ABSTRACT
The aggregation of huntingtin fragments with expanded polyglutamine repeat regions (HttpolyQ) that cause Huntington’s disease depends on the presence of a prion with an amyloid conformation in yeast. As a result of this relationship, HttpolyQ aggregation indirectly depends on Hsp104 due to its essential role in prion propagation. We find that HttQ103 aggregation is directly affected by Hsp104 with and without the presence of [RNQ+] and [PSI+] prions. When we inactivate Hsp104 in the presence of prion, yeast cells have only one or a few large HttQ103 aggregates rather than numerous smaller aggregates. When we inactivate Hsp104 in the absence of prion, there is no significant aggregation of HttQ103, whereas with active Hsp104, HttQ103 aggregates accumulate slowly due to the severing of spontaneously nucleated aggregates by Hsp104. We do not observe either effect with HttQ103P, which has a polyproline-rich region downstream of the polyglutamine region, because HttQ103P does not spontaneously nucleate and Hsp104 does not efficiently sever the prion-nucleated HttQ103P aggregates. Therefore, the only role of Hsp104 in HttQ103P aggregation is to propagate yeast prion. In conclusion, because Hsp104 efficiently severs the HttQ103 aggregates but not HttQ103P aggregates, it has a marked effect on the aggregation of HttQ103 but not HttQ103P.
INTRODUCTION
Huntington’s disease, caused by the polyglutamine (polyQ) expansion repeat region of the Huntingtin (Htt) protein, is a neurodegenerative disease affecting muscle coordination. The Htt protein with an expanded polyglutamine repeat region is cleaved to produce fragments that form amyloid plaques in the cerebral cortex and corpus striatum (Citation1, Citation2). The longer the polyQ repeat region was, the faster the formation of the amyloid fibers was and, perhaps in a related effect, the younger the age of onset of Huntington’s disease was (Citation3Citation–Citation5). The Htt fragments have an amyloid polyQ core, flanked by an N-terminal 17 amino acids, which promote aggregation (Citation3, Citation6Citation–Citation10), and a C-terminal polyproline region, which inhibits aggregation (Citation10Citation–Citation14). In addition to in vitro studies using purified proteins, tissue culture studies and animal models of Huntington’s disease have provided insights into the conditions that affect aggregation and the causes of the pathophysiology (Citation15Citation–Citation18). Yeast is a uniquely useful model system for studying the aggregation involved in Huntington’s disease due to its ease of culture and genetic manipulation (Citation19Citation–Citation21).
Unlike in other organisms, prions in yeast play a critical role in aggregating Htt fragments with expanded polyglutamine repeats (Citation22Citation–Citation26). The Sherman laboratory found that the aggregation of HttpolyQ fragments depends on the presence of [RNQ+] prion (Citation22). They observed numerous HttQ103 aggregates in [RNQ+] cells, whereas either in yeast cured of the [RNQ+] prion or in an rnq1 deletion strain, there were far fewer cells with aggregates. Furthermore, since prion propagation and, thus, [RNQ+] propagation are dependent on the severing activity of Hsp104, an hsp104 deletion yeast strain also showed a marked reduction in the number of cells with aggregates. The Lindquist laboratory extended this observation by looking at the aggregation of different Htt fragments in yeast (Citation23). They observed that the aggregation of HttpolyQP fragments, which have a polyproline region downstream of the polyglutamine region, depends on the presence of the [RNQ+] prion (Citation26). Regardless of the amino acids flanking the HttQ103 fragment, they found markedly fewer HttQ103 aggregates in both yeast cured of [RNQ+] and an rnq1 deletion strain than in [RNQ+] yeast. Interestingly, not only does prion induce HttpolyQ aggregation, but inversely, the expression of extended polyQ repeat regions also induces the formation of [RNQ+] and [PSI+] prions (Citation27Citation–Citation29).
While in yeast, the aggregation of HttpolyQ and HttpolyQP fragments depends on the presence of a prion with an amyloid conformation, these fragments differ in their morphologies and toxicities (Citation22, Citation23, Citation30). In [RNQ+] yeast, the HttQ103 fragments form multiple relatively small aggregates that are toxic to the yeast, while the HttpolyQP fragments tend to form a large aggregate that is not toxic (Citation30). The difference in aggregation for HttQ103 and HttQ103P is attributed to the polyproline-rich region of HttQ103P binding to microtubules, recruiting the HttQ103P aggregates into a large aggresome that colocalizes with the spindle body. Disruption of the microtubule machinery causes HttQ103P to form numerous smaller aggregates (Citation30).
In the present study, we examined the role(s) of Hsp104 in the aggregation of the HttpolyQ fragments. Although previous studies reported the requirement of prion with an amyloid conformation for Htt aggregation in yeast, the only clearly defined role of Hsp104 in Htt aggregation has been an indirect one: to propagate the yeast prion by severing the prion seeds. Formerly, there was no evidence that Hsp104 can sever the polyQ fragments, and in fact, several studies previously suggested that the HttpolyQ fragments are poor substrates for the severing activity of Hsp104. For example, the Weissman laboratory showed that a polyQ62-Sup35 chimera was not a substrate for Hsp104 unless the chimera included the Sup35 oligopeptide region, which spans residues 40 to 112 and is adjacent to the Q/N-rich tract (Citation25). Similarly, the Kushnirov laboratory found that only when hydrophobic residues, particularly tyrosine, are interspersed within the extended polyQ repeat region was the chimera efficiently severed by Hsp104 (Citation29).
In contrast, we now find that Hsp104 efficiently severs HttQ103 aggregates nucleated by prions with an amyloid conformation or spontaneously nucleated in cells without prion. Although, in contrast to HttQ103, Hsp104 poorly severs HttQ103P aggregates nucleated by prion, it forms a large aggregate in yeast with prion regardless of Hsp104 activity. Furthermore, unlike what occurs with HttQ103, there is no significant spontaneous aggregation of HttQ103P in the absence of prion. Thus, the primary function of Hsp104 in the aggregation of the HttQ103P fragment is to propagate the yeast prion. Therefore, the ability of Hsp104 to sever the spontaneously or prion-nucleated HttQ103 aggregates, but not the prion-nucleated HttQ103P aggregates, has a marked effect on the aggregation of these fragments.
RESULTS
Effect of Hsp104 on HttQ103 aggregates in yeast with prion.
To examine whether Hsp104 has a role other than prion propagation in aggregating HttpolyQ fragments, green fluorescent protein (GFP)-labeled HttQ103 under the control of the GAL1 promoter was integrated into the genome of the 779-6a Saccharomyces cerevisiae yeast strain to obtain uniform expression of HttQ103 throughout the yeast population. We first verified that, as shown previously in other yeast strains (Citation22, Citation23), the aggregation of HttQ103 is dependent on the presence of yeast prion in our engineered yeast strain containing the [RNQ+] [PSI+] prions. After growth overnight in galactose medium, confocal imaging showed that all cells had hundreds of small HttQ103 aggregates (). Most of these aggregates were below the diffraction limit of the microscope, indicating that their size was ≤0.15 μm. Consistent with previous studies, HttQ103 was diffuse with no detectable aggregates in >97% of the cells cured of the [RNQ+] [PSI+] prions with guanidine (). Cells with and without prion grew at the same rate in galactose medium, indicating no toxic effects from expressing the HttQ103 fragment in [RNQ+] yeast. In contrast, the Sherman laboratory found that HttQ103 was toxic when expressed from the GAL1 promoter on a 2μm plasmid (Citation22). This is likely due to the much higher expression levels of HttQ103 in their experiments.
FIG 1 Maximized z-stack confocal images of the HttQ103 and HttQ103P fragments expressed under different conditions. (a and A) HttQ103 and HttQ103P fragments in [RNQ+] [PSI+] yeast following growth overnight in galactose medium. (b and B) HttQ103 and HttQ103P fragments in yeast cured of prions by guanidine following growth overnight in galactose medium. (c and C) HttQ103 and HttQ103P fragments in [RNQ+] [PSI+] yeast expressed for 9 h (∼3 generations) in galactose medium. (d and D) HttQ103 and HttQ103P fragments in [RNQ+] [PSI+] yeast expressed for 9 h in galactose and guanidine medium. (e and E) HttQ103 and HttQ103P fragments in yeast grown for 6 h with galactose and guanidine, spun, and resuspended in galactose medium for 3 h. (f and F) HttQ103 and HttQ103P fragments in [RNQ+] [PSI+] yeast grown overnight at a relatively high density (starting OD of ∼0.4) in galactose medium with guanidine. The insets are an enlargement of the profile drawn through an aggregate. Images of HttQ103 and HttQ103P are shown in panels a to f and A to F, respectively.
![FIG 1 Maximized z-stack confocal images of the HttQ103 and HttQ103P fragments expressed under different conditions. (a and A) HttQ103 and HttQ103P fragments in [RNQ+] [PSI+] yeast following growth overnight in galactose medium. (b and B) HttQ103 and HttQ103P fragments in yeast cured of prions by guanidine following growth overnight in galactose medium. (c and C) HttQ103 and HttQ103P fragments in [RNQ+] [PSI+] yeast expressed for 9 h (∼3 generations) in galactose medium. (d and D) HttQ103 and HttQ103P fragments in [RNQ+] [PSI+] yeast expressed for 9 h in galactose and guanidine medium. (e and E) HttQ103 and HttQ103P fragments in yeast grown for 6 h with galactose and guanidine, spun, and resuspended in galactose medium for 3 h. (f and F) HttQ103 and HttQ103P fragments in [RNQ+] [PSI+] yeast grown overnight at a relatively high density (starting OD of ∼0.4) in galactose medium with guanidine. The insets are an enlargement of the profile drawn through an aggregate. Images of HttQ103 and HttQ103P are shown in panels a to f and A to F, respectively.](/cms/asset/fee7f03c-2e05-4c15-aa12-d1d114bb35df/tmcb_a_12277302_f0001.jpg)
Since the severing activity of Hsp104 is necessary to propagate prions in yeast, to examine whether Hsp104 had another function in the presence of prion, galactose and guanidine were added at the same time to [RNQ+] [PSI+] yeast. This induces HttQ103 expression while simultaneously inactivating Hsp104 activity. Although aggregates were detected both in the control and guanidine-treated cells after 9 h (∼3 generations) in galactose medium, there were many more aggregates per cell in the control than in cells incubated with guanidine. The results, quantified in , are clearly visible in the images in and of yeast grown in the presence of galactose and galactose plus guanidine, respectively. After 3 generations in galactose and guanidine, the prions have not yet cured; i.e., the yeast cells still have the [RNQ+] [PSI+] prions. As expected, this effect of guanidine on HttQ103 aggregation was reversible. When we grew cells for 6 h (∼2 generations) in guanidine, followed by washing out the guanidine and then growing the cells for another 3 h in galactose, the majority of cells had numerous aggregates (). These results show that the nucleation of HttQ103 by the [RNQ+] [PSI+] prions is independent of Hsp104 activity, but the number of aggregates per cell is dependent on Hsp104 activity.
TABLE 1 Effect of Hsp104 on the aggregation of HttpolyQ fragmentsTable Footnotea
We observe an even more pronounced effect of guanidine on HttQ103 aggregation after growth overnight. In contrast to the control cells, where the aggregates significantly proliferated overnight (), we noticed a notable reduction in the number of aggregates per cell with increasing incubation time in galactose and guanidine. As shown in , after overnight growth of cells at a relatively high density to minimize the number of cell divisions and the loss of prion seeds, most guanidine-treated cells had only a few HttQ103 aggregates along with a diffuse pool of HttQ103. Quantification of the aggregates in the guanidine-treated cells showed that 80% of the total cells (n = 150) had HttQ103 aggregates. However, unlike the control cells, 75% of the cells with aggregates had <5 aggregates per cell, and one-third of the guanidine-treated cells had a large aggregate of >500 nm (average size of 770 nm ± 164 nm [n = 52]). The large aggregates form from the coalescence of smaller aggregates, as shown by the multiple peaks in the profile line drawn through a large aggregate in the inset of . Therefore, HttQ103 still forms aggregates in cells with prion in the absence of active Hsp104, but there are marked reductions in the number of aggregates and the extent of aggregation. As expected, with continued growth in medium with guanidine, there is a reduction in the number of cells with HttQ103 aggregates until none of the cells have detectable aggregates.
Effect of Hsp104 on HttQ103P aggregates in yeast with prion.
Next, we examined whether Hsp104 affects the aggregation of HttQ103P, which has a polyproline region downstream of the polyglutamine region. Similar to HttQ103, previous reports suggest that HttQ103P aggregation in yeast is also dependent on the presence of a prion with an amyloid conformation (Citation23), making it indirectly dependent on Hsp104 to propagate the prion. As shown previously (Citation23, Citation30, Citation31), the expression of HttQ103P tends to form one large inclusion or aggresome, unlike the numerous aggregates of HttQ103. When we integrate this fragment on the GAL1 promoter into the genome of [RNQ+] [PSI+] 779-6a yeast (), the large HttQ103P aggregate is visible after growth overnight in galactose medium. Interestingly, in addition to a large aggregate, almost all the cells had smaller aggregates, and some cells had only small aggregates in a pool of diffuse HttQ103P. These cells, with small aggregates, are evidently the newly budded daughter cells since it takes time to form a large aggregate, and the preexisting large aggregates cannot pass through the bud neck when yeast cells divide. As expected, when cells were in late log phase, all cells contained large aggregates. In contrast, >98% of the cells did not have a detectable HttQ103P aggregate in yeast cured of prion (), consistent with previous results (Citation23, Citation30, Citation31).
As described above, to determine the role of Hsp104, the [RNQ+] [PSI+] yeast cells were grown in galactose medium in both the presence and absence of guanidine to determine whether Hsp104 severs the HttQ103P aggregates. After inducing HttQ103P expression for 9 h, there were small aggregates and a large diffuse pool of HttQ103P present in the yeast cells regardless of the presence of guanidine. However, the cells grown in guanidine had markedly fewer aggregates per cell (). In fact, unlike the control (), most of the guanidine-treated cells had only one or two aggregates per cell (). When we washed out the guanidine after 6 h and the cells were grown for another 3 h in galactose medium, there was an increase in the number of aggregates per cell (), indicating that the effect of inactivating Hsp104 is reversible. However, compared to the guanidine washout experiment with HttQ103, far fewer cells accumulated numerous aggregates of HttQ103P (). Therefore, the HttQ103 aggregates are a much better substrate for the severing activity of Hsp104 than the HttQ103P aggregates.
With continued growth in medium containing galactose with guanidine overnight using yeast in late log phase to inhibit cell division and, in turn, the loss of prion seeds, >90% of the cells had one large aggregate (). These large aggregates form from the coalescence of the smaller aggregates, as shown by the profile line drawn in the inset of , in agreement with previous results (Citation31). Since HttQ103P formed aggregates in the presence of [RNQ+] [PSI+] prions even when Hsp104 was inactivated by guanidine, this shows that only prion is necessary for aggregate formation.
Factors that affect the aggregation of the HttpolyQ fragments in yeast with prion.
Since [RNQ+] [PSI+] yeast was used in the above-described experiments, we next examined whether the [RNQ+] and [PSI+] prions are equally effective in nucleating these HttpolyQ fragments. The yeast cells with the integrated HttpolyQ constructs were cured of either the [RNQ+] or [PSI+] prion (as described in Materials and Methods) to examine the nucleation of the individual prions. First, we examined the aggregation of HttpolyQ in [RNQ+] yeast. As shown in and , after growth overnight in galactose medium, the aggregation of both the HttQ103 and HttQ103P fragments in [RNQ+] yeast was similar to that observed in [RNQ+] [PSI+] yeast. However, the aggregation of HttQ103 and HttQ103P fragments in yeast with only [PSI+] prion was very different from that in yeast with both [RNQ+] and [PSI+] prions. After growth overnight in galactose medium to induce the expression of the HttpolyQ fragments, 75% and 98% of the [PSI+] yeast cells had no HttQ103 and HttQ103P aggregates, respectively ( and ). Therefore, the [RNQ+] prion is much better than the [PSI+] prion in nucleating both the HttQ103 and the HttQ103P fragments. Since previous studies, including one from our laboratory, showed that the [PSI+] prion effectively nucleates both HttQ103 and HttQ103P when these Htt fragments are expressed at much higher levels from a 2μm plasmid (Citation32Citation–Citation34), this indicates a concentration dependence of the nucleation of the HttpolyQ fragments by the [PSI+] prion.
FIG 2 Effects of different prions and concentrations of Htt fragments on the aggregation of HttpolyQ fragments. (A to D) Htt fragments on the GAL1 promoter, which were integrated into the chromosome, were induced overnight with galactose medium. (A and B) Maximized z-stack projections of HttQ103 (A) and HttQ103P (B) in [RNQ+] yeast. (C and D) maximized z-stack projections of HttQ103 (C) and HttQ103P (D) in [PSI+] yeast. (E and F) HttQ103 (E) and HttQ103P (F) fragments were expressed in [RNQ+] yeast from the CUP1 promoter grown in copper-free medium.
![FIG 2 Effects of different prions and concentrations of Htt fragments on the aggregation of HttpolyQ fragments. (A to D) Htt fragments on the GAL1 promoter, which were integrated into the chromosome, were induced overnight with galactose medium. (A and B) Maximized z-stack projections of HttQ103 (A) and HttQ103P (B) in [RNQ+] yeast. (C and D) maximized z-stack projections of HttQ103 (C) and HttQ103P (D) in [PSI+] yeast. (E and F) HttQ103 (E) and HttQ103P (F) fragments were expressed in [RNQ+] yeast from the CUP1 promoter grown in copper-free medium.](/cms/asset/485d64f4-32af-4e45-83e0-13335cf70696/tmcb_a_12277302_f0002.jpg)
We next examined whether there was a similar dependence on the Htt concentration in yeast with the [RNQ+] prion when we expressed the Htt constructs at much lower levels. The HttpolyQ fragments were expressed from a centromeric plasmid with the copper-inducible CUP1 promoter to reduce the expression level of the Htt fragments below that of the GAL1 promoter. This promoter expresses at very low levels in the absence of copper since the CUP1 promoter is leaky (Citation35). When yeast cells were grown in the absence of copper, the expression level of the Htt fragments was only 10% of the level obtained with the integrated constructs on the GAL1 promoter based on quantifying the GFP intensity. After growing the [RNQ+] yeast cells in the absence of copper, >50% of the cells had no detectable HttQ103 aggregates (), and this percentage was even higher in cells expressing HttQ103P, where >90% had no detectable aggregates (). Therefore, we observe a marked reduction in the nucleation of HttQ103 and HttQ103P by [RNQ+] when we express these fragments at very low levels. Furthermore, this effect is greater for HttQ103P than for HttQ103, which indicates that the polyproline-rich region, which forms a helical structure (Citation13), inhibits prion nucleation of the HttpolyQ fragments just as it inhibits spontaneous nucleation. Apparently, the structure of the polyproline region inhibits nucleation, the accessibility of antibody binding to the polyQ amyloid core (Citation36), and severing by Hsp104.
Surprisingly, when HttQ103 was expressed in [RNQ+] yeast in the absence of copper, most cells with aggregates had only a few or even just one or two aggregates (). Based on the above-described results obtained with [PSI+] yeast (), we expected that once HttQ103 was nucleated by prion, the cells would all accumulate numerous aggregates due to the severing activity of Hsp104. However, the lack of numerous aggregates suggests that at low HttQ103 expression levels, there is slow growth of the HttQ103 fibers nucleated by [RNQ+]. This behavior, in turn, produces short fibers that are severed very slowly, and then, because cell division also dilutes the fibers, there are only a few fibers or aggregates per cell. Collectively, these results show that prion nucleation of both HttQ103 and HttQ103P depends on the concentration of these fragments and the specific prion present in the yeast.
Hsp104 and aggregation of HttpolyQ fragments in yeast cured of prions.
The fact that active Hsp104 proliferates the HttQ103 aggregates that were nucleated by the [RNQ+] [PSI+] prions, perhaps by severing them, raises the possibility that Hsp104 also proliferates the HttQ103 aggregates formed by spontaneous nucleation in the absence of prion. To test this possibility, we examined the aggregation of HttQ103 in cells cured of prion over an extended period. As shown in , after growth overnight in galactose medium, <3% of these cured cells had HttQ103 aggregates. However, with continued growth, the percentage of cells with aggregates and the number of aggregates per cell markedly increased ( to ). After 4 days in galactose medium, approximately one-half of the cells had numerous aggregates (). This percentage increased to >90% after 1 week in galactose medium. Similarly, when cured cells were directly resuspended from galactose plates and grown overnight in galactose, >90% of the cells had numerous aggregates (). The images of HttQ103 aggregates in yeast cured of prions are indistinguishable from those of the aggregates in yeast with the [RNQ+] [PSI+] prions. As expected, when we added guanidine to the cured cells with aggregates, the inactivation of Hsp104 caused the numerous aggregates to coalesce into a few large aggregates (). With continued growth in galactose and guanidine, there was a decrease in the percentage of cells as the cells divided until only an occasional cell was left with an aggregate. Together, these results suggest that Hsp104 severs the HttQ103 aggregates, whether nucleated spontaneously or by prion.
FIG 3 Maximized z-stack images of HttQ103 fragments expressed in yeast cured of prions by guanidine as a function of time in galactose medium. (A to D) Yeast cells were grown in galactose medium for 1 to 4 days. (E and F) A colony from a galactose plate was resuspended in galactose medium and grown overnight in galactose medium (E), followed by growing cells overnight in 5 mM guanidine (F).
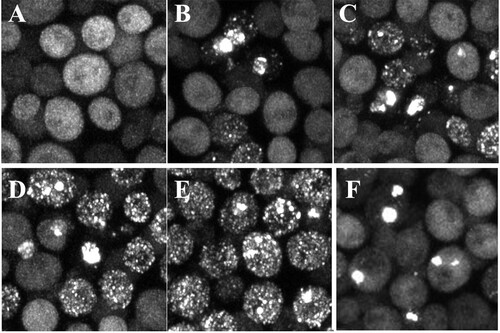
In contrast to HttQ103, cured yeast did not accumulate HttQ103P aggregates even after growth for a week in galactose medium. This phenomenon was also the case when yeast with the integrated HttQ103P construct was grown on galactose plates and resuspended overnight in galactose medium. Only ∼2% of the cured cells had HttQ103P aggregates, probably due to the polyproline region inhibiting the spontaneous nucleation with HttQ103 (Citation10Citation–Citation14). Therefore, these results show that prion is necessary for the nucleation of HttQ103P, and the only role of Hsp104 is to propagate the yeast prion.
Role of Hsp104 in severing HttQ103 aggregates in cured yeast.
To further validate that Hsp104 is severing the spontaneously nucleated HttQ103 aggregates, we examined the effect of different Hsp104 mutants on the proliferation of HttQ103 aggregates. Previous studies show that substrate threading through the Hsp104 channel is essential for several Hsp104 activities, particularly its ability to sever and thus proliferate the [PSI+] prion (Citation37, Citation38). Furthermore, Hsp104 retains severing activity when the Y662 residue located in the central Hsp104 channel is mutated with conserved but not nonconserved residues. This result predicts that if Hsp104 is indeed severing the HttQ103 aggregates, only Hsp104 with conserved channel mutants would be able to sever and, thus, proliferate the spontaneously nucleated HttQ103 aggregates.
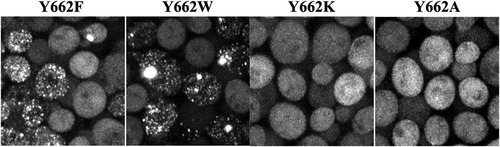
To test this prediction, we first crossed the yeast strain with the integrated HttQ103 construct with an hsp104 deletion strain to eliminate the expression of Hsp104 (see Materials and Methods). This deletion strain was then transformed with plasmids to express different Hsp104(Y662) channel mutants from the HSP104 promoter. After inducing HttQ103 expression for 4 days with galactose, imaging showed that cells expressing Hsp104 with the conserved point mutation Y662F or Y662W had numerous HttQ103 aggregates (). In contrast, there was no significant HttQ103 aggregation in yeast expressing Hsp104 with the nonconserved point mutation Y662K or Y622A (). These results confirm that just as Hsp104 severs prions having an amyloid conformation, it also severs spontaneously nucleated HttQ103 aggregates, which, in turn, markedly increases both the number of HttQ103 aggregates and their transmission to daughter cells when the yeast cells divide.
FIG 4 Maximized z-stack images of the effect of expressing different Hsp104(Y662) channel mutants on the aggregation of HttQ103. HttQ103 was expressed from the GAL1 promoter for 4 days in an hsp104 deletion strain transformed to express the different Hsp104(Y662) point mutants from the HSP104 promoter, as indicated.
Does Hsp104 sever the spontaneously nucleated aggregates as rapidly as it severs the aggregates nucleated by the [RNQ+] [PSI+] prions? Based on our imaging data, numerous aggregates accumulated over a period of 2 generations (∼6 h) after the induction of HttQ103 expression in [RNQ+] [PSI+] yeast with active Hsp104. This time period includes a generation before the levels of HttQ103 are sufficiently high to detect aggregates and then another generation for numerous aggregates to accumulate. To determine whether spontaneously nucleated aggregates are severed with a similar time course, we first transformed an hsp104 deletion strain with HttQ103 on the GPD promoter. In the absence of Hsp104 expression, >95% of the cells had no aggregates. In the occasional cell in which there was spontaneous nucleation, there was an aggregate in a pool of diffuse cytosolic HttQ103. When we imaged a microcolony over the course of 5 h, there was an increase in the number of cells but not aggregates ().
FIG 5 Time course of the formation of HttQ103 aggregates in the absence and presence of Hsp104. (A) Time series obtained in an hsp104 deletion strain constitutively expressing HttQ103. (B) Time series obtained in an hsp104 deletion strain constitutively expressing HttQ103 following the induction of Hsp104 expression from the GAL1 promoter. (Top) No detectable aggregate was present in the cell at the start of the experiment; (bottom) a large aggregate was present in the cell at the start of the experiment. The arrow in the cell at the 30-min time point in the bottom panel indicates a flare-like projection coming off the aggregate. (See Movies S1 and S2 in the supplemental material for images acquired at 10-min intervals.) In these experiments, the yeast cells were immobilized on an agarose pad to acquire z-stack images of the same field at various times. Images are all maximized z-stacks.
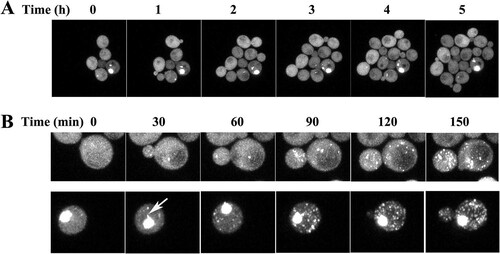
Consistent with the role of Hsp104 in proliferating aggregates, when the hsp104 deletion strain was transformed with a plasmid to express Hsp104, cells accumulated numerous HttQ103 aggregates. To determine the time course of this aggregation, cells constitutively expressing HttQ103 were transformed with a plasmid to express Hsp104 from the GAL1 promoter. Following the induction of Hsp104 expression, we imaged a cell with no detectable aggregates at 10-min intervals. Maximized z-stack images taken every 30 min (, top) show a marked proliferation of aggregates, followed by their transmission to the daughter cells in 2.5 h (see Movie S1 in the supplemental material). We also imaged a cell with one large aggregate following the induction of Hsp104 expression. As shown in (bottom), Hsp104 rapidly severed the HttQ103 aggregates in this cell just as it severed the aggregates in the cell that did not initially have a detectable aggregate (Movie S2). Comparing images from cells with and those without an initial aggregate, it appears as if there is a more rapid proliferation of aggregates in the former than in the latter cells. This difference may be due to aggregates being produced from Hsp104 severing the rim of the large aggregate since projections are emanating at the edge of the aggregate (, arrow). However, the large aggregate is still present after 10 h, suggesting that Hsp104 cannot completely disassemble it. These results show that the rate of severing of aggregates that were spontaneously nucleated is comparable to the rate of severing of aggregates nucleated by [RNQ+] [PSI+] prions. Furthermore, regardless of the presence of a large aggregate, once Hsp104 is induced, aggregate proliferation in a cell is relatively fast compared to the many days required for all cells to accumulate numerous aggregates. Therefore, the low rate of HttQ103 aggregation in cured cells is due to a low rate of spontaneous nucleation and not to a low rate of severing by Hsp104. Once a nucleus forms, it is rapidly severed by Hsp104 so that the cell quickly accumulates many HttQ103 aggregates.
Biochemical properties of spontaneously and prion-nucleated HttQ103 aggregates.
Even though the nucleation rates are markedly different between the prion-nucleated and spontaneously nucleated HttQ103 aggregates, fluorescence images show no differences in their structures. However, it is not clear whether these aggregates have the same biochemical properties, i.e., whether they both are typical polyglutamine-type aggregates that are heat stable and SDS resistant. Unlike the polyglutamine-type aggregates, yeast amyloid-based prion aggregates are readily solubilized when boiled in SDS. To determine if the spontaneously and prion-nucleated HttQ103 aggregates shared or differed in these characteristics, we used a standard filtration retention assay to retain the polyglutamine aggregates present in heat-denatured lysates on a 0.2-μm filter (Citation39, Citation40). This assay has been used to detect the polyglutamine aggregates in cell lysates, homogenates of tissues prepared from Caenorhabditis elegans and mice, and yeast expressing HttQ103 on a 2μm plasmid (Citation41Citation–Citation47). Surprisingly, regardless of whether the lysates were prepared from yeast in which the HttQ103 aggregates were prion nucleated or spontaneously nucleated, HttQ103 was not detected on the filter after denaturation of the lysates (). We observed retention of HttQ103 in a lysate from yeast expressing HttQ103 made from a 2μm plasmid, in agreement with previous studies (Citation42, Citation43). The 2μm plasmid produces more protein and larger aggregates than the chromosomally integrated HttQ103 construct, which we used to obtain uniform expression in this study. Since the confocal images showed that most of the HttQ103 aggregates were diffraction limited in size (≤0.15 μm) with the integrated construct, it is not clear whether their lack of retention is due to the small size of the aggregates or the solubilization of the aggregates after boiling in SDS buffer.
FIG 6 Dot blot and Western blot assays of HttQ103. (A) Dot blot of HttQ103 from yeast lysates that were either not treated with SDS buffer or boiled in 2% SDS. Lane 1, lysate prepared from [PRION+] yeast cells expressing HttQ103 from the GAL1 promoter, which was integrated into the chromosome; lane 2, lysate prepared from [prion−] yeast cells expressing HttQ103 from the GAL1 promoter integrated into the chromosome that were cultured on galactose plates and then grown on galactose medium; lane 3, lysate prepared from [PRION+] yeast cells expressing HttQ103 from the GAL1 promoter on a 2μm plasmid. The same concentration of total protein was added to each well. (B) Western blotting of HttQ103 from yeast lysates grown under different conditions. Lanes 1 and 3, total lysates that were boiled in SDS buffer; lanes 2 and 4, lysates treated with formic acid before boiling in SDS buffer. Lanes 1 and 2 are lysates prepared from [PRION+] yeast cells expressing HttQ103 from the GAL1 promoter integrated into the chromosome. Lanes 3 and 4 are lysates prepared from [prion−] yeast cells expressing HttQ103 from the GAL1 promoter integrated into the chromosome that were cultured on galactose plates and then grown on galactose medium. [PRION+] yeast contains both the [RNQ+] and [PSI+] prions. [prion−] yeast cells are cured of all prions.
![FIG 6 Dot blot and Western blot assays of HttQ103. (A) Dot blot of HttQ103 from yeast lysates that were either not treated with SDS buffer or boiled in 2% SDS. Lane 1, lysate prepared from [PRION+] yeast cells expressing HttQ103 from the GAL1 promoter, which was integrated into the chromosome; lane 2, lysate prepared from [prion−] yeast cells expressing HttQ103 from the GAL1 promoter integrated into the chromosome that were cultured on galactose plates and then grown on galactose medium; lane 3, lysate prepared from [PRION+] yeast cells expressing HttQ103 from the GAL1 promoter on a 2μm plasmid. The same concentration of total protein was added to each well. (B) Western blotting of HttQ103 from yeast lysates grown under different conditions. Lanes 1 and 3, total lysates that were boiled in SDS buffer; lanes 2 and 4, lysates treated with formic acid before boiling in SDS buffer. Lanes 1 and 2 are lysates prepared from [PRION+] yeast cells expressing HttQ103 from the GAL1 promoter integrated into the chromosome. Lanes 3 and 4 are lysates prepared from [prion−] yeast cells expressing HttQ103 from the GAL1 promoter integrated into the chromosome that were cultured on galactose plates and then grown on galactose medium. [PRION+] yeast contains both the [RNQ+] and [PSI+] prions. [prion−] yeast cells are cured of all prions.](/cms/asset/891d3c2d-4f4c-44cf-9467-06fd3bf2a148/tmcb_a_12277302_f0006.jpg)
To further characterize the biophysical properties of the HttQ103 aggregates in the lysates, we took advantage of the property that polyglutamine-type aggregates are not solubilized by boiling in SDS buffer but are solubilized by heating in formic acid (Citation48). Therefore, lysates were divided in half, with one fraction being directly boiled in SDS buffer, which solubilizes the heat-labile SDS-sensitive HttQ103 in the lysate, and the other fraction being centrifuged to sediment the HttQ103 aggregates, followed by resuspension of the pellet in formic acid, which solubilizes all the aggregated HttQ103 (see Materials and Methods). The differentially treated lysates were then run on SDS gels, followed by Western blotting. shows that treating the pellet with formic acid increased the intensity of the HttQ103 band 3-fold for both the prion-nucleated and spontaneously nucleated HttQ103 aggregates. These results show that at least two-thirds of the prion-nucleated and spontaneously nucleated HttQ103 aggregates are heat stable and SDS resistant, indicating they are both typical polyglutamine-type HttQ103 aggregates and are biochemically similar.
DISCUSSION
The molecular chaperone Hsp104, a member of the AAA ATPase family, has disaggregase activity functioning to dissolve protein aggregates and amyloids in yeast (Citation49Citation–Citation51). Although this chaperone is not present in higher eukaryotes, the purified protein has been shown to disaggregate many proteins that cause neurodegenerative diseases, including Tau, Aβ,α-synuclein, TDP43, and polyglutamine fragments (Citation52, Citation53). Based on this finding, it has been proposed that Hsp104 has therapeutic potential, which has been examined by expressing Hsp104 in both tissue culture cells and animals (Citation53Citation–Citation56). Regarding Huntington’s disease, Hsp104 has been shown to reduce aggregate formation and protect against cell death in tissue culture cells (Citation56). Furthermore, Hsp104 has been shown to inhibit aggregate formation and reduce toxicity in models of Huntington’s disease in C. elegans and mice (Citation54, Citation55, Citation57). Given the therapeutic interest in Hsp104, it is important to understand the effect of Hsp104 on HttpolyQ aggregation in yeast, an organism that endogenously expresses this chaperone.
Our results show that Hsp104 severs the HttQ103 aggregates regardless of whether they are spontaneously nucleated or prion nucleated. The severing of aggregates results in their proliferation and, in turn, enhances their transmission to the daughter cells. In this regard, the role of Hsp104 in proliferating HttQ103 aggregates is very similar to its role in propagating prion. However, unlike prion, in which the soluble properly folded conformation is stable, with HttQ103, the soluble form has slow spontaneous nucleation. Once the nuclei form, they are severed by Hsp104, enhancing their transmission to the daughter cells. Since Hsp104 proliferates the HttQ103 aggregates, this suggests that with some pathological polyglutamine constructs, the expression of Hsp104 may be detrimental instead of protective. In fact, this has been observed in a Drosophila model of the polyglutamine disease spinocerebellar ataxia type 3 (Citation58).
Although it has not been previously examined whether the HttQ103 fragment is a substrate for the severing activity of Hsp104, this question was indirectly examined by the Kushnirov laboratory using chimeras of Sup35 in which the prion domain of Sup35 was swapped with polyQ residues of various lengths (Citation29). The propagation and fragmentation of the chimeras were analyzed by determining their size on partially denaturing SDS gels. In cured yeast grown for an extended length of time, the Kushnirov laboratory observed fragmentation of a chimera containing extended polyQ repeat regions. However, based on the large size of the partially denatured fragments, they concluded that the polyQ-Sup35 chimera was a poor substrate for the severing activity of Hsp104 unless tyrosine residues were interspersed among the polyQ repeats, in which case the size of the fragments was reduced.
In agreement with previous studies, the aggregation properties of HttQ103P are very different from those of HttQ103 (Citation23, Citation30, Citation59). In yeast with prions, HttQ103P tends to form a large aggregate that comes from the coalescence of smaller aggregates, as suggested in a previous study (Citation31). In contrast to the latter study, we find that aggregate formation depends only on prion and not directly on Hsp104. Instead, HttQ103P aggregation is indirectly dependent on Hsp104 since Hsp104 is essential for prion propagation. Interestingly, in [RNQ+] [PSI+] yeast expressing HttQ103 with inactive Hsp104, HttQ103 forms a large aggregate, similar to that observed in cells expressing HttQ103P. This behavior suggests that the absence of severing leads to the formation of large HttpolyQ aggregates in yeast cells.
Surprisingly, our results show a marked difference in the abilities of [RNQ+] and [PSI+] prions to nucleate the HttpolyQ fragments. The expression of these constructs on the GAL1 promoter, when integrated into the chromosome, showed that the [RNQ+] but not the [PSI+] prion nucleated both HttQ103 and HttQ103P fragments despite the [RNQ+] prion having far fewer seeds than the [PSI+] prion in our yeast strain based on the rate of curing of these prions in guanidine (Citation60). This indicates that the prion conformation rather than the prion seed number is the primary factor involved in the nucleation of Htt fragments. Since prions exist as variants with different conformations, this suggests that the ability of a given prion to nucleate Htt fragments may also depend on the prion variant.
As expected, the nucleation of HttpolyQ fragments also depends on their level of expression. When these fragments were expressed from the CUP1 promoter in the absence of copper in [RNQ+] yeast, there was a marked reduction in the aggregation of both the HttQ103 and HttQ103P fragments. On the other hand, both HttQ103 and HttQ103P have been reported to form aggregates in the absence of prion when expressed at very high levels from the GAL1 promoter using a 2μm plasmid (Citation23, Citation61). Notably, it is unknown whether the Htt aggregates formed at low and high expression levels have the same structure. Recent cryo-electron microscopy (cryo-EM) studies showed that HttpolyQ fragments expressed at high levels in yeast form assorted structures, including amyloid fibers, amyloid aggregates, and amorphous structures (Citation44, Citation61). It has been shown that the disaggregase activity of Hsp104 works on both amorphous protein aggregates and amyloid fibrils (Citation52). However, since biochemical analysis showed that the HttQ103 aggregates were heat stable and SDS resistant, in contrast to amorphous aggregates (Citation31, Citation42, Citation43, Citation62, Citation63), this suggests that Hsp104 is severing aggregates with an amyloid structure.
Interestingly, the prion-nucleated and spontaneously nucleated HttQ103 aggregates in lysates prepared from yeast expressing the integrated HttQ103 construct were too small to be retained on a 0.2-μm filter, the standard assay for detecting polyglutamine aggregates. The small size of the aggregates is consistent with imaging that showed that the majority of HttQ103 aggregates were diffraction limited in size, indicating that they were ≤0.15 μm. Diffraction-limited aggregates have also been detected in tissue culture cells, but they are typically obscured by the large inclusion body (Citation64). HttpolyQ aggregates that were too small to be retained by the filter assay were in brain homogenates from a mouse Huntington’s disease model, but they were detected using a fluorescence resonance energy transfer (FRET)-based seeding assay (Citation47).
In conclusion, the paradigm that prion is essential for the aggregation of different Htt fragments with an expanded polyQ repeat region in yeast is an oversimplification. Our results show that when integrated HttQ103 is expressed from the GAL1 promoter, the extent to which this fragment forms aggregates is dependent on active Hsp104 to propagate the aggregates and not prion. Thus, the role of prion is to significantly accelerate the rate of nucleation, as suggested previously (Citation19). In contrast, the aggregation of the integrated HttQ103P fragment on the GAL1 promoter is dependent on prion, which in turn makes it only indirectly dependent on Hsp104. Thus, instead of yeast providing a simple model for HttpolyQ aggregation, many factors affect aggregation in yeast, including the primary sequence, specific prion, and expression level. Finally, considering the complexity of HttpolyQ aggregation, this may affect the therapeutic potential of Hsp104 in treating different polyglutamine diseases.
MATERIALS AND METHODS
Strains and plasmids.
The yeast strains were derived from the parental [RNQ+] [PSI+] 779-6a strain (MATa kar1-1 SUQ5 ade2-1 his3Δ202 leu2Δ1 trp1Δ63 ura3-520) (Citation65). For galactose-induced expression of HttQ103 (strain 1786) or HttQ103P (1876), the genes were cloned into pNC1124 (Citation66), and the SmaI fragment was used for integration into the LYS2 chromosomal locus of 779-6a. His+ transformants identified as lys− were confirmed for proper integration by PCR. The hsp104 deletion strain 1854-5D came from a backcross of 1786 with the hsp104 deletion strain J104NC (Citation65). After mating the strains, the yeast cells were sporulated, and the colonies were then analyzed to obtain the hsp104 deletion strain with the integrated HttQ103 construct. Yeast strains were cured of all prion by growing the cells in 5 mM guanidine. To cure yeast of the [PSI+] prion, yeast cells were cured by Hsp104 overexpression by expressing Hsp104 on a pRS316 plasmid. After plating cells on one-half-strength yeast extract-peptone-dextrose (1/2 YPD) plates, a cured colony was selected by its red color. To cure yeast of the [RNQ+] prion, cells were grown overnight in 5 mM guanidine, followed by plating on 1/2 YPD plates. Several of the colonies that plated white, indicating that they were [PSI+], were transformed with the pRS316-RNQ1-Rnq1-GFP plasmid. The transformed colonies were imaged, and a colony with diffuse GFP was selected. After having obtained colonies with the desired prion phenotype, cells were treated with 5-fluoroorotic acid to eliminate the pRS316 plasmids.
HttQ103 and HttQ103P were subcloned from the pYES2 vector (Citation22, Citation67) to be integrated into the yeast genome as described above. In addition, the HttQ103 plasmids were used for subcloning into a pRS316 centromeric plasmid with either a GPD promoter or a CUP1 promoter. All the HttpolyQ constructs had a FLAG epitope tag at the N terminus and GFP at the C terminus. pRS315 Hsp104 with the GAL1 promoter and the Hsp104(Y662) point mutants with the HSP104 promoter were described previously (Citation65, Citation68).
Standard yeast media and growth conditions were used (Citation69). YPAD contains 1% yeast extract, 2% peptone, 2% dextrose, and 400 mg/liter adenine. 1/2 YPD is similar but lacks adenine and contains 0.5% yeast extract. Synthetic defined (SD) glucose media contain 6.7% yeast nitrogen base (Difco), 2% glucose, and the required nutrients. SD plates with limiting adenine contain 8 mg/liter adenine. SGal is medium with galactose (Sunrise Science) in place of glucose. Cells were grown at 30°C unless indicated otherwise.
Analysis of HttQ103 in yeast lysates.
When cells in an actively growing culture reached an optical density at 600 nm (OD600) of 0.7 to 0.8, they were pelleted, washed in phosphate-buffered saline (PBS), and then suspended in an equal volume of lysis buffer (50 mM Tris-HCl [pH 7.4], 200 mM NaCl, 0.5 mM EDTA [pH 8.0], 0.5 mM dithiothreitol [DTT], and one tablet of a protease inhibitor cocktail [Roche, Indianapolis, IN]). Cells were broken with 0.5-mm glass beads in a bead beater, followed by low-speed centrifugation to remove the debris. The protein concentration was determined using the Bradford reagent to load identical amounts on either gels or cellulose filters for the filtration assay.
Biochemical analysis of lysates.
Lysates were either not treated or boiled in 2% SDS prior to being filtered through a cellulose acetate membrane (Whatman OE 66, with a 0.2-μm pore size) as described previously (Citation39, Citation40). SDS gels (4 to 12%) were run on samples that were boiled in Laemmli buffer. An aliquot of the sample was centrifuged, treated with formic acid at 37°C for 1 h followed by vacuum drying, and then resuspended in additional Laemmli buffer (Citation59). Samples were run on SDS-PAGE gels, followed by transferring the samples to a nitrocellulose membrane (Invitrogen, Carlsbad, CA). The following antibodies were used: anti-GFP polyclonal antibody (Abcam) and anti-Pgk1 monoclonal antibody (Molecular Probes, Carlsbad, CA).
Microscopy.
Images of the GFP fluorescence in live cells were obtained with the Zeiss LSM 880 Airyscan confocal microscope equipped with a 63× 1.4-numerical-aperture (NA) objective. z-stack confocal images were taken of all cells and then maximized. The yeast cells were routinely imaged in 8-well 25-mm2 chambered coverslips (Lab-Tek, Rochester, NY). Imaging overnight was performed by placing the yeast cells on an agarose pad slide. z-stack confocal images were obtained at 10-min intervals.
SUPPLEMENTAL FILE 1 Legends for Movies S1 and S2. Download TMCB_A_12277302_SM0001.pdf, MOV file, 0.7 MB.
Download PDF (94.8 KB)SUPPLEMENTAL FILE 2 Movie S1. Download TMCB_A_12277302_SM0002.mov, MOV file, 0.6 MB.
Download QuickTime Video (724.6 KB)SUPPLEMENTAL FILE 3 Movie S2. Download TMCB_A_12277302_SM0003.mov, PDF file, 0.1 MB.
Download QuickTime Video (627.4 KB)ACKNOWLEDGMENTS
We thank Xufeng Wu for microscopic assistance and the NHLBI microscopy core.
This work was supported by the Intramural Program of the NIH, The National Heart, Lung, and Blood Institute, and The National Institute of Diabetes and Digestive and Kidney Diseases.
SUPPLEMENTAL MATERIAL
Supplemental material is available online only.
REFERENCES
- de la Monte SM, Vonsattel JP, Richardson EP, Jr. 1988. Morphometric demonstration of atrophic changes in the cerebral cortex, white matter, and neostriatum in Huntington’s disease. J Neuropathol Exp Neurol 47:516–525. https://doi.org/10.1097/00005072-198809000-00003.
- Fennema-Notestine C, Archibald SL, Jacobson MW, Corey-Bloom J, Paulsen JS, Peavy GM, Gamst AC, Hamilton JM, Salmon DP, Jernigan TL. 2004. In vivo evidence of cerebellar atrophy and cerebral white matter loss in Huntington disease. Neurology 63:989–995. https://doi.org/10.1212/01.wnl.0000138434.68093.67.
- Jayaraman M, Kodali R, Sahoo B, Thakur AK, Mayasundari A, Mishra R, Peterson CB, Wetzel R. 2012. Slow amyloid nucleation via alpha-helix-rich oligomeric intermediates in short polyglutamine-containing huntingtin fragments. J Mol Biol 415:881–899. https://doi.org/10.1016/j.jmb.2011.12.010.
- Landrum E, Wetzel R. 2014. Biophysical underpinnings of the repeat length dependence of polyglutamine amyloid formation. J Biol Chem 289:10254–10260. https://doi.org/10.1074/jbc.C114.552943.
- Finkbeiner S. 2011. Huntington’s disease. Cold Spring Harb Perspect Biol 3:a007476. https://doi.org/10.1101/cshperspect.a007476.
- Arndt JR, Chaibva M, Legleiter J. 2015. The emerging role of the first 17 amino acids of huntingtin in Huntington’s disease. Biomol Concepts 6:33–46. https://doi.org/10.1515/bmc-2015-0001.
- Pandey NK, Isas JM, Rawat A, Lee RV, Langen J, Pandey P, Langen R. 2018. The 17-residue-long N terminus in huntingtin controls stepwise aggregation in solution and on membranes via different mechanisms. J Biol Chem 293:2597–2605. https://doi.org/10.1074/jbc.M117.813667.
- Crick SL, Ruff KM, Garai K, Frieden C, Pappu RV. 2013. Unmasking the roles of N- and C-terminal flanking sequences from exon 1 of huntingtin as modulators of polyglutamine aggregation. Proc Natl Acad Sci USA 110:20075–20080. https://doi.org/10.1073/pnas.1320626110.
- Sivanandam VN, Jayaraman M, Hoop CL, Kodali R, Wetzel R, van der Wel PC. 2011. The aggregation-enhancing huntingtin N-terminus is helical in amyloid fibrils. J Am Chem Soc 133:4558–4566. https://doi.org/10.1021/ja110715f.
- Khoshnan A, Ko J, Patterson PH. 2002. Effects of intracellular expression of anti-huntingtin antibodies of various specificities on mutant huntingtin aggregation and toxicity. Proc Natl Acad Sci USA 99:1002–1007. https://doi.org/10.1073/pnas.022631799.
- Chen M, Wolynes PG. 2017. Aggregation landscapes of huntingtin exon 1 protein fragments and the critical repeat length for the onset of Huntington’s disease. Proc Natl Acad Sci USA 114:4406–4411. https://doi.org/10.1073/pnas.1702237114.
- Bhattacharyya A, Thakur AK, Chellgren VM, Thiagarajan G, Williams AD, Chellgren BW, Creamer TP, Wetzel R. 2006. Oligoproline effects on polyglutamine conformation and aggregation. J Mol Biol 355:524–535. https://doi.org/10.1016/j.jmb.2005.10.053.
- Darnell G, Orgel JP, Pahl R, Meredith SC. 2007. Flanking polyproline sequences inhibit beta-sheet structure in polyglutamine segments by inducing PPII-like helix structure. J Mol Biol 374:688–704. https://doi.org/10.1016/j.jmb.2007.09.023.
- Slepko N, Bhattacharyya AM, Jackson GR, Steffan JS, Marsh JL, Thompson LM, Wetzel R. 2006. Normal-repeat-length polyglutamine peptides accelerate aggregation nucleation and cytotoxicity of expanded polyglutamine proteins. Proc Natl Acad Sci USA 103:14367–14372. https://doi.org/10.1073/pnas.0602348103.
- Rubinsztein DC. 2002. Lessons from animal models of Huntington’s disease. Trends Genet 18:202–209. https://doi.org/10.1016/S0168-9525(01)02625-7.
- Heintz N, Zoghbi HY. 2000. Insights from mouse models into the molecular basis of neurodegeneration. Annu Rev Physiol 62:779–802. https://doi.org/10.1146/annurev.physiol.62.1.779.
- Li SH, Li XJ. 2004. Huntingtin and its role in neuronal degeneration. Neuroscientist 10:467–475. https://doi.org/10.1177/1073858404266777.
- Labbadia J, Morimoto RI. 2013. Huntington’s disease: underlying molecular mechanisms and emerging concepts. Trends Biochem Sci 38:378–385. https://doi.org/10.1016/j.tibs.2013.05.003.
- Sherman MY, Muchowski PJ. 2003. Making yeast tremble: yeast models as tools to study neurodegenerative disorders. Neuromolecular Med 4:133–146. https://doi.org/10.1385/NMM:4:1-2:133.
- Khurana V, Lindquist S. 2010. Modelling neurodegeneration in Saccharomyces cerevisiae: why cook with baker’s yeast? Nat Rev Neurosci 11:436–449. https://doi.org/10.1038/nrn2809.
- Mason RP, Giorgini F. 2011. Modeling Huntington disease in yeast: perspectives and future directions. Prion 5:269–276. https://doi.org/10.4161/pri.18005.
- Meriin AB, Zhang X, He X, Newnam GP, Chernoff YO, Sherman MY. 2002. Huntingtin toxicity in yeast model depends on polyglutamine aggregation mediated by a prion-like protein Rnq1. J Cell Biol 157:997–1004. https://doi.org/10.1083/jcb.200112104.
- Duennwald ML, Jagadish S, Muchowski PJ, Lindquist S. 2006. Flanking sequences profoundly alter polyglutamine toxicity in yeast. Proc Natl Acad Sci USA 103:11045–11050. https://doi.org/10.1073/pnas.0604547103.
- Gokhale KC, Newnam GP, Sherman MY, Chernoff YO. 2005. Modulation of prion-dependent polyglutamine aggregation and toxicity by chaperone proteins in the yeast model. J Biol Chem 280:22809–22818. https://doi.org/10.1074/jbc.M500390200.
- Osherovich LZ, Cox BS, Tuite MF, Weissman JS. 2004. Dissection and design of yeast prions. PLoS Biol 2:E86. https://doi.org/10.1371/journal.pbio.0020086.
- Duennwald ML, Jagadish S, Giorgini F, Muchowski PJ, Lindquist S. 2006. A network of protein interactions determines polyglutamine toxicity. Proc Natl Acad Sci USA 103:11051–11056. https://doi.org/10.1073/pnas.0604548103.
- Derkatch IL, Uptain SM, Outeiro TF, Krishnan R, Lindquist SL, Liebman SW. 2004. Effects of Q/N-rich, polyQ, and non-polyQ amyloids on the de novo formation of the [PSI+] prion in yeast and aggregation of Sup35 in vitro. Proc Natl Acad Sci USA 101:12934–12939. https://doi.org/10.1073/pnas.0404968101.
- Urakov VN, Vishnevskaya AB, Alexandrov IM, Kushnirov VV, Smirnov VN, Ter-Avanesyan MD. 2010. Interdependence of amyloid formation in yeast: implications for polyglutamine disorders and biological functions. Prion 4:45–52. https://doi.org/10.4161/pri.4.1.11074.
- Alexandrov IM, Vishnevskaya AB, Ter-Avanesyan MD, Kushnirov VV. 2008. Appearance and propagation of polyglutamine-based amyloids in yeast: tyrosine residues enable polymer fragmentation. J Biol Chem 283:15185–15192. https://doi.org/10.1074/jbc.M802071200.
- Wang Y, Meriin AB, Zaarur N, Romanova NV, Chernoff YO, Costello CE, Sherman MY. 2009. Abnormal proteins can form aggresome in yeast: aggresome-targeting signals and components of the machinery. FASEB J 23:451–463. https://doi.org/10.1096/fj.08-117614.
- Aktar F, Burudpakdee C, Polanco M, Pei S, Swayne TC, Lipke PN, Emtage L. 2019. The huntingtin inclusion is a dynamic phase-separated compartment. Life Sci Alliance 2:e201900489. https://doi.org/10.26508/lsa.201900489.
- Zhao X, Park YN, Todor H, Moomau C, Masison D, Eisenberg E, Greene LE. 2012. Sequestration of Sup35 by aggregates of huntingtin fragments causes toxicity of [PSI+] yeast. J Biol Chem 287:23346–23355. https://doi.org/10.1074/jbc.M111.287748.
- Gong H, Romanova NV, Allen KD, Chandramowlishwaran P, Gokhale K, Newnam GP, Mieczkowski P, Sherman MY, Chernoff YO. 2012. Polyglutamine toxicity is controlled by prion composition and gene dosage in yeast. PLoS Genet 8:e1002634. https://doi.org/10.1371/journal.pgen.1002634.
- Kochneva-Pervukhova NV, Alexandrov AI, Ter-Avanesyan MD. 2012. Amyloid-mediated sequestration of essential proteins contributes to mutant huntingtin toxicity in yeast. PLoS One 7:e29832. https://doi.org/10.1371/journal.pone.0029832.
- Peng B, Williams TC, Henry M, Nielsen LK, Vickers CE. 2015. Controlling heterologous gene expression in yeast cell factories on different carbon substrates and across the diauxic shift: a comparison of yeast promoter activities. Microb Cell Fact 14:91. https://doi.org/10.1186/s12934-015-0278-5.
- Lin HK, Boatz JC, Krabbendam IE, Kodali R, Hou Z, Wetzel R, Dolga AM, Poirier MA, van der Wel PCA. 2017. Fibril polymorphism affects immobilized non-amyloid flanking domains of huntingtin exon1 rather than its polyglutamine core. Nat Commun 8:15462. https://doi.org/10.1038/ncomms15462.
- Lum R, Tkach JM, Vierling E, Glover JR. 2004. Evidence for an unfolding/threading mechanism for protein disaggregation by Saccharomyces cerevisiae Hsp104. J Biol Chem 279:29139–29146. https://doi.org/10.1074/jbc.M403777200.
- Hung GC, Masison DC. 2006. N-terminal domain of yeast Hsp104 chaperone is dispensable for thermotolerance and prion propagation but necessary for curing prions by Hsp104 overexpression. Genetics 173:611–620. https://doi.org/10.1534/genetics.106.056820.
- Wanker EE, Scherzinger E, Heiser V, Sittler A, Eickhoff H, Lehrach H. 1999. Membrane filter assay for detection of amyloid-like polyglutamine-containing protein aggregates. Methods Enzymol 309:375–386. https://doi.org/10.1016/s0076-6879(99)09026-6.
- Ast A, Schindler F, Buntru A, Schnoegl S, Wanker EE. 2018. A filter retardation assay facilitates the detection and quantification of heat-stable, amyloidogenic mutant huntingtin aggregates in complex biosamples. Methods Mol Biol 1780:31–40. https://doi.org/10.1007/978-1-4939-7825-0_3.
- Muchowski PJ, Schaffar G, Sittler A, Wanker EE, Hayer-Hartl MK, Hartl FU. 2000. Hsp70 and Hsp40 chaperones can inhibit self-assembly of polyglutamine proteins into amyloid-like fibrils. Proc Natl Acad Sci USA 97:7841–7846. https://doi.org/10.1073/pnas.140202897.
- Johnson BS, McCaffery JM, Lindquist S, Gitler AD. 2008. A yeast TDP-43 proteinopathy model: exploring the molecular determinants of TDP-43 aggregation and cellular toxicity. Proc Natl Acad Sci USA 105:6439–6444. https://doi.org/10.1073/pnas.0802082105.
- Kryndushkin D, Wickner RB, Shewmaker F. 2011. FUS/TLS forms cytoplasmic aggregates, inhibits cell growth and interacts with TDP-43 in a yeast model of amyotrophic lateral sclerosis. Protein Cell 2:223–236. https://doi.org/10.1007/s13238-011-1525-0.
- Bauerlein FJB, Saha I, Mishra A, Kalemanov M, Martinez-Sanchez A, Klein R, Dudanova I, Hipp MS, Hartl FU, Baumeister W, Fernandez-Busnadiego R. 2017. In situ architecture and cellular interactions of polyQ inclusions. Cell 171:179–187.e10. https://doi.org/10.1016/j.cell.2017.08.009.
- Sin O, Mata-Cabana A, Seinstra RI, Nollen EAA. 2018. Filter retardation assay for detecting and quantifying polyglutamine aggregates using Caenorhabditis elegans lysates. Bio Protoc 8:e3042. https://doi.org/10.21769/BioProtoc.3042.
- Waelter S, Boeddrich A, Lurz R, Scherzinger E, Lueder G, Lehrach H, Wanker EE. 2001. Accumulation of mutant huntingtin fragments in aggresome-like inclusion bodies as a result of insufficient protein degradation. Mol Biol Cell 12:1393–1407. https://doi.org/10.1091/mbc.12.5.1393.
- Ast A, Buntru A, Schindler F, Hasenkopf R, Schulz A, Brusendorf L, Klockmeier K, Grelle G, McMahon B, Niederlechner H, Jansen I, Diez L, Edel J, Boeddrich A, Franklin SA, Baldo B, Schnoegl S, Kunz S, Purfurst B, Gaertner A, Kampinga HH, Morton AJ, Petersen A, Kirstein J, Bates GP, Wanker EE. 2018. mHTT seeding activity: a marker of disease progression and neurotoxicity in models of Huntington’s disease. Mol Cell 71:675–688.e6. https://doi.org/10.1016/j.molcel.2018.07.032.
- Hazeki N, Tukamoto T, Goto J, Kanazawa I. 2000. Formic acid dissolves aggregates of an N-terminal huntingtin fragment containing an expanded polyglutamine tract: applying to quantification of protein components of the aggregates. Biochem Biophys Res Commun 277:386–393. https://doi.org/10.1006/bbrc.2000.3682.
- Glover JR, Lindquist S. 1998. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell 94:73–82. https://doi.org/10.1016/s0092-8674(00)81223-4.
- Gates SN, Yokom AL, Lin J, Jackrel ME, Rizo AN, Kendsersky NM, Buell CE, Sweeny EA, Mack KL, Chuang E, Torrente MP, Su M, Shorter J, Southworth DR. 2017. Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104. Science 357:273–279. https://doi.org/10.1126/science.aan1052.
- Parsell DA, Kowal AS, Singer MA, Lindquist S. 1994. Protein disaggregation mediated by heat-shock protein Hsp104. Nature 372:475–478. https://doi.org/10.1038/372475a0.
- DeSantis ME, Leung EH, Sweeny EA, Jackrel ME, Cushman-Nick M, Neuhaus-Follini A, Vashist S, Sochor MA, Knight MN, Shorter J. 2012. Operational plasticity enables hsp104 to disaggregate diverse amyloid and nonamyloid clients. Cell 151:778–793. https://doi.org/10.1016/j.cell.2012.09.038.
- Jackrel ME, DeSantis ME, Martinez BA, Castellano LM, Stewart RM, Caldwell KA, Caldwell GA, Shorter J. 2014. Potentiated Hsp104 variants antagonize diverse proteotoxic misfolding events. Cell 156:170–182. https://doi.org/10.1016/j.cell.2013.11.047.
- Vacher C, Garcia-Oroz L, Rubinsztein DC. 2005. Overexpression of yeast hsp104 reduces polyglutamine aggregation and prolongs survival of a transgenic mouse model of Huntington’s disease. Hum Mol Genet 14:3425–3433. https://doi.org/10.1093/hmg/ddi372.
- Perrin V, Regulier E, Abbas-Terki T, Hassig R, Brouillet E, Aebischer P, Luthi-Carter R, Deglon N. 2007. Neuroprotection by Hsp104 and Hsp27 in lentiviral-based rat models of Huntington’s disease. Mol Ther 15:903–911. https://doi.org/10.1038/mt.sj.6300141.
- Carmichael J, Chatellier J, Woolfson A, Milstein C, Fersht AR, Rubinsztein DC. 2000. Bacterial and yeast chaperones reduce both aggregate formation and cell death in mammalian cell models of Huntington’s disease. Proc Natl Acad Sci USA 97:9701–9705. https://doi.org/10.1073/pnas.170280697.
- Satyal SH, Schmidt E, Kitagawa K, Sondheimer N, Lindquist S, Kramer JM, Morimoto RI. 2000. Polyglutamine aggregates alter protein folding homeostasis in Caenorhabditis elegans. Proc Natl Acad Sci USA 97:5750–5755. https://doi.org/10.1073/pnas.100107297.
- Cushman-Nick M, Bonini NM, Shorter J. 2013. Hsp104 suppresses polyglutamine-induced degeneration post onset in a drosophila MJD/SCA3 model. PLoS Genet 9:e1003781. https://doi.org/10.1371/journal.pgen.1003781.
- Dehay B, Bertolotti A. 2006. Critical role of the proline-rich region in huntingtin for aggregation and cytotoxicity in yeast. J Biol Chem 281:35608–35615. https://doi.org/10.1074/jbc.M605558200.
- Cox B, Ness F, Tuite M. 2003. Analysis of the generation and segregation of propagons: entities that propagate the [PSI+] prion in yeast. Genetics 165:23–33. https://doi.org/10.1093/genetics/165.1.23.
- Peskett TR, Rau F, O’Driscoll J, Patani R, Lowe AR, Saibil HR. 2018. A liquid to solid phase transition underlying pathological huntingtin exon1 aggregation. Mol Cell 70:588–601.e6. https://doi.org/10.1016/j.molcel.2018.04.007.
- Capitini C, Conti S, Perni M, Guidi F, Cascella R, De Poli A, Penco A, Relini A, Cecchi C, Chiti F. 2014. TDP-43 inclusion bodies formed in bacteria are structurally amorphous, non-amyloid and inherently toxic to neuroblastoma cells. PLoS One 9:e86720. https://doi.org/10.1371/journal.pone.0086720.
- Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, Pozniakovski A, Poser I, Maghelli N, Royer LA, Weigert M, Myers EW, Grill S, Drechsel D, Hyman AA, Alberti S. 2015. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162:1066–1077. https://doi.org/10.1016/j.cell.2015.07.047.
- Sahl SJ, Weiss LE, Duim WC, Frydman J, Moerner WE. 2012. Cellular inclusion bodies of mutant huntingtin exon 1 obscure small fibrillar aggregate species. Sci Rep 2:895. https://doi.org/10.1038/srep00895.
- Jung G, Masison DC. 2001. Guanidine hydrochloride inhibits Hsp104 activity in vivo: a possible explanation for its effect in curing yeast prions. Curr Microbiol 43:7–10. https://doi.org/10.1007/s002840010251.
- Houser JR, Ford E, Chatterjea SM, Maleri S, Elston TC, Errede B. 2012. An improved short-lived fluorescent protein transcriptional reporter for Saccharomyces cerevisiae. Yeast 29:519–530. https://doi.org/10.1002/yea.2932.
- Meriin AB, Zhang X, Alexandrov IM, Salnikova AB, Ter-Avanesian MD, Chernoff YO, Sherman MY. 2007. Endocytosis machinery is involved in aggregation of proteins with expanded polyglutamine domains. FASEB J 21:1915–1925. https://doi.org/10.1096/fj.06-6878com.
- Zhao X, Rodriguez R, Silberman RE, Ahearn JM, Saidha S, Cummins KC, Eisenberg E, Greene LE. 2017. Heat shock protein 104 (Hsp104)-mediated curing of [PSI(+)] yeast prions depends on both [PSI(+)] conformation and the properties of the Hsp104 homologs. J Biol Chem 292:8630–8641. https://doi.org/10.1074/jbc.M116.770719.
- Sherman F. 2002. Getting started with yeast. Methods Enzymol 350:3–41. https://doi.org/10.1016/s0076-6879(02)50954-x.
