice Research Institute .
- Jia , Z , Ishihara , R , Nakajima , Y , Asakawa , S and Kimura , M . 2007 . Molecular characterization of T4-like bacteriophages in rice fields . Environ. Microbiol. , 9 : 1091 – 1096 .
- Kimura , M , Jia , Z , Nakayama , N and Asakawa , S . 2008 . Viral ecology in soil: Past, present, and future perspectives . Soil Sci. Plant Nutr. , 53 : 1 – 32 .
- Kyuma , K . 2004 . Paddy Soil Science , Melbourne : Trans Pacific Press .
- Moebus , K . 1992 . Further investigations on the concentration of marine bacteriophages in the water around Helgoland, with reference to the phage–host systems encountered . Helgolander Meeresun. , 46 : 275 – 292 .
- Nakayama , N , Asakawa , S and Kimura , M . 2009 . Comparison of g23gene sequence diversity between Novosphingobiumand Sphingomonasphages and phage communities in the floodwater of a Japanese paddy field . Soil Biol. Biochem. , 41 : 179 – 185 .
- Page , RDM . 1996 . TreeView:An application to display phylogenetic trees on personal computers . Comput. Appl. Biosci. , 12 : 357 – 358 .
- Parker , ML , Christensen , AC , Boosman , A , Stockard , J , Young , ET and Doermann , AH . 1984 . Nucleotide sequence of bacteriophage T4 gene 23 and the amino acid sequence of its product . J. Mol. Biol. , 180 : 399 – 416 .
- Takahashi , Z , Takahashi , S and Oka , N . 1978 . Rice-straw compost: A new formula . Mushroom J. , 71 : 348 – 351 .
- Tétart , F , Desplats , C , Kutateladze , K , Monod , C , Ackermann , HW and Krisch , HM . 2001 . Phylogeny of the major head and tail genes of the wide-ranging T4-type bacteriophages . J. Bacteriol. , 183 : 358 – 366 .
- Thompson , JD , Gibson , TJ , Plewniak , F , Jeanmougin , F and Higgins , DG . 1997 . The ClustalX windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools . Nucleic Acids Res. , 24 : 4876 – 4882 .
- Wang , G , Hayashi , M , Saito , M , Tsuchiya , K , Asakawa , S and Kimura , M . 2009a . Diversity of major capsid genes (g23) of T4-type bacteriophages in Japanese paddy field soils . Soil Biol. Biochem. , 41 : 13 – 20 .
- Wang , G , Jin , J , Asakawa , S and Kimura , M . 2009b . Survey of major capsid genes (g23) of T4-type bacteriophages in rice fields in Northeast China . Soil Biol. Biochem. , 41 : 423 – 427 .
- Wang , G , Murase , J , Taki , K , Ohasi , Y , Yoshikawa , N , Asakawa , S and Kimura , M . 2009c . Changes in major capsid genes (g23) of T4-type bacteriophages with soil depth in two Japanese rice fields . Biol. Fertil. Soils , 45 : 521 – 529 .
- Weinbauer , MG . 2004 . Ecology of prokaryotic viruses . FEMS Microbiol. Rev. , 28 : 127 – 181 .
- Weinbauer , MG and Rassoulzadegan , F . 2004 . Are viruses driving microbial diversification and diversity? . Environ. Microbiol. , 6 : 1 – 11 .
- Yu , MX , Slater , MR and Ackermann , HW . 2006 . Isolation and characterization of Thermusbacteriophages . Arch. Virol. , 151 : 663 – 679 .