Abstract
Changes in nuclear Ca2+ homeostasis activate specific gene expression programs and are central to the acquisition and storage of information in the brain. DREAM (downstream regulatory element antagonist modulator), also known as calsenilin/KChIP-3 (K+ channel interacting protein 3), is a Ca2+-binding protein that binds DNA and represses transcription in a Ca2+-dependent manner. To study the function of DREAM in the brain, we used transgenic mice expressing a Ca2+-insensitive/CREB-independent dominant active mutant DREAM (daDREAM). Using genome-wide analysis, we show that DREAM regulates the expression of specific activity-dependent transcription factors in the hippocampus, including Npas4, Nr4a1, Mef2c, JunB, and c-Fos. Furthermore, DREAM regulates its own expression, establishing an autoinhibitory feedback loop to terminate activity-dependent transcription. Ablation of DREAM does not modify activity-dependent transcription because of gene compensation by the other KChIP family members. The expression of daDREAM in the forebrain resulted in a complex phenotype characterized by loss of recurrent inhibition and enhanced long-term potentiation (LTP) in the dentate gyrus and impaired learning and memory. Our results indicate that DREAM is a major master switch transcription factor that regulates the on/off status of specific activity-dependent gene expression programs that control synaptic plasticity, learning, and memory.
INTRODUCTION
A major challenge for neuroscience is to identify the regulatory molecules underpinning the storage of information in neurons. Activity-dependent gene expression underlies neuronal plasticity and adaptive responses to different environmental stimuli in the central nervous system (CNS) and is determinant in the formation and storage of memories. Diverse signaling pathways participate in these processes. Among them, changes in intracellular free calcium concentration are the most universal signal, and the final output, in terms of adapted gene expression, is given by a specific set of proteins that decode the calcium signal according to its frequency, subcellular location, and intensity (Citation1Citation–Citation3). A nuclear tool kit of Ca2+-dependent effectors modifies the activity or the properties of specific transcription factors to regulate gene expression in response to the Ca2+ signal (for reviews, see references Citation4 and Citation5). Despite extensive investigation, a detailed mechanistic description of Ca2+-dependent signaling in the expression of the late, transcription-dependent component of long-term potentiation (LTP) is far from been complete (reviewed in reference Citation6). Here, we examine the role of the Ca2+-dependent transcriptional repressor DREAM (downstream regulatory element antagonist modulator) in the control of activity-dependent transcription and the expression of LTP, as well as in learning and memory.
The DREAM/calsenilin/KChIP-3 gene belongs to a group of four genes (encoding K+ channel interacting proteins 1 to 4 [KChIP-1 to -4]) that regulate the membrane expression and gating of Kv4 potassium channels (reviewed in reference Citation4) and also encode structurally and functionally related calcium sensors able to repress transcription in a Ca2+-dependent manner (Citation7, Citation8). The transcriptional activity of DREAM is effected by its binding to DNA and by specific interactions with other nucleoproteins, including CREM and CREB (Citation9Citation–Citation11). The high-affinity binding of DREAM to DRE sequences in DNA is regulated by the level of nuclear Ca2+ and requires DREAM oligomerization (Citation7, Citation12, Citation13). Unbinding of DREAM from DRE results in transcriptional derepression of target genes, as shown for prodynorphin (Citation7, Citation14, Citation15). Mutation of the EF-hands in DREAM results in a Ca2+-insensitive repressor that also blocks CREB binding protein recruitment by phosphoCREB, impairing CRE-dependent transcription, since the DREAM-CREB interaction is also Ca2+ dependent (Citation9). The mutation of a leucine-charged residue-rich domain (LCD) motif at the N-terminal end of DREAM prevents the interaction with CREB and releases CREB-dependent transcription from basal repression by DREAM (Citation9). A combination of LCD and EF-hand mutations generates a calcium-insensitive double mutant, here called daDREAM (dominant active DREAM), that actively represses DREAM target genes, preventing DREAM-mediated derepression in the presence of Ca2+ and cyclic AMP (cAMP) stimulation. Of note, the double mutant daDREAM does not impair transcription after CREB phosphorylation. Consistent with the notion of DREAM binding to the DNA as homo- or heterooligomers with other KChIP proteins (Citation7, Citation13, Citation16), daDREAM acts as a cross-dominant active mutant in vivo (Citation17, Citation18). However, since trafficking and gating of Kv4 channels by KChIP proteins does not require KChIP oligomerization, daDREAM does not behave as a dominant mutant for Kv4 channel function as shown in transgenic spinal cord neurons (Citation15).
Interestingly, despite the potential role for a Ca2+-dependent repressor such as DREAM in the regulation of synaptic plasticity and learning and memory processes, DREAM knockout mice are not different from wild-type mice in paired pulse facilitation, resting membrane potential, or the input-output relation of field excitatory postsynaptic potentials (fEPSPs) (Citation19). Furthermore, DREAM knockout mice do not show an obvious phenotype in a place-learning version of the Morris water maze test (Citation14) and have only a slight increase in LTP in the dentate gyrus of the hippocampal formation (Citation19) and slight improvement in memory in a contextual fear test (Citation20). This is likely due to the functional redundancy among DREAM/KChIP proteins and their overlapping expression patterns (Citation8, Citation16). Supporting this idea, genetic ablation of KChIP-2 (Citation21) or KChIP-1 (Citation22) also resulted in mild phenotypes.
In the present study, we used transgenic mice expressing a Ca2+-insensitive/CREB-independent dominant active mutant DREAM (daDREAM) to reveal the functional involvement of DRE-mediated transcription in hippocampal function. We found that the expression of several activity-dependent transcription factors is affected in daDREAM mice and the expression of their downstream targets is modified in transgenic hippocampal neurons. Notably, downregulation of Npas4 in daDREAM mice resulted in a severe alteration of GABAergic transmission, leading to a reduction in recurrent inhibition and enhanced LTP in the dentate gyrus in vivo. Associated with these physiological abnormalities, DREAM transgenic mice showed significant impairments in learning and memory.
MATERIALS AND METHODS
Mice.
The generation of DREAM transgenic mice has been reported previously (Citation15, Citation17). Of the different transgenic lines generated, in this study we used transgenic line 26, which shows specific expression of the transgene in the telencephalon. The expression of the transgene in different brain areas was quantified by real-time quantitative PCR (qPCR) as described previously (Citation16). DREAM knockout mice have been reported previously (Citation14).
Microarray.
RNA from the whole hippocampus from wild-type and transgenic mice was prepared using TRIzol (Invitrogen) and the RNAeasy Minikit (Qiagen). The RNA was quantified and the quality was assessed with a 2100 Bioanalyzer (Agilent technologies). cDNA was synthesized from 4 μg of total RNA using one-cycle target labeling and control reagents (Affymetrix) to produce biotin-labeled cRNA. The cRNA preparation (15 μg) was fragmented at 94°C for 35 min into segments 35 to 200 bases in length. Labeled cRNAs were hybridized to Affymetrix chips (GeneChip mouse genome 430 2.0 array). Each sample was added to a hybridization solution containing 100 mM 2-(N-morpholino)ethanesulfonic acid, 1 M Na+, and 20 mM EDTA in the presence of 0.01% Tween 20 to a final cRNA concentration of 0.05 μg/ml. Hybridization was performed for 16 h at 45°C. Each microarray was washed and stained with streptavidin-phycoerythrin in a Fluidics station 450 (Affymetrix) and scanned at 1.56-μm resolution in a GeneChip Scanner 3000 7G system (Affymetrix).
Microarray data analysis.
Three biological replicates were independently hybridized for each transcriptomic comparison. The GeneChip intensities were background-corrected, normalized, and summarized by the robust multiarray average (RMA) method (Citation23) using the affy package (Citation24) from Bioconductor. For each comparison, the moderated t test was applied to identify differentially expressed genes as implemented in the limma package (Citation25) from Bioconductor. Raw P values were adjusted for multiple hypothesis testing using the false discovery rate (FDR) method (Citation26). Genes with an FDR of <0.1 and a fold change in expression of >1.6 or <−1.6 were included in the list of induced or repressed candidates, respectively. The FIESTA viewer was used to facilitate the application of these numerical filters and the selection of candidate genes in each comparison (http://bioinfogp.cnb.csic.es/tools/FIESTA; J. C. Oliveros). For the biological classification of candidate genes, gene ontology (GO) terms included in the generic GO slim set (http://www.geneontology.org/GO.slims.shtml; S. Mundodi and A. Ireland) were used to group candidate genes into a reduced number of biological categories. Terms from the Molecular function name space were taken into account. The GO terms associated with the genes were obtained from the original Affymetrix annotation files.
Primary neuronal culture.
Serum-free corticohippocampal neurons from mouse E14 wild-type, homozygous transgenic, or DREAM-deficient embryos were cultured as described previously (Citation27) in Neurobasal medium supplemented with B27, 2 mM Glutamax, and 100 μg/ml penicillin-streptomycin (Invitrogen). After 10 days in culture, fully differentiated neurons were stimulated with 60 mM KCl and harvested after 30 or 90 min for RNA or chromatin preparation, respectively. For knockdown of KChIP proteins, neuronal cultures were transduced 24 h after plating using lentiviral vectors encoding antisense KChIP-2 or green fluorescent protein (GFP) or empty vector prepared as described previously (Citation17). The virus concentration was estimated by measuring the amount of p24 protein (Perkin-Elmer). The cultures received 5 μg of p24/106 neurons, and the medium was changed 8 h after the addition of the virus. Neurons were harvested 7 days after infection. The typical infection efficiency was about 90% as assessed using viral delivery of GFP.
Real-time quantitative PCR.
RNA was isolated from whole tissues or cell suspensions using TRIzol (Invitrogene), treated with DNase (Ambion), and reverse transcribed using hexamer primer and Moloney murine leukemia virus reverse transcriptase. To confirm the absence of genomic DNA, each sample was processed in parallel without reverse transcriptase. Real-time qPCR for endogenous DREAM and daDREAM was performed as described previously (Citation16). Validation of up- or downregulated genes in the microarray was done with specific primers and TaqMan MGB probes (Applied Biosystems) or SYBR green-based technology (see Table S1 in the supplemental material) and with specific assays from Applied Biosystems (see Table S2 in the supplemental material). The results (triplicates) were normalized by parallel amplification of hypoxanthine phosphoribosyltransferase (HPRT) or β-actin as indicated (see Table S1).
Chromatin immunoprecipitation.
Chromatin immunoprecipitation was performed as described previously (Citation28). Briefly, nuclei from primary cultured neurons were collected and chromatin was sonicated to an average length of 500 to 1,000 bp. The sheared chromatin was precleared with blocked protein A/G-Sepharose beads (Pierce) and used for immunoprecipitation with 6 μg of affinity-purified polyclonal antibody Ab731 against amino acids (aa) 22 to 42 in the DREAM protein (Citation29). The immunoprecipitated DNA was subjected to semiquantitative PCR with specific primers (see Table S3 in the supplemental material). The PCR was performed within the linear range using 20 to 25 cycles of amplification and trace amounts of [32P]dCTP. Scanned autoradiograms were quantified using the QuantityOne software (Bio-Rad).
Western blot.
Whole-cell extracts from neuronal cell cultures were prepared by incubating cell pellets on ice for 45 min with occasional vortexing with lysis buffer (50 mM Tris HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.5% deoxycholate, 0.1% SDS) supplemented with protease inhibitor cocktail (Roche). The extracts were cleared by centrifugation at 14,000 × g for 20 min, and the protein content quantified with the DC protein assay (Bio-Rad). The affinity-purified rabbit polyclonal antibodies (Ab) used were the DREAM-specific Ab730 (Citation29) or the pan-DREAM/KChIP Ab1014 (Citation16). Other antibodies used were for PSD95 (7E3-1B8; Affinity BioReagents), GABAA receptor β3 (ab 4046; Abcam), and β-actin (AC-15; Sigma). Proteins were visualized with horseradish peroxidase (HRP)-conjugated secondary antibody (Jackson) followed by enhanced chemiluminescence (ECL) (SuperSignal West Femto; Thermo Scientific). Blots were quantified using Quantity One software (Bio-Rad).
Seizures.
Wild-type and transgenic adult mice were injected intraperitoneally with pentylenetetrazol (45 mg/kg of body weight) or with PBS. The behavioral score according to a modified Racine's scale (0 to 6, where 0 is no response and 6 is generalized tonic-clonic convulsions) (Citation30) was recorded every 5 min during the 30 min following drug administration. The median score for each animal was calculated, and the data were analyzed using the Mann-Whitney U test for differences between groups.
Electrophysiology in vivo.
Adult male mice homozygous for the transgene and wild-type littermates aged 3 to 6 months were anesthetized with urethane (1.8 g/kg intraperitoneally [i.p.]). Experiments were performed under the Animals (Scientific Procedures) Act (1986), United Kingdom. Animals were placed in a head holder, and a glass micropipette was positioned in the ipsilateral hilus of the dentate gyrus to record the field responses evoked by stimuli delivered through a concentric bipolar stimulating electrode placed in the ipsilateral perforant path. In order to assess changes in feedforward and feedback inhibition, pairs of stimuli were delivered at interpulse intervals varying from 10 to 500 ms at an intensity sufficient to evoke a population spike of ∼1 mV. For each animal, the average of three responses was obtained at each of nine intervals between 10 and 50 ms. For LTP experiments, single test stimuli were delivered at a frequency of 0.033 Hz, intensity of 70 to 300 μA, and pulse width of 60 μs. The pulse width was doubled during the tetanus (six series of six trains of six pulses at 400 Hz, with 200 ms between trains and 20 s between series). The slope of the field excitatory postsynaptic potentials (fEPSPs) was normalized with respect to the mean response in the 10 min before the tetanus. The data are presented as the means ± standard errors of the means (SEM). For comparisons of the magnitude of LTP between groups, the normalized values of the fEPSPs 50 to 60 min after the tetanus were averaged for each animal and the groups were compared using Student's unpaired two-tailed t test.
Electrophysiology in vitro.
Transgenic and wild-type littermates were anesthetized by intraperitoneal injection of a mixture of medetomidine (1 mg/kg) and ketamine (76 mg/kg) before being killed by cervical dislocation. The brains were rapidly removed and chilled (<3°C). Parasagital slices were cut from the dorsal hippocampus using a Vibroslice (Campden Instruments, Loughborough, United Kingdom). A detailed description of the protocols used for in vitro electrophysiology can be found in the supplemental material.
Behavioral analysis.
Behavioral experiments were performed with adult male mice homozygous for the transgene and wild-type littermates. Mice were initially housed five per cage in a temperature (21 ± 1°C [mean ± standard deviation]) and humidity (65% ± 10%) controlled room with a 12-h/12-h light/dark cycle (lights on from 0800 to 2000 h) with ad libitum food and water. The experiments took place during the light phase. Behavioral tests and animal care were conducted in accordance with the standard ethical guidelines (European Communities Directive 86/609 EEC and National Institutes of Health 1995) and approved by the local ethical committee (CEEA-PRBB). All behavior experiments were carried out under blinded conditions. Detailed descriptions of the protocols used for the Morris water maze, open field, active avoidance, object recognition task, and fear conditioning experiments can be found in the supplemental material.
Microarray accession number.
Microarray data are available under Gene Expression Omnibus accession number GSE17844.
RESULTS
Genome-wide analysis in daDREAM hippocampus.
daDREAM has been shown to function as a dominant active mutant for the transcriptional repressor function of endogenous DREAM/KChIP proteins (Citation16, Citation17, Citation29). We used transgenic mice expressing daDREAM in telencephalic areas of the brain (), where endogenous DREAM is expressed (Citation31), to search for DREAM transcriptional targets that could reveal a potential role for DREAM in hippocampal synaptic plasticity, learning, and memory. Comparison of basal gene expression in the wild-type and transgenic adult hippocampus, using cDNA microarrays, identified more than 250 genes whose expression was altered in daDREAM transgenic mice (Gene Expression Omnibus accession number GSE17844; see also Fig. S1 in the supplemental material). Up- and downregulated genes were categorized according to GO terms and are presented in Tables S4 to S6 in the supplemental material.
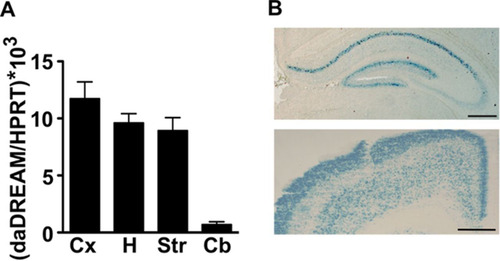
FIG 1 Expression of daDREAM in transgenic brain. (A) Comparative real-time qPCR analysis of the expression levels of daDREAM in cerebral cortex (Cx), hippocampus (H), striatum (St), and cerebellum (Cb) from transgenic mice. Values are normalized with respect to HPRT mRNA content. Results are the means ± SEM from 8 to 12 mice in two independent experiments. (B) Coronal brain sections from daDREAM mice showing the distribution of β-galactosidase activity in hippocampus (top) and cerebral cortex (bottom). Bars represent 250 μm.
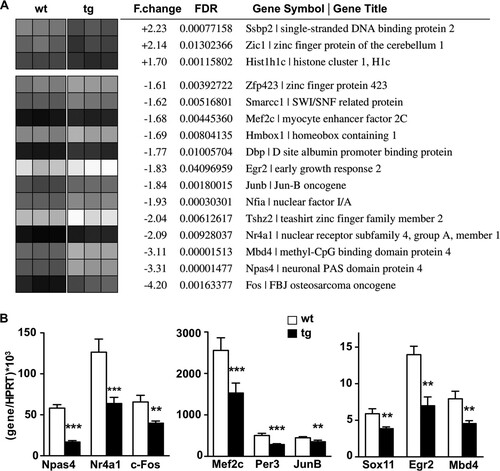
Among the genes with modified expression, we focused our attention on those predicted to encode transcription factors that could initiate gene expression programs governing changes in synaptic plasticity, learning, and memory. We found that several genes encoding early-induced, activity-dependent transcription factors, including Npas4, Nr4a1, Mef2C, JunB, and c-Fos, were downregulated in the transgenic hippocampus (). For these targets, the microarray results were confirmed in the transgenic hippocampus by real-time qPCR (). Importantly, the expression of other factors known to mediate activity-dependent changes in gene expression, such as SRF, Atf3, and c-Jun (Citation32Citation–Citation34), was not modified in the transgenic hippocampus, supporting the specificity of the changes induced by daDREAM. Taken together, these results implicate Ca2+-dependent, DREAM-mediated derepression of specific immediate early gene transcription in the function of hippocampal neurons.
FIG 2 Genome-wide analysis in daDREAM and wild-type hippocampus. (A) Functional clustering (molecular function, gene ontology) of up- and downregulated genes encoding transcription factors or DNA binding proteins. Biological triplicates of wild-type (wt) and transgenic (tg) mice are presented as a heat map. F.change, differential expression based on the ratio between averaged intensities (linear scale); FDR, adjusted P values; Gene Symbol | Gene Title, probe annotations as provided by Affymetrix. (B) Real-time qPCR analysis of indicated transcripts in the hippocampus from wild-type and transgenic mice. Values are normalized with respect to HPRT mRNA content. Results are the means ± SEM from 8 to 12 mice in two independent experiments. **, P < 0.01, and ***, P < 0.001, relative to wt (two-tailed, unpaired t test).
DREAM directly downregulates transcription from the Npas4 promoter.
Among target proteins with a function in the nucleus, we focused our attention on Npas4, a basic helix-loop-helix–PAS transcriptional activator (Citation35). Contrary to the products of other immediate early genes, such as c-Fos, JunB, Egr2, or Nr4a1, Npas4 is selectively induced by rises in nuclear Ca2+, binds to numerous enhancers in an activity-dependent manner (Citation37), and has been shown to transactivate the brain-derived neuotrophic factor (BDNF) gene and to control the formation of GABAergic inhibitory synapses (Citation38). A reduced basal level of Npas4 mRNA was found in the transgenic hippocampus () but also in other areas with transgene expression, such as the cerebral cortex and striatum, and was not modified in the cerebellum, where daDREAM is not expressed (see Fig. S2 in the supplemental material).
Sequence analysis of the Npas4 proximal promoter identified several DRE sites for DREAM binding (see Fig. S3A in the supplemental material). To determine whether Npas4 is a direct target for DREAM repression, we used chromatin immunoprecipitation with DNA isolated from primary cultured corticohippocampal neurons, a system in which DREAM and other KChIP proteins are expressed (). The basal expression of Npas4 was reduced in primary daDREAM neuronal cultures compared to its basal expression in wild-type cultures (), confirming the results obtained in adult tissues. Importantly, membrane depolarization by stimulation with 60 mM K+ resulted in strong induction of Npas4 expression in wild-type neurons, while only a slight induction was obtained in transgenic neurons (). Chromatin immunoprecipitation with a DREAM-specific antibody showed that under basal conditions, the binding of DREAM to the Npas4 promoter was stronger in transgenic cultures, while potassium depolarization resulted in the Ca2+-dependent unbinding of DREAM from the Npas4 promoter in wild-type neurons but had no effect in transgenic cultures (). As negative controls for these assays, omission of the DREAM antibody or chromatin immunoprecipitation from cultured DREAM-deficient neurons resulted in failure to amplify the Npas4 promoter (). These results indicate that DREAM directly binds to and controls the expression of the Npas4 gene in neurons. As a positive control for these experiments, we analyzed the expression of c-Fos, a bona fide target for DREAM (Citation7). The basal and activity-dependent expression levels of c-Fos were reduced in cultured daDREAM neurons compared to the results for wild-type neurons (), and chromatin immunoprecipitation confirmed the direct binding of DREAM to the c-Fos promoter (; see also Fig. S3B in the supplemental material).
FIG 3 DREAM directly regulates Npas4-mediated transcription. (A) Western blot analysis of cortex-hippocampal neuronal cultures from wild-type (wt), transgenic daDREAM (tg), and DREAM knockout (ko) embryos. A DREAM-specific immunoreactive band (empty arrowhead), detected by the peptide-specific antibody Ab730 against DREAM, was increased in transgenic daDREAM neurons and absent in knockout cultures. A nonspecific band is marked with an asterisk. Use of the pan-KChIP antibody Ab1014 on the same samples showed different immunoreactive bands (full arrowheads) corresponding to the different KChIP isoforms expressed in these cultured neurons. Quantification of the specific DREAM and the total KChIP immunoreactivities as relative ratios versus loading control β-actin is shown at the bottom. (B) Real-time qPCR analysis of Npas4 in corticohippocampal primary cultured neurons from wild-type (wt) and transgenic (tg) embryos under basal conditions and 30 min after stimulation with 60 mM K+. Values are normalized with respect to HPRT mRNA content. Results are the means ± SEM from three separate cultures in triplicates. ****, P < 0.0001 (one-way analysis of variance [ANOVA], Tukey's multiple comparison). (C) Chromatin immunoprecipitation assay of the Npas4 promoter using chromatin isolated from primary cultured neurons from wild-type (wt) and transgenic (tg) embryos before and after potassium depolarization. Cultured neurons from DREAM knockout (ko) embryos and exclusion of the antibody (-Ab) in the immunoprecipitation of wild-type chromatin were included as negative controls. Autoradiogram of the semiquantitative PCR is shown. After densitometric quantification, the results shown are the means ± SEM from four experiments. Input is 1%. ****, P < 0.0001 versus wt nonstimulated (n = 8, two-tailed, unpaired t test). (D and E) Real-time qPCR analysis of c-Fos mRNA and chromatin immunoprecipitation assay of the c-Fos promoter, respectively, as described for panels B and C. For qPCR, ****, P < 0.0001 (one-way ANOVA, Tukey's multiple comparison). For chromatin immunoprecipitation, *, P = 0.0380 versus wt nonstimulated (n = 5, two-tailed, unpaired t test).
![FIG 3 DREAM directly regulates Npas4-mediated transcription. (A) Western blot analysis of cortex-hippocampal neuronal cultures from wild-type (wt), transgenic daDREAM (tg), and DREAM knockout (ko) embryos. A DREAM-specific immunoreactive band (empty arrowhead), detected by the peptide-specific antibody Ab730 against DREAM, was increased in transgenic daDREAM neurons and absent in knockout cultures. A nonspecific band is marked with an asterisk. Use of the pan-KChIP antibody Ab1014 on the same samples showed different immunoreactive bands (full arrowheads) corresponding to the different KChIP isoforms expressed in these cultured neurons. Quantification of the specific DREAM and the total KChIP immunoreactivities as relative ratios versus loading control β-actin is shown at the bottom. (B) Real-time qPCR analysis of Npas4 in corticohippocampal primary cultured neurons from wild-type (wt) and transgenic (tg) embryos under basal conditions and 30 min after stimulation with 60 mM K+. Values are normalized with respect to HPRT mRNA content. Results are the means ± SEM from three separate cultures in triplicates. ****, P < 0.0001 (one-way analysis of variance [ANOVA], Tukey's multiple comparison). (C) Chromatin immunoprecipitation assay of the Npas4 promoter using chromatin isolated from primary cultured neurons from wild-type (wt) and transgenic (tg) embryos before and after potassium depolarization. Cultured neurons from DREAM knockout (ko) embryos and exclusion of the antibody (-Ab) in the immunoprecipitation of wild-type chromatin were included as negative controls. Autoradiogram of the semiquantitative PCR is shown. After densitometric quantification, the results shown are the means ± SEM from four experiments. Input is 1%. ****, P < 0.0001 versus wt nonstimulated (n = 8, two-tailed, unpaired t test). (D and E) Real-time qPCR analysis of c-Fos mRNA and chromatin immunoprecipitation assay of the c-Fos promoter, respectively, as described for panels B and C. For qPCR, ****, P < 0.0001 (one-way ANOVA, Tukey's multiple comparison). For chromatin immunoprecipitation, *, P = 0.0380 versus wt nonstimulated (n = 5, two-tailed, unpaired t test).](/cms/asset/82285502-8f3d-4a6a-93e5-d0e6f631ea09/tmcb_a_12275120_f0003_oc.jpg)
GABAergic transmission is affected in daDREAM mice.
Npas4 regulates the establishment of GABAergic synapses (Citation38). Thus, to assess whether Npas4 repression in the daDREAM hippocampus has functional consequences, we measured by real-time qPCR the expression of several postsynaptic GABAergic markers. The expression of the α1, α2, β2, and β3 GABA receptor subunits was markedly reduced in the hippocampus of daDREAM mice (). Of note, downregulation of the α2 subunit was already noticeable in the results of the genome-wide analysis (see Tables S4 to S6 in the supplemental material). The downregulation of GABA receptor subunits in the daDREAM hippocampus was specific and did not affect the expression of presynaptic markers of inhibitory synapses, such as GABA transporters (GAT1 and VIAAT), glutamic acid decarboxylase (GAD), or calbindin, which did not show any difference between genotypes (). Immunostaining for parvalbumin or GAT1 did not show cellular deficits in hippocampal GABAergic interneurons in transgenic mice (see Fig. S4 in the supplemental material). Furthermore, the levels of PSD95 protein (), a marker of excitatory synapses, were also not affected. For comparison, the reduced level of β3 GABA receptor protein in the transgenic hippocampus is shown ().
FIG 4 GABAergic inhibitory transmission is reduced in daDREAM hippocampus. (A and B) Real-time qPCR analysis of indicated GABAR subunits (A) and GABAergic markers (B) in wild-type (wt) and transgenic (tg) hippocampus. Results are the means ± SEM from 8 to 12 mice in two independent experiments. *, P < 0.05, and ***, P < 0.001 (one-way ANOVA, Tukey's multiple comparison). (C) Western blot analysis of PSD95 and β3 GABA receptor proteins in wild-type and transgenic hippocampus. β-Actin content was used as loading control. Representative autoradiograms are shown. Densitometric quantification of the specific immunoreactive band versus loading control β-actin, from a total of 4 (PSD95) or 8 (GABARβ3) samples from each genotype, is shown. **, P = 0.0039 (two-tailed, unpaired t test). (D) Seizure severity after convulsant administration (PTZ, 45 mg/kg) in wild-type (wt, n = 7) and transgenic mice (tg, n = 8). **, P = 0.0083 (Mann-Whitney test).
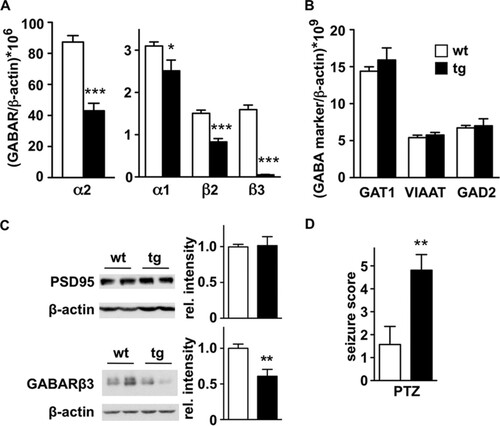
The impairment of GABA receptor signaling associated with specific mutations in GABAA receptor subunits is linked to epilepsy in humans, and genetic ablation of specific subunits reproduces a phenotype prone to spontaneous or pharmacologically induced seizures (Citation39). To investigate whether reduced mRNA levels of specific GABAA receptor subunits had a functional correlate, we analyzed the susceptibility to convulsants of daDREAM mice. The administration of subconvulsive doses of pentylenetetrazol or kainate induced a significantly increased response in transgenic mice compared to the response in the wild type (). These results indicate that the reduction in specific GABAA receptor subunits in daDREAM mice has a functional correlate at the receptor protein level that, in turn, may anticipate changes in the electrophysiological properties of transgenic neurons.
Paired pulse inhibition is abolished and LTP is enhanced in daDREAM mice.
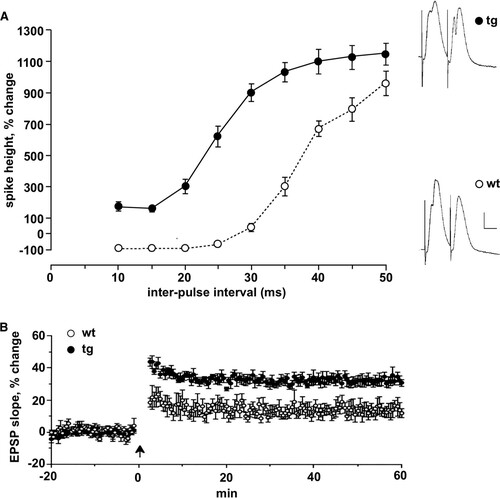
To investigate changes in synaptic plasticity in daDREAM mice, we analyzed paired-pulse inhibition and long-term potentiation (LTP) in vivo. The simultaneous discharge of granule cells evoked by a strong stimulus to perforant path fibers sets up a powerful recurrent inhibition mediated by inhibitory interneurons, particularly basket cells (Citation40). As a result, the amplitude of the population spike elicited by the second of a pair of strong stimuli is greatly reduced when the interstimulus interval is less than ∼30 ms. Indeed, in wild-type mice, the population spike in response to the second stimulus was abolished at intervals of less than 20 ms (). Conversely, in daDREAM mice, due to the reduced GABAergic synaptic strength, the feedback inhibition was completely absent and, even at the shortest intervals, the population spike to the second pulse was enhanced rather than inhibited (). At longer intervals, when paired pulse depression gives way to potentiation and a massive enhancement of the population spike, no significant differences were observed between wild type and daDREAM mice (). The impairment of paired pulse inhibition was replicated in vitro, where whole-cell recording confirmed the substantial weakening of inhibition (see Fig. S5A and B in the supplemental material). The inhibitory postsynaptic currents (IPSCs) recorded from wild-type neurons with a holding potential of 0 mV were evoked by stimuli as weak as 1 mA and grew progressively with increasing stimulus strength. IPSCs were also evoked in transgenic cells but were substantially and significantly smaller for all but the largest stimuli (see Fig. S5B). To localize the source of the weakening of inhibition, we measured the frequency and amplitude of spontaneous IPSCs at a holding potential of 0 mV (see Fig. S5C). The mean amplitude of the spontaneous IPSCs did not differ between genotypes, nor did the time constants and half width, but the frequency of the spontaneous IPSCs was significantly lower in transgenic mice, about half the frequency in the wild-type cells (see Fig. S5C). To test the strength of excitatory input to granule cells, plots of the mean slope of the EPSP as a function of stimulus strength were collected for wild-type and transgenic mice. No difference was seen between the two genotypes, suggesting that the excitatory drive was not grossly affected in transgenic mice (see Fig. S5D). Spontaneous EPSCs were not statistically significant different, but there was a tendency to increased frequency and amplitude (see Fig. S5E). The magnitude of early LTP, measured 50 to 60 min after induction, was greater in transgenic mice than in their wild-type littermates (), presumably as a result of impaired recurrent inhibition at the time of induction, leading to greater depolarization and enhanced activation of N-methyl-d-aspartate (NMDA) receptors. Importantly, the modified synaptic plasticity in daDREAM mice was not related to changes in A-type currents, which were identical in transgenic and wild-type hippocampal neurons (see Fig. S6 in the supplemental material). Similar results were previously shown in spinal cord neurons (Citation15).
FIG 5 Loss of paired-pulse inhibition and enhanced LTP in DREAM mutant mice. (A) For paired-pulse studies, the amplitude of the population spike evoked by the second stimulus, normalized to the amplitude of the population spike evoked by the first stimulus, is plotted as a function of the interstimulus interval for transgenic (tg, n = 6) and corresponding wild-type littermates (wt, n = 11). Note the complete suppression of the second population spike at short intervals in wild-type mice and its absence at all intervals in transgenic mice. Stimulus intensity was set so that the first stimulus of the pair evoked a population spike of ∼1 mV. Representative responses to paired stimuli for each group are displayed on the right. Calibration, 3 mV, 4 ms. (B) LTP was measured at perforant path-granule cell synapses in anesthetized transgenic and wild-type mice. LTP, measured 50 to 60 min after the tetanus (arrow), was significantly enhanced. The magnitude of LTP was 32.3% ± 2.6% for transgenic and 13.7% ± 4.0% for wild-type mice (P < 0.005).
DREAM directly regulates BDNF expression.
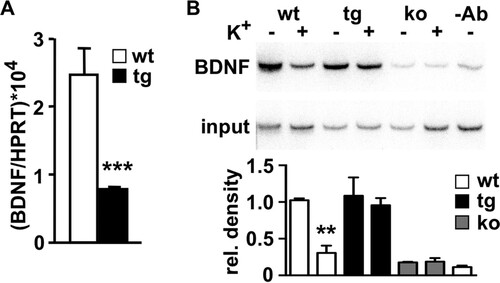
The activity-dependent expression of the mouse BDNF gene is induced by several mechanisms. These include transactivators, such as CREB, Npas4, and CaRF, which bind to promoter regions located immediately before exons II and IV (see Fig. S3C in the supplemental material), and derepression mechanisms operated by REST, MeCP2, and DREAM (Citation15, Citation41Citation–Citation43). Consistent with the expression of daDREAM and the reduced levels of the Npas4, a significant decrease in BDNF mRNA () was noticeable in the hippocampus of daDREAM mice. The binding of DREAM to DRE sites in the rat BDNF promoter has been shown in vitro, and reduced levels of BDNF mRNA and protein have been reported in the spinal cord from a different transgenic line that has ubiquitous daDREAM expression (Citation15). To investigate direct regulation of BDNF expression by DREAM, we performed chromatin immunoprecipitation. Using specific primers for mouse promoter IV, we could detect the Ca2+-dependent binding of wild-type DREAM to this promoter and the stronger interaction of daDREAM both under basal conditions and after depolarization (). Since BDNF expression has been related to structural plasticity, learning, and memory, the blockade of activity-dependent BDNF expression in the daDREAM hippocampus may predict impaired cognition in these mice.
FIG 6 DREAM directly regulates activity-dependent BDNF gene expression. (A) Real-time qPCR of BDNF levels in wild-type (wt) and transgenic (tg) hippocampus. Values are normalized with respect to HPRT mRNA content. Results are the means ± SEM of 8 to 12 mice. ***, P < 0.001 (one-way ANOVA, Tukey's multiple comparison). (B) Chromatin immunoprecipitation assay of promoter IV of the BDNF gene using chromatin isolated from primary cultured neurons from wild-type (wt) and transgenic (tg) embryos before and after potassium depolarization. Cultured neurons from DREAM knockout (ko) embryos or exclusion of the antibody (-Ab) in the immunoprecipitation of wild-type chromatin were included as negative controls. Autoradiogram of the semiquantitative PCR is shown. The densitometric quantification results shown at the bottom are the means ± SEM from four experiments. **, P = 0.0022 versus wt nonstimulated (n = 3, two-tailed, unpaired t test).
Impaired learning and memory in daDREAM mice.
Ca2+-dependent gene expression underpins changes in synaptic plasticity and learning and memory processes. To assess a potential phenotype in daDREAM mice, we used three learning and memory tests, the Morris water maze, the object recognition test, and an active avoidance task. In the water maze, transgenic mice showed a clear defect in hippocampal learning. During the acquisition sessions (A1 to A10), performance was impaired (; see also Fig. S7A in the supplemental material). Swimming speed is unlikely to contribute to this phenotype, since daDREAM mice showed no differences with respect to the swimming speed of wild-type mice (, inset). However, in the removal session, no impairment was detected in reference memory, once learning was established after extensive training (see Fig. S7B). In the reversal learning session, daDREAM mice had difficulties in learning the new platform position (see Fig. S8 in the supplemental material). Thus, a repeated reversal learning experiment was performed, which showed that transgenic mice have a significant deficit in both reference (odd trials) (; see also Table S7 in the supplemental material) and working memory (even trials) (; see also Table S7), being less efficient than wild-type mice. In the object recognition task, novelty recognition was not affected when analyzed 1 h after training, and transgenic mice exhibited a discrimination index similar to that in the wild type (63.7 ± 4.1 and 62.5 ± 3.6, respectively). Conditioned learning in the active avoidance procedure showed similar results for learning performance during the first five training sessions in both genotypes (see Fig. S9 in the supplemental material). In the last sessions, however, the performance in daDREAM mice was significantly decreased (see Fig. S9), suggesting deficits in associative learning in these mice.
FIG 7 Impaired learning and memory in daDREAM mice. (A) Escape latency across acquisition sessions (A1 to A10) is increased in transgenic (tg, n = 11) with respect to wild-type (wt, n = 10) mice [repeated-measures ANOVA, F(1,19) = 9.052, P = 0.007]. (Inset) No difference in swimming speed was detected between genotypes [repeated-measures ANOVA, F(1,19) = 3.633, P = 0.072]. (B and C) In the repeated reversal learning paradigm, transgenic mice (n = 10) showed increases in escape latency in odd (B) and even (C) trials, related to impairment in reference and working memory, respectively, across the eight acquisition sessions (A1 to A8), compared to the results for wild-type mice (n = 10). Data are expressed as means ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001. For statistical details, see Table S7 in the supplemental material.
![FIG 7 Impaired learning and memory in daDREAM mice. (A) Escape latency across acquisition sessions (A1 to A10) is increased in transgenic (tg, n = 11) with respect to wild-type (wt, n = 10) mice [repeated-measures ANOVA, F(1,19) = 9.052, P = 0.007]. (Inset) No difference in swimming speed was detected between genotypes [repeated-measures ANOVA, F(1,19) = 3.633, P = 0.072]. (B and C) In the repeated reversal learning paradigm, transgenic mice (n = 10) showed increases in escape latency in odd (B) and even (C) trials, related to impairment in reference and working memory, respectively, across the eight acquisition sessions (A1 to A8), compared to the results for wild-type mice (n = 10). Data are expressed as means ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001. For statistical details, see Table S7 in the supplemental material.](/cms/asset/93345cfd-2ded-4f97-adbb-be5bfea74c19/tmcb_a_12275120_f0007_oc.jpg)
Gene compensation accounts for the mild phenotype in DREAM−/− mice.
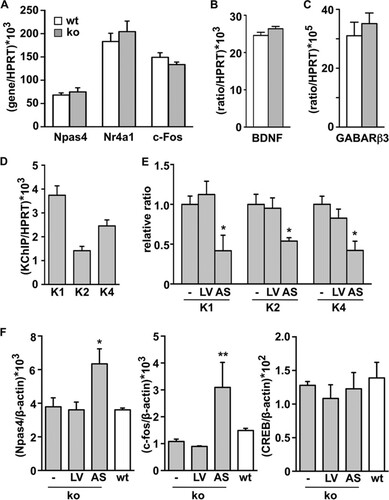
The strong phenotype due to daDREAM expression in the hippocampus contrasts with the mild phenotype of DREAM knockout mice (Citation14, Citation19). Functional redundancy among DREAM/KChIP proteins and their overlapping expression patterns in different organs and brain areas, including the hippocampus (Citation8), might account for the lack of phenotype. Supporting this idea, genetic ablation of KChIP-2 (Citation21) or KChIP-1 (Citation22) also resulted in mild phenotypes. In fact, we found that the expression levels of the transcription factors Npas4, Mef 2C, Nr4a1, Jun B, and c-Fos were not significantly modified in the DREAM−/− hippocampus (). Moreover, we observed no change in BDNF or in GABAergic postsynaptic markers in the DREAM−/− hippocampus ( and ). These data are in agreement with the lack of increase in prodynorphin gene expression originally reported in the hippocampus of DREAM knockout mice (Citation14). To assess whether gene compensation by other KChIP family members is responsible for the absence of changes in gene expression in the DREAM−/− hippocampus, we aimed to knock down the expression of other KChIPs expressed in primary cultured corticohippocampal neurons from DREAM−/− embryos (), using antisense lentiviral expression vectors. As expected, the transduction of primary cultured DREAM−/− neurons with the antisense RNA for full-length KChIP-2 resulted in a reduction in KChIP-2 mRNA levels (). Unexpectedly, however, the levels of KChIP-1 and KChIP-4 mRNA were also reduced after antisense KChIP-2 expression (). Importantly, a further reduction in KChIP levels in the DREAM−/− background was associated with increased mRNA levels of the activity-dependent Npas4 and c-Fos target genes compared to the KChIP expression in untransduced DREAM−/− or wild-type neurons (). The infection with the antisense KChIP-2 lentiviral vector and the knockdown of KChIP expression did not produce a nonspecific generalized effect on the cultured neurons, since we observed no effect on the mRNA level of CREB (), an activity-dependent transcription factor whose activity is mostly controlled at the posttranslational level and which did not appear as a target for DREAM in the genome-wide analysis. These results support the notion of a compensatory effect by other KChIPs as an explanation for the absence of a transcriptional phenotype in DREAM−/− mice.
FIG 8 Activity-dependent gene expression is not modified in the hippocampus of DREAM knockout mice. (A to C) Real-time qPCR analysis of indicated transcripts in the hippocampus from wild-type (wt, n = 10) and DREAM-deficient (ko, n = 10) mice. Values are normalized with respect to HPRT mRNA content. Results are the means ± SEM of two separate experiments. No statistically significant differences were found. (D) Real-time qPCR analysis of KChIP transcripts (K1, -2, and -4, KChIP-1, -2, and -4) in primary cultured corticohippocampal DREAM−/− neurons under basal conditions. (E) Real-time qPCR analysis of KChIP transcripts in primary cultured corticohippocampal DREAM−/− neurons under basal conditions or after transduction with empty (LV) or antisense KChIP-2 lentiviral vector (AS). The results are expressed relative to the basal expression level of the respective KChIP. *, P < 0.05 versus respective basal expression level (one-way ANOVA, Dunnett's multiple comparison test). (F) Real-time qPCR analysis of Npas4, c-fos, and CREB mRNA in corticohippocampal primary cultured neurons from DREAM−/− embryos under basal conditions (-) or 6 days after infection with the indicated lentiviral vector. The increases in Npas4 and c-fos levels following KChIP knockdown are not observed in CREB mRNA levels. For comparison, no significant changes in Npas4, c-Fos, or CREB mRNA levels were observed under basal conditions in DREAM−/− versus wt cultured neurons. Values are normalized with respect to HPRT or β-actin mRNA content. Results are the means ± SEM from three separate cultures in triplicates. *, P < 0.05, and **, P < 0.01, versus the respective basal expression level (one-way ANOVA, Dunnett's multiple comparison test).
DREAM regulates its own expression.
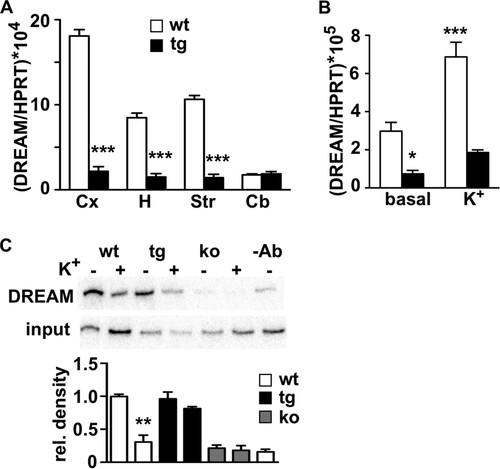
To further understand the mechanism of DREAM regulation of activity-dependent gene expression, we sought evidence for a mechanism that could switch off activity-dependent transcriptional cascades once initiated. Of note, the microarray results showed a significant downregulation of endogenous DREAM expression in the daDREAM hippocampus, and qPCR analysis confirmed reduced levels of endogenous DREAM in the daDREAM transgenic hippocampus (). The reduction was also significant in other telencephalic areas where the transgene is expressed, as well as in cultured transgenic neurons ( and ). Thus, we investigated whether DREAM could regulate its own expression and in this way close a self-regulatory loop. Chromatin immunoprecipitation analysis substantiated this hypothesis and disclosed the Ca2+-dependent binding of DREAM to DRE sites present in the promoter region of the DREAM gene (; see also Fig. S3D in the supplemental material). These results indicate that activity-dependent derepression of endogenous DREAM leads to an increase in DREAM levels, simultaneous with the increase in other DREAM targets, that in turn will contribute to restore basal repression of DREAM and its target genes in the wild-type hippocampus, in this way leading to the transient induction of many activity-dependent genes.
FIG 9 DREAM directly regulates its own activity-dependent expression. (A) Real-time qPCR analysis of endogenous DREAM mRNA in different telencephalic areas from wild-type (wt) and transgenic (tg) mice. Values are normalized with respect to HPRT mRNA content. Results are the means ± SEM of 8 to 12 mice. ***, P < 0.001 (unpaired t test). Cx, cerebral cortex; H, hippocampus; St, striatum; Cb, cerebellum. (B) Real-time qPCR analysis of endogenous DREAM mRNA in primary corticohippocampal cultures from wild-type and transgenic embryos under basal conditions and after potassium depolarization. Values are normalized with respect to HPRT mRNA content. Results are the means ± SEM from three separate cultures in triplicates. *, P < 0.05, and ***, P < 0.0001 (one-way ANOVA, Tukey's multiple comparison). (C) Chromatin immunoprecipitation assay of the mouse DREAM promoter using a DREAM-specific antibody and chromatin isolated from primary cultured neurons from wild-type (wt) and transgenic (tg) embryos before and after potassium depolarization. Cultured neurons from DREAM knockout (ko) embryos or exclusion of the antibody (-Ab) in the immunoprecipitation of wild-type chromatin were included as negative controls. Autoradiogram of the semiquantitative PCR is shown. Densitometric quantification results shown at the bottom are the means ± SEM from four experiments. **, P = 0.0031 versus wt nonstimulated (n = 3, two-tailed, unpaired t test).
DISCUSSION
The activation of calcium-dependent kinases and phosphatases has been proposed as a universal mechanism driving activity-dependent gene expression (reviewed in references Citation41 and Citation4). The identification of the calcium-dependent transcriptional repressor DREAM disclosed an alternative mechanism to link changes in nuclear calcium concentration directly with enhanced expression of the immediate early c-Fos gene (Citation7). In the present work, a genome-wide analysis of transgenic and wild-type mice has identified several additional immediate early genes as targets for DREAM derepression, indicating that DREAM is a permissive transcriptional switch for activity-dependent transcription. Furthermore, the self-regulatory function of DREAM identifies a loop to shut down the transcription of immediate early genes shortly after initiation by the upregulation of DREAM protein.
Genome-wide expression analysis was performed using the whole hippocampus, and therefore, the results do not discriminate between hippocampal subareas where DREAM could differentially regulate early transcriptional activation in different neuronal subpopulations. This limitation of the present study is particularly relevant since the results showed that DREAM controls activity-regulated transcription factors that contribute in opposite directions to experience-dependent synapse development and remodeling. Thus, the downregulation of Mef2c in DREAM transgenic mice may result in a deficient elimination of the excess of excitatory synapses during development (Citation44) in a subset of neurons, whereas the repression of Npas4 in other subareas results in a reduction of the number of GABAergic synapses that form on excitatory neurons (Citation38). Nevertheless, expression analysis of postsynaptic markers in the whole hippocampus confirmed the downregulation of GABA receptor subunits participating in inhibitory mechanisms, while no change was observed either in the levels of PSD95, a marker of excitatory synapses, or in the expression of several glutamate receptors (Citation45).
An analysis of activity-induced genes in cultured hippocampal neurons (Citation34) revealed a set of 169 genes that are more than 2-fold upregulated in response to bicuculline-induced action potential bursting and whose induction is blocked by at least 40% in the presence of CaMBP4, a nuclear fusion protein containing four repeats of the M13 calmodulin binding peptide that binds to and inactivates the nuclear calcium/calmodulin complex (Citation46). To categorize activity-dependent genes according to the mechanism of induction, we compared the results from Zhang et al. (Citation34) with the results from our transcriptomic analysis. Although these two studies used different experimental conditions, in vivo versus cultured neurons and basal repression versus activity-dependent induction, the comparison established three useful gene categories. The first includes genes that are significantly downregulated in the daDREAM hippocampus but are not or only slightly affected by inhibition of the nuclear calcium/calmodulin complex in hippocampal neurons. The most relevant genes from the point of view of transcriptional cascades are shown in Table S8A in the supplemental material and correspond to specific DREAM targets, whose activity-dependent early induction in wild-type neurons is mediated mainly by DREAM derepression. The precise molecular mechanisms leading to gene transactivation once DREAM has detached from the promoter of each target gene are not yet fully understood. The second category encompasses genes that are significantly downregulated in the daDREAM hippocampus and strongly dependent on the nuclear calcium/calmodulin complex. The most important genes are shown in Table S8B and correspond to genes in which DREAM has a permissive function at the initial on/off switch but whose full induction is dependent on the activity of calcium/calmodulin-regulated kinases. The third category includes genes whose activity-dependent induction is strictly independent of DREAM. A short list (see Table S8C) includes those genes induced in a first or a second wave of gene expression after membrane depolarization and that are not modified in the daDREAM hippocampus. Thus, under normal conditions, endogenous DREAM has a permissive effect on the early activation of a set of genes, some of which will be further activated by parallel pathways involving the nuclear calcium/calmodulin complex. Importantly, it has been shown that posttranslational modifications of DREAM, such as sumoylation or a change in the redox state, which are associated with increased nuclear localization and repressor capability, respectively (Citation36, Citation47), could render a DREAM protein with increased repressor activity to block to a greater extent activity-dependent learning and memory formation. In this scenario, it is tempting to speculate that changes in the posttranslational processing of DREAM could participate in the cognitive decline associated with aging or pathological conditions of the brain.
Ablation of the DREAM gene in DREAM−/− mice resulted in minor or no changes in learning and memory (Citation14, Citation19), possibly due to compensation by other KChIPs expressed in the hippocampus. Our results after knockdown of KChIP proteins in cultured neurons supported this idea and confirmed the repressor activity of DREAM/KChIP proteins on the expression of the transcription factors Npas4, Mef 2C, Nr4a1, Jun B, and c-Fos. Interestingly, the hypoalgesic phenotype described in DREAM−/− mice (Citation14) is associated with increased expression of the prodynorphin gene, a bona fide target for DREAM/KChIP repression, in the spinal cord. These data suggest that there is no gene compensation at this level, which could be related to the low expression levels of KChIP-1 and KChIP-2 in the spinal cord (J. R. Naranjo, unpublished data) or to additional mechanisms not yet characterized. Previous work using a mammalian expression vector for full-length antisense KChIP-2 successfully reduced KChIP-2 expression without affecting DREAM mRNA levels, after transfection in lymphocytes (Citation16). Here, using a lentiviral vector for antisense KChIP-2, we observed the downregulation of KChIP-2 but also of KChIP-1 and -4. Their high degree of sequence homology might account for cross-downregulation among KChIP mRNAs, and the higher infection rates and transduction efficiencies of lentiviral vectors than of mammalian expression vectors might contribute to manifest the cross-activity observed in the present study.
In contrast, the expression of daDREAM in transgenic hippocampal neurons results in a tonic basal repression of important transcriptional cascades, with traceable consequences at the electrophysiological level. Thus, the reduced Npas4 content in daDREAM hippocampal neurons resulted in a reduction of the mean frequency, i.e., an increase in the mIPSC interevent interval, of the spontaneous IPSC to about half that in wild-type neurons, as previously shown after transient or permanent Npas4 ablation (Citation38). The mean amplitudes, time constants, and half widths of the spontaneous IPSCs did not differ between genotypes (see Fig. S5C in the supplemental material), as reported for the hippocampus of Npas4 knockout mice (Citation38), while these parameters were decreased only after transient Npas4 knockdown (Citation38). Permanent versus transient Npas4 downregulation might account for developmental compensatory mechanisms responsible for the distinct effects on IPSC amplitudes. Compensatory events during development in the daDREAM hippocampus might mask changes in miniature IPSC (mIPSC) amplitude despite a net reduction in the expression of GABA receptors through different mechanisms involving, for instance, the insertion and clustering of membrane receptors as shown in mice deficient for the scaffolding protein gephyrin at hippocampal inhibitory synapses. In these mice, the frequency of mIPSCs is the same as in wild-type mice but the amplitude is significantly reduced (Citation48). In addition, it has recently been shown that the Npas4-mediated transcriptional program differentially regulates inhibitory inputs in distinct neuronal compartments (Citation49). Thus, increased somatic inhibition or decreased dendritic inhibition could be recorded upon Npas4 activation. Whether these or yet other unknown mechanisms are operating in the daDREAM hippocampus is not presently known. Reduced expression of GABAA receptors and decreased GABAergic transmission could account for the reduction in recurrent inhibition and enhanced LTP in the transgenic dentate gyrus.
To relate specific deficits in transcriptional cascades in daDREAM hippocampal neurons to changes in behavior is more complex. Nevertheless, the reduction in the activity-dependent response in transgenic neurons in culture is in line with the impaired ability in learning and memory tests shown by daDREAM mice. Overall, the cognitive profile suggests a complex defect probably specific for the corticohippocampal circuit, which mainly affects spatial and associative learning but also working and reference memories. These results agree with the impairment in long-term recognition memory shown in Npas4-deficient mice (Citation50, Citation51). Conversely, short-term object recognition memory is not affected in daDREAM mice. Interestingly, daDREAM mice showed increases in early LTP in the hippocampus, which is considered to be a cellular analogue of learning. This is more likely determined by the level of inhibition rather than by other factors, such as the level of BDNF, which nevertheless was markedly reduced in daDREAM mice. BDNF has a role in activity-dependent neuroplasticity in the hippocampus that could explain the spatial learning defects in daDREAM mice with a BDNF deficiency. In this regard, heterozygous BDNF knockout mice reportedly underexpress BDNF and have reduced late LTP, but their spatial memory and search strategy assessed with the Morris water maze (Citation52) is preserved, suggesting that the relationship between LTP and memory is not always straightforward. Since it is always the case that early LTP transits smoothly into late LTP, daDREAM may also affect long-lasting forms of memory that are generally believed to be mediated by new protein synthesis, leading to synaptic growth and a requirement of gene transcription and new protein synthesis (Citation53, Citation54). In a broader view, the present results also suggest a role of DREAM throughout development in the establishment and refinement of neuronal circuitries which, indisputably, will result in changes in brain function in the adult animal.
In conclusion, the results from the genome-wide analysis of the hippocampus from daDREAM mice indicate that Ca2+-dependent unbinding from DNA of DREAM/KChIP proteins is an initial regulatory step necessary to switch on and off gene expression cascades that control the inhibition-excitation balance and ultimately allow learning and memory to occur. Central to these cascades and immediately downstream from DREAM, the Npas4 protein emerges as a critical early regulator of activity-dependent learning and memory. The master-switch regulatory role of DREAM makes this repressor protein a novel target for drug development of new-generation molecules, not based on phosphodiesterase inhibition and independent of the CREB pathway, which could improve cognition.
tmcb_a_12275120_sm0001.pdf
Download PDF (3.6 MB)ACKNOWLEDGMENTS
This work was supported by grants from Spanish Ministry of Health and Science, Madrid Community, La Marató, La Caixa, Reina Sofía and Areces Foundations, the EU 6th Framework Program (NeuroNE, CureFXS), the ERA-NET Program (Neuron and E-Rare), and the Medical Research Council. S.K. has a postdoctoral contract from the Ramón y Cajal Program of the Ministry of Science and Innovation.
We thank Xose M. Dopazo for technical assistance.
We have no conflicts of interest.
SUPPLEMENTAL MATERIAL
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00360-13.
REFERENCES
- Dolmetsch RE, Lewis RS, Goodnow CC, Healy JI. 1997. Differential activation of transcription factors induced by Ca2+ response amplitude and duration. Nature 386:855–858. http://dx.doi.org/10.1038/386855a0.
- Hardingham GE, Fukunaga Y, Bading H. 2002. Extrasynaptic NMDARs oppose synaptic NMDARs by triggering CREB shut-off and cell death pathways. Nat. Neurosci. 5:405–414. http://dx.doi.org/10.1038/nn835.
- Li W, Llopis J, Whitney M, Zlokarnik G, Tsien RY. 1998. Cell-permeant caged InsP3 ester shows that Ca2+ spike frequency can optimize gene expression. Nature 392:936–941. http://dx.doi.org/10.1038/31965.
- Mellstrom B, Savignac M, Gomez-Villafuertes R, Naranjo JR. 2008. Ca2+-operated transcriptional networks: molecular mechanisms and in vivo models. Physiol. Rev. 88:421–449. http://dx.doi.org/10.1152/physrev.00041.2005.
- Naranjo JR, Mellstrom B. 2012. Ca2+-dependent transcriptional control of Ca2+ homeostasis. J. Biol. Chem. 287:31674–31680. http://dx.doi.org/10.1074/jbc.R112.384982.
- Cavazzini M, Bliss T, Emptage N. 2005. Ca2+ and synaptic plasticity. Cell Calcium. 38:355–367. http://dx.doi.org/10.1016/j.ceca.2005.06.013.
- Carrion AM, Link WA, Ledo F, Mellstrom B, Naranjo JR. 1999. DREAM is a Ca2+-regulated transcriptional repressor. Nature 398:80–84. http://dx.doi.org/10.1038/18044.
- Link WA, Ledo F, Torres B, Palczewska M, Madsen TM, Savignac M, Albar JP, Mellstrom B, Naranjo JR. 2004. Day-night changes in downstream regulatory element antagonist modulator/potassium channel interacting protein activity contribute to circadian gene expression in pineal gland. J. Neurosci. 24:5346–5355. http://dx.doi.org/10.1523/JNEUROSCI.1460-04.2004.
- Ledo F, Kremer L, Mellstrom B, Naranjo JR. 2002. Ca2+-dependent block of CREB-CBP transcription by repressor DREAM. EMBO J. 21:4583–4592. http://dx.doi.org/10.1093/emboj/cdf440.
- Ledo F, Link WA, Carrion AM, Echeverria V, Mellstrom B, Naranjo JR. 2000. The DREAM-DRE interaction: key nucleotides and dominant negative mutants. Biochim. Biophys. Acta 1498:162–168. http://dx.doi.org/10.1016/S0167-4889(00)00092-6.
- Rivas M, Mellstrom B, Naranjo JR, Santisteban P. 2004. Transcriptional repressor DREAM interacts with thyroid transcription factor-1 and regulates thyroglobulin gene expression. J. Biol. Chem. 279:33114–33122. http://dx.doi.org/10.1074/jbc.M403526200.
- Osawa M, Dace A, Tong KI, Valiveti A, Ikura M, Ames JB. 2005. Mg2+ and Ca2+ differentially regulate DNA binding and dimerization of DREAM. J. Biol. Chem. 280:18008–18014. http://dx.doi.org/10.1074/jbc.M500338200.
- Osawa M, Tong KI, Lilliehook C, Wasco W, Buxbaum JD, Cheng HY, Penninger JM, Ikura M, Ames JB. 2001. Calcium-regulated DNA binding and oligomerization of the neuronal calcium-sensing protein, calsenilin/DREAM/KChIP3. J. Biol. Chem. 276:41005–41013. http://dx.doi.org/10.1074/jbc.M105842200.
- Cheng HY, Pitcher GM, Laviolette SR, Whishaw IQ, Tong KI, Kockeritz LK, Wada T, Joza NA, Crackower M, Goncalves J, Sarosi I, Woodgett JR, Oliveira-dos-Santos AJ, Ikura M, van der Kooy D, Salter MW, Penninger JM. 2002. DREAM is a critical transcriptional repressor for pain modulation. Cell 108:31–43. http://dx.doi.org/10.1016/S0092-8674(01)00629-8.
- Rivera-Arconada I, Benedet T, Roza C, Torres B, Barrio J, Krzyzanowska A, Avendano C, Mellstrom B, Lopez-Garcia JA, Naranjo JR. 2010. DREAM regulates BDNF-dependent spinal sensitization. Mol. Pain 6:95. http://dx.doi.org/10.1186/1744-8069-6-95.
- Savignac M, Pintado B, Gutierrez-Adan A, Palczewska M, Mellstrom B, Naranjo JR. 2005. Transcriptional repressor DREAM regulates T-lymphocyte proliferation and cytokine gene expression. EMBO J. 24:3555–3564. http://dx.doi.org/10.1038/sj.emboj.7600810.
- Gomez-Villafuertes R, Torres B, Barrio J, Savignac M, Gabellini N, Rizzato F, Pintado B, Gutierrez-Adan A, Mellstrom B, Carafoli E, Naranjo JR. 2005. Downstream regulatory element antagonist modulator regulates Ca2+ homeostasis and viability in cerebellar neurons. J. Neurosci. 25:10822–10830. http://dx.doi.org/10.1523/JNEUROSCI.3912-05.2005.
- Savignac M, Mellstrom B, Naranjo JR. 2007. Calcium-dependent transcription of cytokine genes in T lymphocytes. Pflugers Arch. 454:523–533. http://dx.doi.org/10.1007/s00424-007-0238-y.
- Lilliehook C, Bozdagi O, Yao J, Gomez-Ramirez M, Zaidi NF, Wasco W, Gandy S, Santucci AC, Haroutunian V, Huntley GW, Buxbaum JD. 2003. Altered Abeta formation and long-term potentiation in a calsenilin knock-out. J. Neurosci. 23:9097–9106.
- Alexander JC, McDermott CM, Tunur T, Rands V, Stelly C, Karhson D, Bowlby MR, An WF, Sweatt JD, Schrader LA. 2009. The role of calsenilin/DREAM/KChIP3 in contextual fear conditioning. Learn Mem. 16:167–177. http://dx.doi.org/10.1101/lm.1261709.
- Kuo HC, Cheng CF, Clark RB, Lin JJ, Lin JL, Hoshijima M, Nguyen-Tran VT, Gu Y, Ikeda Y, Chu PH, Ross J, Giles WR, Chien KR. 2001. A defect in the Kv channel-interacting protein 2 (KChIP2) gene leads to a complete loss of I(to) and confers susceptibility to ventricular tachycardia. Cell 107:801–813. http://dx.doi.org/10.1016/S0092-8674(01)00588-8.
- Xiong H, Xia K, Li B, Zhao G, Zhang Z. 2009. KChIP1: a potential modulator to GABAergic system. Acta Biochim. Biophys. Sin (Shanghai) 41:295–300. http://dx.doi.org/10.1093/abbs/gmp013.
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4:249–264. http://dx.doi.org/10.1093/biostatistics/4.2.249.
- Gautier L, Cope L, Bolstad BM, Irizarry RA. 2004. affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20:307–315. http://dx.doi.org/10.1093/bioinformatics/btg405.
- Smyth GK. 2005. Limma: linear models for microarray data, p 397–420. In Gentleman R, Carey VJ, Huber W, Irizarry RA, Dudoit S (ed), Bioinformatics and computational biology solutions using R and Bioconductor. Springer, New York, NY.
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 57:289–300.
- Lucas JJ, Mellstrom B, Colado MI, Naranjo JR. 1993. Molecular mechanisms of pain: serotonin1A receptor agonists trigger transactivation by c-fos of the prodynorphin gene in spinal cord neurons. Neuron 10:599–611. http://dx.doi.org/10.1016/0896-6273(93)90163-L.
- Takahashi Y, Rayman JB, Dynlacht BD. 2000. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 14:804–816. http://dx.doi.org/10.1101/gad.14.7.804.
- Savignac M, Mellstrom B, Bebin AG, Oliveros JC, Delpy L, Pinaud E, Naranjo JR. 2010. Increased B cell proliferation and reduced Ig production in DREAM transgenic mice. J. Immunol. 185:7527–7536. http://dx.doi.org/10.4049/jimmunol.1000152.
- Racine RJ, Gartner JG, Burnham WM. 1972. Epileptiform activity and neural plasticity in limbic structures. Brain Res. 47:262–268. http://dx.doi.org/10.1016/0006-8993(72)90268-5.
- Rhodes KJ, Carroll KI, Sung MA, Doliveira LC, Monaghan MM, Burke SL, Strassle BW, Buchwalder L, Menegola M, Cao J, An WF, Trimmer JS. 2004. KChIPs and Kv4 alpha subunits as integral components of A-type potassium channels in mammalian brain. J. Neurosci. 24:7903–7915. http://dx.doi.org/10.1523/JNEUROSCI.0776-04.2004.
- Knöll B, Kretz O, Fiedler C, Alberti S, Schutz G, Frotscher M, Nordheim A. 2006. Serum response factor controls neuronal circuit assembly in the hippocampus. Nat. Neurosci. 9:195–204. http://dx.doi.org/10.1038/nn1627.
- Tischmeyer W, Grimm R, Schicknick H, Brysch W, Schlingensiepen KH. 1994. Sequence-specific impairment of learning by c-jun antisense oligonucleotides. Neuroreport 5:1501–1504. http://dx.doi.org/10.1097/00001756-199407000-00023.
- Zhang SJ, Zou M, Lu L, Lau D, Ditzel DA, Delucinge-Vivier C, Aso Y, Descombes P, Bading H. 2009. Nuclear calcium signaling controls expression of a large gene pool: identification of a gene program for acquired neuroprotection induced by synaptic activity. PLoS Genet. 5:e1000604. http://dx.doi.org/10.1371/journal.pgen.1000604.
- Ooe N, Saito K, Mikami N, Nakatuka I, Kaneko H. 2004. Identification of a novel basic helix-loop-helix-PAS factor, NXF, reveals a Sim2 competitive, positive regulatory role in dendritic-cytoskeleton modulator drebrin gene expression. Mol. Cell. Biol. 24:608–616. http://dx.doi.org/10.1128/MCB.24.2.608-616.2004.
- Palczewska M, Casafont I, Ghimire K, Rojas AM, Valencia A, Lafarga M, Mellstrom B, Naranjo JR. 2011. Sumoylation regulates nuclear localization of repressor DREAM. Biochim. Biophys. Acta 1813:1050–1058. http://dx.doi.org/10.1016/j.bbamcr.2010.11.001.
- Kim JJ, Fanselow MS. 1992. Modality-specific retrograde amnesia of fear. Science 256:675–677. http://dx.doi.org/10.1126/science.1585183.
- Lin Y, Bloodgood BL, Hauser JL, Lapan AD, Koon AC, Kim TK, Hu LS, Malik AN, Greenberg ME. 2008. Activity-dependent regulation of inhibitory synapse development by Npas4. Nature 455:1198–1204. http://dx.doi.org/10.1038/nature07319.
- Galanopoulou AS, Moshe SL. 2009. The epileptic hypothesis: developmentally related arguments based on animal models. Epilepsia 50(Suppl 7):37–42. http://dx.doi.org/10.1111/j.1528-1167.2009.02217.x.
- Andersen P, Eccles JC, Loyning Y. 1963. Recurrent inhibition in the hippocampus with identification of the inhibitory cell and its synapses. Nature 198:540–542. http://dx.doi.org/10.1038/198540a0.
- Greer PL, Greenberg ME. 2008. From synapse to nucleus: calcium-dependent gene transcription in the control of synapse development and function. Neuron 59:846–860. http://dx.doi.org/10.1016/j.neuron.2008.09.002.
- Mellstrom B, Torres B, Link WA, Naranjo JR. 2004. The BDNF gene: exemplifying complexity in Ca2+-dependent gene expression. Crit. Rev. Neurobiol. 16:43–49. http://dx.doi.org/10.1615/CritRevNeurobiol.v16.i12.40.
- Pruunsild P, Sepp M, Orav E, Koppel I, Timmusk T. 2011. Identification of cis-elements and transcription factors regulating neuronal activity-dependent transcription of human BDNF gene. J. Neurosci. 31:3295–3308. http://dx.doi.org/10.1523/JNEUROSCI.4540-10.2011.
- Flavell SW, Cowan CW, Kim TK, Greer PL, Lin Y, Paradis S, Griffith EC, Hu LS, Chen C, Greenberg ME. 2006. Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science 311:1008–1012. http://dx.doi.org/10.1126/science.1122511.
- Wu LJ, Mellstrom B, Wang H, Ren M, Domingo S, Kim SS, Li XY, Chen T, Naranjo JR, Zhuo M. 2010. DREAM (downstream regulatory element antagonist modulator) contributes to synaptic depression and contextual fear memory. Mol. Brain 3:3. http://dx.doi.org/10.1186/1756-6606-3-3.
- Wang J, Campos B, Jamieson GAJr, Kaetzel MA, Dedman JR. 1995. Functional elimination of calmodulin within the nucleus by targeted expression of an inhibitor peptide. J. Biol. Chem. 270:30245–30248. http://dx.doi.org/10.1074/jbc.270.51.30245.
- Rivas M, Aurrekoetxea K, Mellstrom B, Naranjo JR. 2011. Redox signaling regulates transcriptional activity of the Ca(2+)-dependent repressor DREAM. Antioxid. Redox Signal 14:1237–1243. http://dx.doi.org/10.1089/ars.2010.3385.
- Levi S, Logan SM, Tovar KR, Craig AM. 2004. Gephyrin is critical for glycine receptor clustering but not for the formation of functional GABAergic synapses in hippocampal neurons. J. Neurosci. 24:207–217. http://dx.doi.org/10.1523/JNEUROSCI.1661-03.2004.
- Bloodgood BL, Sharma N, Browne HA, Trepman AZ, Greenberg ME. 2013. The activity-dependent transcription factor NPAS4 regulates domain-specific inhibition. Nature 503:121–125. http://dx.doi.org/10.1038/nature12743.
- Coutellier L, Beraki S, Ardestani PM, Saw NL, Shamloo M. 2012. Npas4: a neuronal transcription factor with a key role in social and cognitive functions relevant to developmental disorders. PLoS One 7:e46604. http://dx.doi.org/10.1371/journal.pone.0046604.
- Ramamoorthi K, Fropf R, Belfort GM, Fitzmaurice HL, McKinney RM, Neve RL, Otto T, Lin Y. 2011. Npas4 regulates a transcriptional program in CA3 required for contextual memory formation. Science 334:1669–1675. http://dx.doi.org/10.1126/science.1208049.
- Montkowski A, Holsboer F. 1997. Intact spatial learning and memory in transgenic mice with reduced BDNF. Neuroreport 8:779–782. http://dx.doi.org/10.1097/00001756-199702100-00040.
- Minichiello L. 2009. TrkB signalling pathways in LTP and learning. Nat. Rev. Neurosci. 10:850–860. http://dx.doi.org/10.1038/nrn2738.
- Pang PT, Lu B. 2004. Regulation of late-phase LTP and long-term memory in normal and aging hippocampus: role of secreted proteins tPA and BDNF. Ageing Res. Rev. 3:407–430. http://dx.doi.org/10.1016/j.arr.2004.07.002.
