ABSTRACT
The three conserved core subunits of the cytochrome c oxidase are encoded by mitochondria in close to all eukaryotes. The Cox2 subunit spans the inner membrane twice, exposing the N and C termini to the intermembrane space. For this, the N terminus is exported cotranslationally by Oxa1 and subsequently undergoes proteolytic maturation in Saccharomyces cerevisiae. Little is known about the translocation of the C terminus, but Cox18 has been identified to be a critical protein in this process. Here we find that the scaffold protein Cox20, which promotes processing of Cox2, is in complex with the ribosome receptor Mba1 and translating mitochondrial ribosomes in a Cox2-dependent manner. The Mba1-Cox20 complex accumulates when export of the C terminus of Cox2 is blocked by the loss of the Cox18 protein. While Cox20 engages with Cox18, Mba1 is no longer present at this stage. Our analyses indicate that Cox20 associates with nascent Cox2 and Mba1 to promote Cox2 maturation cotranslationally. We suggest that Mba1 stabilizes the Cox20-ribosome complex and supports the handover of Cox2 to the Cox18 tail export machinery.
INTRODUCTION
The mitochondrial oxidative phosphorylation system generates the bulk of cellular ATP. During this process, electrons are transferred from NADH and FADH2 to the mitochondrial respiratory chain, resulting in the generation of a proton gradient across the inner mitochondrial membrane that drives ATP synthesis by the F1Fo ATP synthase. Four multisubunit protein complexes constitute the respiratory chain. The terminal electron transfer enzyme is cytochrome c oxidase, which reduces molecular oxygen to water. The enzymatic activity of the oxidase resides in the highly conserved central subunits Cox1 and Cox2, which coordinate the heme and copper cofactors responsible for electron transport (Citation1–Citation5). Twelve polypeptides of dual genetic origin build the cytochrome c oxidase of the yeast Saccharomyces cerevisiae. The majority of these subunits are encoded by the nucleus and need to be imported into the organelle. In addition, the enzyme contains three core subunits encoded by the mitochondria, Cox1, Cox2, and Cox3, which are cotranslationally inserted into the inner mitochondrial membrane by the coordinated action of mitochondrial ribosomes and the protein export machinery (Citation6, Citation7).
After membrane insertion of the core subunits, assembly of the enzyme complex is initiated on Cox1. Concomitantly, specific factors, so-called assembly factors, assist with the maturation and incorporation of cytochrome c oxidase subunits into the complex. The current concept of the assembly process proposes a stepwise process through a series of assembly intermediates (Citation8–Citation10).
However, how the core subunits encoded by the mitochondria are handed over from the export machinery to the assembly intermediates is not understood.
Studies in the yeast Saccharomyces cerevisiae have led to the identification of several components involved in the biogenesis of the Cox2 core subunit. Cox2 is embedded in the inner membrane with two transmembrane spans and exposes its N- and C-terminal domains to the intermembrane space. In S. cerevisiae and in plants, Cox2 is expressed with a cleavable N-terminal presequence of 12 to 15 amino acids (Citation11, Citation12). Prior to presequence cleavage, Cox2 targeting of the membrane begins with the action of the membrane-associated translational activator Pet111, which recognizes the 5′ untranslated region of COX2 mRNA (Citation13–Citation16). Cotranslational insertion of the Cox2 N-terminal tail together with the first transmembrane span depends on the export machinery, Oxa1 (Citation17–Citation19). The peripheral inner membrane protein Mba1 cooperates with Oxa1 in the insertion of mitochondrial translation products (Citation20–Citation25). Mba1 is thought to align the ribosomal exit tunnel with the export machinery (Citation23). Once it is exported into the intermembrane space, the Cox2 N terminus is processed by the Imp1/Imp2/Som1 protease complex (Citation26–Citation28) in a reaction requiring the Cox2-specific chaperone, Cox20 (Citation12, Citation29). Cox20 and its human homolog (COX20) comprise two transmembrane spans exposing the N and C termini into the intermembrane space (Citation30, Citation31). In addition, Cox20 participates in the C-terminal export of Cox2 (Citation12). Cox18, which is dedicated to Cox2 C-terminal translocation, associates with Cox20 in a Cox2-dependent manner, suggesting a possible role for Cox20 in handing over the processed Cox2 to Cox18 (Citation32–Citation35). However, Cox20 is also present in organisms that lack a Cox2 presequence. This fact is indicative of an additional function for Cox20, aside from Cox2 processing. In humans, a mutation in COX20 has been linked to muscle hypotonia and ataxia with cytochrome c oxidase deficiency (Citation31, Citation36).
Here, we carried out a comprehensive analysis of the Cox20 interaction network. The characterization of Cox20-containing complexes defined Cox20 to be a scaffold protein that interacts with the mitochondrial ribosome, linking its function to Cox2 translation. Interestingly, we demonstrate that the ribosome-binding protein Mba1 associates with Cox20 in a defined complex in a Cox2-dependent manner. In addition, our studies show that Mba1 associates with the ribosome and Cox2 assemblies in a dynamic manner. Based on these findings, we propose that Mba1 escorts newly synthesized Cox2 from the insertion machinery to maturing assembly intermediates in a Cox20-dependent manner.
MATERIALS AND METHODS
Yeast strains and growth conditions.
The S. cerevisiae strains used in this study are listed in . All strains with the exception of the imp1Δ strain are congenic to strain YPH499; the imp1Δ strain is derived from BY4741 (Citation37, Citation38). Deletions of MBA1, PET111, COX18, IMP1, and the sequence corresponding to the N terminus of COX20 were generated by introduction of HIS3MX6, klTRP1, and natNT2 cassettes. Protein A (ProtA)-tagged versions of COX20, MBA1, and COX18 were created by homologous recombination using PCR-derived cassettes amplified from plasmids pYM10 and pYM9 (Citation39).Yeast strains were grown on nonfermentable medium, 1% yeast extract, 2% peptone, 3% glycerol (YPG), or fermentable medium, 1% yeast extract, 2% peptone, 2% glucose (YPD) or 1% yeast extract, 2% peptone, 2% galactose (YPGal). Unless otherwise indicated, yeast cells were grown at 30°C with shaking. Yeast growth tests were performed by adjusting precultures to an optical density at 600 nm of 0.3, spotting serial 1:10 dilutions onto YPD and YPG agar plates, and incubating the yeast cells for 3 days at the temperatures indicated below. Mitochondria were isolated from yeast cells grown on YPG or YPGal medium at 30°C as previously described (Citation40). Mitochondrial translation inhibition treatment was performed as follows: yeast cells were grown until mid-log-phase and incubated with 6 mM chloramphenicol for 3 h before mitochondrial isolation (Citation41).
TABLE 1 Yeast strains used in this study
In organello labeling of mitochondrial translation products.
Mitochondrial translation products were radiolabeled for 20 min with 20 μM [35S]methionine (10 mCi/ml) at 30°C, as described previously (Citation42). Labeling reactions were stopped by addition of excess methionine (15 mM). Prior to IgG chromatography, mitochondria were reisolated and washed with SEM buffer (250 mM saccharose, 1 mM EDTA, 10 mM MOPS [morpholinepropanesulfonic acid]). To detect radioactively labeled proteins, storage phosphor screens (GE Healthcare) were used, and signals were digitized using a scanner (Storm820; GE Healthcare).
IgG affinity chromatography.
CNBr-activated Sepharose (GE Healthcare) was coupled to human IgG (Sigma-Aldrich) according to the producer's specifications. Protein complexes were purified from mitochondria isolated from strains expressing the ProtA-tagged proteins of interest. For this purpose, mitochondria were solubilized in 1% digitonin buffer containing 150 mM NaCl, 50 mM Tris-HCl, pH 7.5, 10% glycerol, and 2 mM phenylmethylsulfonyl fluoride (PMSF) for 30 min on ice. Solubilized material was clarified by centrifugation at 20,000 × g and 4°C for 10 min and added to IgG-Sepharose beads at a ratio of 10 μl beads per 1 mg protein. Binding was carried out for a period of 2 h at 4°C with mild agitation. The beads were then washed 10 times with washing buffer containing 0.3% digitonin. Bound proteins were eluted with 0.1 M glycine, pH 2.8, or cleaved overnight at 4°C with 0.4 mg/ml acetylated tobacco etch virus (AcTEV; Thermo Fisher Scientific) protease. Eluates were mixed with loading dye and subsequently analyzed by SDS-PAGE, blue native (BN)-PAGE, or mass spectrometry (MS).
Miscellaneous.
Standard methods were used for SDS-PAGE and Western blotting. Detection of primary antibody-protein complexes was performed using horseradish peroxidase-coupled secondary antibodies (Jackson ImmunoResearch). Signals were detected using an enhanced chemiluminescence system (Thermo Scientific) and exposed on X-ray films (GE Healthcare). BN-PAGE analysis was performed as previously described (Citation43). Isolated mitochondria were solubilized for 10 min in digitonin buffer (1% digitonin, 150 mM NaCl, 50 mM Tris-HCl, pH 7.5, 10% glycerol, 2 mM PMSF), and extracts were clarified by centrifugation for 15 min at 4°C and 20,000 × g. Loading dye (10×; 5% Coomassie G-250, 500 mM 6-aminohexanoic acid, 0.1 M bis-Tris, pH 7.0) was added to the supernatant, and samples were separated on a 4 to 13% polyacrylamide gel.
SILAC labeling and mass spectrometry.
For stable isotope labeling by amino acids in cell culture (SILAC) analysis of Cox20-containing complexes, the ARG4 gene was deleted from wild-type cells (Citation44) and cells expressing TEV-protein A-7His-tagged Cox20 (Cox20ProtA) (see Table S1 in the supplemental material). Yeast cells were cultured on minimal medium (0.67% yeast nitrogen base, appropriate amino acids, 2% galactose) containing either stable isotope-labeled heavy (H) l-arginine (U-13C6, 99%; U-15N4, 99%) and l-lysine (U-13C6, 99%; U-15N2, 99%) (Cambridge Isotope Laboratories) or the nonlabeled light (L) counterparts. Two independent replicates, including a label switch, were performed. Differentially labeled mitochondria (from wild-type cells and cells expressing Cox20ProtA) were isolated, equally pooled, and solubilized. Cox20ProtA complex purification using IgG beads was performed as described above. Eluates were analyzed both by SDS-PAGE and by 4 to 13% gradient BN-PAGE. Gel lanes were cut into 10 slices (SDS gels) or 13 slices (BN gels) of equal sizes. The gel slices were processed for tryptic in-gel digestion, including reduction of disulfide bonds and alkylation of free thiol groups, as described previously (Citation45).
Tryptic peptides were analyzed by liquid chromatography (LC)-MS on an LTQ-Orbitrap XL mass spectrometer (Thermo Scientific, Bremen, Germany) connected to an Ultimate 3000 RSLCnano system (Thermo Scientific, Dreieich, Germany). Peptides were washed and preconcentrated on PepMap C18 μ-precolumns (5 mm by 0.3 mm; Thermo Scientific) and separated using a C18 reversed-phase nano-LC column (50 cm by 75 μm; particle and pore size, 2 μm and 100 Å, respectively; Acclaim PepMap RSLC column; Thermo Scientific). For peptide elution, a linear gradient of a 35-min duration (samples from SDS-PAGE) or a 55-min duration (samples from BN-PAGE) ranging from 0.3% to 19.5% acetonitrile and 0.5% to 32.5% methanol in 4% dimethyl sulfoxide and 0.1% formic acid was applied. The flow rate was 250 nl/min. The mass spectrometer was operated with settings described before (Citation46).
MS/MS data were processed for protein identification and SILAC-based relative quantification using the MaxQuant/Andromeda software tool (version 1.4.1.2) (Citation47, Citation48). Searches against the Saccharomyces Genome Database (SGD; www.yeastgenome.org; download version, 2 March 2011) and relative quantification were performed as described previously (Citation46). For the analysis of samples from SDS gels, all 10 slices of a replicate were defined as a single experiment in the MaxQuant experimental design template in order to determine the overall protein SILAC ratios for each replicate. Ratios are reported as Cox20ProtA-expressing strain/wild-type ratios (i.e., L/H for replicate 1 and H/L for replicate 2) (see Table S1 in the supplemental material). For the analysis of samples derived from BN gels, each slice was defined as an individual experiment in the experimental design template in order to retrieve information about proteins for each individual slice. Normalized abundance profiles of selected proteins were established on the basis of the sum of all peptide MS intensities assigned to a given protein for each slice and normalized to the highest intensity determined across all slices of a replicate. Depending on the SILAC labeling of the Cox20ProtA-expressing cells, the light (replicate 1) or heavy (replicate 2) MS intensity was used (see Table S2).
RESULTS
Cox20 participates in two distinct complexes.
Cytochrome c oxidase assembly occurs through different assembly intermediates, during which Cox1, Cox2, and Cox3 are added to the maturating enzyme in a sequential manner. While Cox1 maturation has been assessed in great detail, the assembly pathway of Cox2 is ill defined. Among Cox2-specific assembly factors, the inner membrane protein Cox20 is critical for its maturation (Citation3, Citation12, Citation29, Citation49). To assess the role of Cox20 in the biogenesis pathway of Cox2, we analyzed the protein composition of Cox20-containing complexes. Therefore, wild-type mitochondria were solubilized in digitonin-containing buffer and analyzed by two-dimensional electrophoresis (BN-PAGE followed by SDS-PAGE) and Western blotting. Cox20 migrated in two distinct complexes at 100 kDa and at 65 kDa, which we termed Cox20100 and Cox2065, respectively (). These complexes did not comigrate with respiratory chain supercomplexes (III2IV2 and III2IV) or with the early assembly intermediates of Cox1 (COA complexes) (Citation41, Citation50, Citation51).
FIG 1 Functionality of a C-terminally tagged Cox20. (A) Endogenous Cox20 is present in two complexes of 65 and 100 kDa. Mitochondria (100 μg) isolated from wild-type cells were solubilized and analyzed by 4 to 13% BN-PAGE followed by two-dimensional SDS-PAGE and Western blotting. (B) Test of Cox20ProtA-expressing cell growth. Wild-type (WT) and Cox20ProtA-expressing cells were spotted on fermentable (glucose, YPD) and nonfermentable (glycerol, YPG) carbon sources in serial 10-fold dilutions and incubated at the indicated temperatures. The respiration-deficient cox20Δ strain was used as a control. (C) Mitochondrial protein synthesis in a Cox20ProtA-expressing strain. Mitochondrial translation products of wild-type and Cox20ProtA-expressing strains were labeled in organello for 20 min. Samples were analyzed by SDS-PAGE and digital autoradiography or Western blotting. As a control, the imp1Δ Cox2 processing-deficient strain was used. (D and E) Protein steady-state levels in cox20Δ and Cox20ProtA-expressing strains. Solubilized mitochondria from wild-type, cox20Δ mutant, and Cox20ProtA-expressing strains were separated by SDS-PAGE (D) or 4 to 13% BN-PAGE (E) and analyzed by Western blotting using specific antibodies, as indicated.
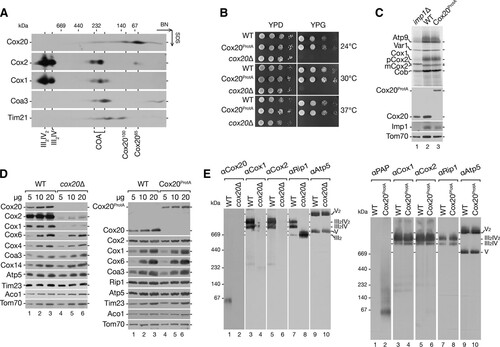
This finding prompted us to investigate the Cox20 function and interaction network in more detail. To this end, we integrated a protein A tag-encoding cassette into the COX20 chromosomal locus, allowing expression of a fusion protein under the control of the endogenous COX20 promoter. The C-terminal protein A tag could be cleaved from the Cox20 portion by tobacco etch virus (TEV) protease treatment. To assess the functionality of the construct, we compared the growth behavior in comparison to that of the wild type and the cox20Δ mutant on fermentable (YPD) and nonfermentable (YPG) carbon sources at different temperatures (). While the cox20Δ strain displayed the expected growth defect (Citation12, Citation29, Citation52), Cox20ProtA-expressing cells exhibited wild-type-like growth, indicating that the fusion protein was functional (). In addition, considering that Cox20 is required for processing of the Cox2 precursor (pCox2) (Citation29), we analyzed if Cox20ProtA allowed the proper processing of Cox2. Therefore, we labeled mitochondrial translation products in mitochondria isolated from wild-type, imp1Δ mutant, and Cox20ProtA-expressing cells. Mitochondria from the imp1Δ mutant failed to process newly synthesized Cox2 and concomitantly accumulate pCox2 (Citation27, Citation28, Citation53) (, lane 1). Cox2 expression and processing were indistinguishable between mitochondria from wild-type and Cox20ProtA-expressing cells, again supporting the functionality of the fusion construct (, lane 2 versus lane 3). When we compared the steady-state levels of selected mitochondrial proteins from wild-type, cox20Δ mutant, and Cox20ProtA-expressing cells by Western blotting, mitochondria from the cox20Δ mutant showed the expected reduction in the amount of Cox1 and Cox2 (, left). In contrast, Cox20ProtA-containing mitochondria displayed wild-type-like protein levels (, right). Lastly, we solubilized mitochondria from wild-type, cox20Δ mutant, and Cox20ProtA-expressing cells and investigated mitochondrial complexes by blue native (BN)-PAGE and Western blot analyses (). In agreement with previously published findings, mitochondria from cox20Δ mutant cells displayed a specific cytochrome c oxidase deficiency (, left). In mitochondria from Cox20ProtA-expressing cells, all tested mitochondrial protein complexes were similar to those in the mitochondria from wild-type cells (, right). Moreover, we were able to detect Cox20ProtA-containing complexes in the expected size range for Cox20100 and Cox2065. In conclusion, Cox20 and Cox20ProtA can be detected in two distinct protein complexes. A Cox20ProtA fusion protein is functional and can be used for further analyses of Cox20-containing complexes.
The mitochondrial ribosome-associated factor Mba1 forms a complex with Cox20.
To define the composition of Cox20-containing complexes, we solubilized Cox20ProtA-containing and wild-type mitochondria and purified complexes by IgG chromatography. Bound Cox20-containing protein complexes were released by TEV protease treatment and analyzed by Western blotting. Among the tested cytochrome c oxidase subunits, only Cox2 coisolated with Cox20, while control proteins, such as Rip1, Tim17, and Tom70, were not detected in the eluate (). To analyze intact Cox20-containing complexes, purified fractions were divided, separated by BN-PAGE, and analyzed for the presence of Cox2 and Cox20 by Western blotting (). Cox20 was detected in the previously described Cox2065 and Cox20100 complexes, as expected. In addition, Cox2 was present in both complexes ().
FIG 2 Characterization of Cox20-containing complexes. (A) Cox20 interacts with Cox2. Mitochondria from Cox20ProtA (TEV-protein A-7His-tagged Cox20)-expressing cells were solubilized in digitonin buffer, and the Cox20ProtA-containing complexes were natively purified by IgG chromatography and subsequent TEV cleavage. Samples were subjected to SDS-PAGE and Western blotting. Total, 1%; eluate, 100%. (B) Purified Cox20 complexes contain Cox2. Native eluted proteins, as described in the legend to panel A, were analyzed by 4 to 13% BN-PAGE and Western blotting using specific antibodies against Cox20 and Cox2. (C) Mass spectrometric analysis of Cox20ProtA complexes isolated after SILAC. Equal amounts of differentially labeled mitochondria from wild-type (WT) and Cox20ProtA-expressing cells were pooled, solubilized, and subjected to IgG chromatography, followed by native elution via TEV cleavage. Eluates were analyzed by SDS-PAGE and LC-MS. Proteins enriched in Cox20ProtA purifications compared to their levels in the wild type with a log2 ratio (light/heavy and heavy/light) of >2 were considered potential candidates. Red, Cox20; green, proteins confirmed by Western blotting. (D) Verification of putative Cox20 interaction partners. Mitochondria isolated from Cox20ProtA-expressing and wild-type strains were solubilized and subjected to IgG chromatography. Samples were analyzed by SDS-PAGE and Western blotting. (E) Yeast cells from the indicated strains were spotted on medium containing glucose (YPD) or glycerol (YPG) and incubated at 30°C. (F) Mitochondria isolated from wild-type and Cox18ProtA-expressing cells were solubilized in digitonin-containing buffer. Protein complexes were isolated via IgG chromatography. Eluates were analyzed by SDS-PAGE and Western blotting.
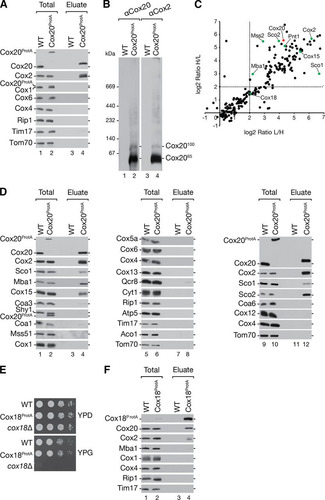
For a global assessment of the Cox20 interaction network, cells were metabolically labeled using stable isotope labeling by amino acids in cell culture (SILAC) analysis (Citation54) (see Table S1 in the supplemental material). For this, Cox20ProtA-containing or control cells were cultured in either heavy or light amino acid-containing medium. In addition to Cox20 and Cox2, mass spectrometric analyses demonstrated an enrichment of proteins implicated in Cox2 biogenesis and the heme A synthase, Cox15 (Citation55–Citation57) (). Unexpectedly, the ribosome receptor Mba1 was also identified within the Cox20 interaction network (). Mba1 has been implicated in the process of insertion of newly synthesized translation products by aligning the mitochondrial ribosome with the membrane insertion machinery (Citation22–Citation25). To support the mass spectrometric data, Cox20ProtA-containing complexes were purified from mitochondria, and the purified fraction was analyzed by Western blotting ( and , lane 5). In addition, Mba1 and Cox15 were confirmed to copurify with Cox20 (, lane 4). Interestingly, the other metallochaperone, Coa6, was not detected (, lane 12). Other inner membrane space twin CX9C motif-containing proteins (e.g., Cmc1, Cox19, Cox23) were not identified in the mass spectrometric analysis. As a control we assessed the presence of Cox1 assembly factors. COA complex components (e.g., Coa1, Shy1, and Mss51) could not be detected in the eluate. Structural subunits of the cytochrome c oxidase (e.g., Cox4, Cox5a, Cox6, Cox12, and Cox13) and subunits of the cytochrome bc1 complex (e.g., Cyt1, Rip1, and Qcr8) were also not present in the eluate ().
FIG 3 Mba1 and Cox20 form a distinct complex at about 100 kDa. (A) SILAC analysis of differentially labeled Cox20ProtA-expressing and control cells. Equal amounts of mitochondria were solubilized and subjected to IgG chromatography, followed by native elution. Eluates were analyzed by BN-PAGE. Gel lanes were cut into 13 slices (slice 1 is at the top, and slice 13 is at the bottom), followed by LC-MS analysis. The normalized intensity profiles of selected proteins copurified with Cox20ProtA are shown. The experiment was repeated with a label swap (see Fig. S1 in the supplemental material). Dashed lines, gel fractions corresponding to the highest-intensity Cox20 peaks; gray box, Cox20100 complex; Rel. Intensity, relative intensity. (B) In organello labeling was performed for 20 min in mitochondria from wild-type (WT), Mba1ProtA-expressing, and mba1Δ mutant cells. Mitochondrial translation products were analyzed by SDS-PAGE and Western blotting or digital autoradiography. (C) Cox20 interacts with Mba1. Protein complexes were isolated from mitochondria from cells expressing Cox20ProtA and Mba1ProtA (TEV-protein A-7His-tagged Mba1) by IgG affinity chromatography and analyzed by SDS-PAGE and Western blotting. Asterisk, a cross-reactive signal; star, Cox20ProtA signal; dot, Mba1ProtA signal. Total, 1%; eluate, 100%. (D) Mba1 associates with the Cox20100 complex. Eluate from the assay whose results are presented in panel C was analyzed by 4 to 13% BN-PAGE for Cox20ProtA- and Mba1ProtA-containing complexes.
Despite an enrichment of Mss2 and Pnt1, proteins implicated in the Cox2 C-terminal translocation machinery, the putative C-terminal Cox2 translocase protein Cox18 was identified in the sample but was less enriched. Due to the lack of a Cox18 antibody, we generated a strain expressing TEV-protein A-7His-tagged Cox18 (Cox18ProtA) to investigate the association of Cox18 with Cox20. To assess the functionality of the tagged protein, growth analysis of the Cox18ProtA-expressing strain was performed (). Strains were analyzed for growth on fermentable and nonfermentable carbon sources. In contrast to the respiration-deficient cox18Δ strain, the Cox18ProtA-expressing strain grew well under all the conditions tested, indicating that tagged Cox18 is functional. We then isolated Cox18ProtA complexes and analyzed them by Western blotting (). Cox20 and Cox2 could be identified in the eluate, in agreement with the results of previous analyses (Citation12). Interestingly, Mba1 was not recovered in the eluate of the Cox18ProtA isolation (). These findings underline the presence of Cox20 in several distinct protein complexes. Furthermore, the association of the Cox2 C-terminal export machinery and Mba1 with Cox20 implicates a close functional connection between the first steps of Cox2 biogenesis, N-terminal maturation, the mitochondrial ribosome, and the C-terminal export machinery.
Mba1 is a constituent of the Cox20100 complex.
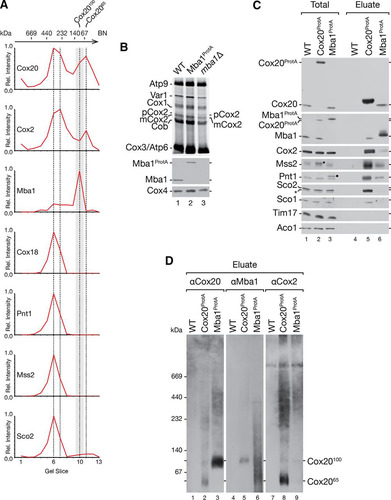
To determine the protein composition of the Cox2065 and Cox20100 complexes, we carried out quantitative mass spectrometric analysis of these protein complexes after separation by BN-PAGE. Therefore, we cultured wild-type and Cox20ProtA-containing cells in SILAC medium and isolated native complexes. The eluates were loaded onto a single BN-polyacrylamide gel lane, which was cut into 13 slices for mass spectrometric analyses (see Table S2 in the supplemental material). The normalized abundance profiles of selected proteins coisolated with Cox20ProtA were plotted against the gel slices (; see also Fig. S1). The abundance distribution of Cox20 along the BN-polyacrylamide gel lane displayed two main peaks in slices 6 and 7 (approximately 400 to 250 kDa) and slices 10 and 11 (approximately 140 to 60 kDa). The Cox2 profile matched the Cox20 profile. Slices 6 and 7 contained proteins involved in the Cox2 C-terminal export machinery (Mss2, Pnt1, and Cox18) and Sco2, which participates in copper insertion into Cox2. Slices 10 and 11 included the Cox2065 and the Cox20100 complexes. Interestingly, Mba1 displayed a maximal abundance in slice 10, while Cox20 and Cox2 peaked in slice 11 ().
To support the finding that Mba1 was selectively present in Cox20100, MBA1 was chromosomally tagged to encode a fusion protein with a C-terminal protein A extension. Using in organello labeling, we confirmed the functionality of tagged Mba1 (). Indeed, TEV-protein A-7His-tagged Mba1 (Mba1ProtA) exhibited a wild-type-like Cox2 processing efficiency, in contrast to its respective deletion strain, in which there was an accumulation of pCox2. Subsequently, Mba1-containing complexes were purified by IgG chromatography and analyzed by Western blotting (). In comparison to the findings with Cox20 purification, smaller amounts of Pnt1, Mss2, and Cox2 coisolated with Mba1. However, the copper chaperones Sco1 and Sco2 did not copurify with Mba1. These results support the idea that Mba1 associates with Cox2 assembly but not the copper insertion machinery. The low level but reproducible coisolation of Cox2 with Mba1 suggests that only a small population of Cox20 complexes contains Mba1. To directly assess the amount of Mba1 in Cox20100, we isolated Cox20ProtA- and Mba1ProtA-containing complexes under native conditions and subjected the eluate to BN-PAGE and Western blotting (). Membrane probing for Cox20 or Mba1 identified Cox20100 as the Mba1-containing complex (, lanes 1 to 6), while the majority of Cox2 was identified in Cox2065 (, lanes 7 to 9). The Cox20 antibody is directed against the C terminus of Cox20. After cleavage of the protein A portion from Cox20ProtA, the remaining spacer amino acids affected recognition by the Cox20 antibody. We conclude that Mba1 and Cox20 form the 100-kDa complex, while Cox2065 comprises Cox2 and Cox20 but lacks Mba1.
Mba1 and Cox20 interact with the mitochondrial ribosome to initiate Cox2 assembly.
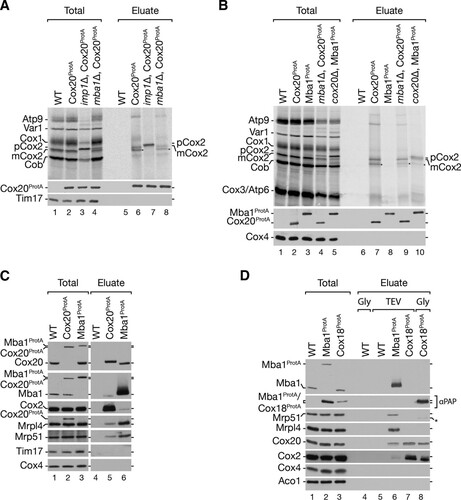
Previous studies defined Mba1 as a mitochondrial ribosome receptor required for respiratory chain biogenesis (Citation20, Citation22, Citation24). The identification of Cox20100 as a Cox20-Mba1-specific complex prompted us to investigate its function in Cox2 translation and maturation. To this end, we deleted IMP1 and MBA1 from a Cox20ProtA-expressing yeast strain and radioactively labeled mitochondrial translation products prior to purification via IgG chromatography (). Cox20ProtA copurified with mature Cox2 (mCox2) in the wild-type background and exclusively with pCox2 in the imp1Δ strain (, lanes 6 and 7). Interestingly, despite the accumulation of pCox2 in mitochondria from mba1Δ and imp1Δ strains (, lanes 3 and 4), pCox2 was not recovered from mba1Δ strain mitochondria by Cox20ProtA (, lane 8). We speculated that Mba1 is required for the efficient transfer of pCox2 to Cox20. To support this hypothesis, we performed a complementary analysis in which we investigated the Mba1-Cox2 association in the absence of Cox20 (). Therefore, we generated an Mba1ProtA-expressing strain and an Mba1ProtA-expressing cox20Δ strain and compared the association of newly synthetized Cox2. For comparison, we purified Cox20ProtA from mba1Δ cells (see above). In agreement with the findings of Preuss et al. (Citation24), no mitochondrial translation products copurified with Mba1ProtA in the absence of a chemical cross-linker (, lane 8). However, in the absence of Cox20, Mba1 was coisolated with mature Cox2 as well as the precursor, pCox2 (, lane 10). Accordingly, when Cox20 was missing, Cox2 accumulated in an Mba1-containing complex. Despite the absence of Cox20, Cox2 underwent proteolytic maturation to some extent. These findings are in agreement with Mba1 acting upstream of Cox20 in the biogenesis of Cox2.
FIG 4 Role of Mba1 in Cox2 synthesis and assembly. (A) Mba1 is required for the association of Cox2 assembly factors with unprocessed Cox2. Mitochondria were isolated from the indicated strains, and mitochondrial translation products were radiolabeled in organello for 20 min. After solubilization of the mitochondria with digitonin, IgG affinity chromatography was performed and samples were analyzed by SDS-PAGE and digital autoradiography or Western blotting. Total, 10%; eluate, 100%. (B) Mba1 binds newly synthesized unprocessed and matured Cox2 in the absence of Cox20. ProtA isolation was performed from the indicated strains after in organello labeling of mitochondrial translation products. Samples were separated by SDS-PAGE and analyzed by digital autoradiography or Western blotting. Asterisks, a Cox2 isoform; pCox2, Cox2 precursor; mCox2, mature Cox2. Total, 10%; eluate, 100%. (C) Cox20 associates with Mba1 and the mitochondrial ribosome. Western blotting was used to analyze the samples for Cox20ProtA and Mba1ProtA. The indicated antibodies against components of the mitochondrial ribosomal large subunit (Mrpl4) and small subunit (Mrp51) were used. Total, 1%; eluate, 100%. (D) Mba1 does not associate with late Cox2 assembly factor Cox18. IgG affinity purification was used to isolate Mba1ProtA and Cox18ProtA from mitochondria. Bound proteins were eluted by TEV cleavage (TEV) or glycine, pH 2.8 (Gly). The Mba1ProtA and the Cox18ProtA signals were detected using anti-ProtA (αPAP) antibody. Asterisk, a cross-reaction. Total, 1%; eluate, 100%.
The observed complex between Cox20 and Mba1 suggested that Cox20 could be in close proximity to the mitochondrial ribosome. To test this directly, we purified Cox20ProtA and Mba1ProtA from mitochondria. Both Cox20ProtA and Mba1ProtA coisolated with Mrpl4, a member of the large ribosomal subunits, and Mrp51, a component of the small ribosomal subunit (). Hence, both Cox20 and Mba1 associate with mitochondrial ribosomes. To investigate the ribosome association along the Cox2 assembly line, we analyzed if Cox18, acting at a later stage in Cox2 maturation, also displayed a ribosome association. Mba1ProtA- and Cox18ProtA-containing complexes were purified and eluted from the column by TEV protease cleavage. In addition, due to the lack of a Cox18 antibody, in a second identical Cox18ProtA isolation, Cox18ProtA was eluted by use of a pH shift to release the tagged protein from the IgG resin. Cox20 was detected in Cox18 and Mba1 isolations (, lanes 6 to 8). In contrast, ribosomal proteins were identified only in the Mba1ProtA isolation. Accordingly, an interaction of Mba1 and Cox20 with the ribosome is restricted to the early steps of Cox2 maturation but is apparently lost when Cox18 engages Cox20 and Cox2 (, lane 6).
Translation of Cox2 is a prerequisite for Mba1-Cox20 interaction with ribosomes.
We hypothesized that actively translating ribosomes are required for the formation of the Mba1-Cox20 complex. To test this, we treated yeast cells with the translational inhibitor chloramphenicol (CAP) and isolated Cox20ProtA- and Mba1ProtA-containing protein complexes under native conditions (). In the presence of chloramphenicol, the Cox20-Mba1 interaction was lost. In addition, ribosomes were no longer purified by Cox20 (, lane 7 versus lane 9). In contrast, chloramphenicol had little impact on the ribosome interaction with Mba1 (, lane 8 versus lane 10). We concluded that mitochondrial translation is required for formation of the Mba1-Cox20 complex with the ribosome.
FIG 5 Defects in processing of Cox2 sequester Mba1 with unassembled Cox2. (A and B) Cox20 and Mba1 interact in a Cox2-dependent manner. Mitochondria were isolated from wild-type and pet111Δ mutant strains expressing Cox20ProtA or Mba1ProtA. Cells were pretreated with 6 mM chloramphenicol (CAP) for 3 h, where indicated. Solubilized mitochondria were subjected to IgG chromatography and analyzed by SDS-PAGE and Western blotting. Total, 1%; eluate, 100%. long exp., long exposure. (C) The Cox20 ribosome association is Mba1 independent. Protein complexes were purified from the indicated deletion mutants expressing Cox20ProtA and Mba1ProtA by IgG affinity chromatography. Total, 1%; eluate, 100%. (D) Cox2 assembly defects lead to a reduce Mba1 association with the ribosome. Mba1ProtA was isolated from the indicated strains. Samples were analyzed by SDS-PAGE and Western blotting. Total, 1%; eluate, 100%. (E) Mitochondria were isolated from the indicated strains, and in organello labeling of mitochondrial translation products was performed for 20 min. Subsequently, the mitochondria were solubilized, subjected to IgG chromatography, and analyzed by SDS-PAGE and Western blot analysis or digital autoradiography. Total, 10%; eluate, 100%. Black star, antibody cross-reaction; asterisk, aberrantly migrating form of mature Cox2.
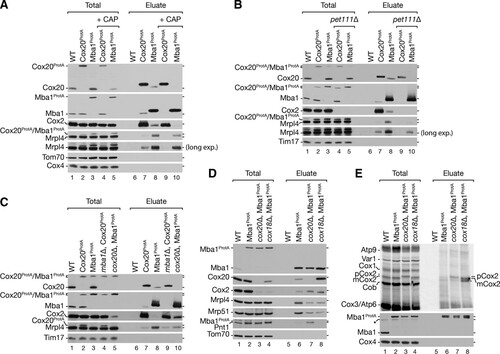
Since CAP is a general mitochondrial protein translation inhibitor, we generated pet111Δ strains with protein A-tagged Cox20 and Mba1. Pet111 is a translational activator for Cox2 mRNA. Deletion of PET111 abolishes Cox2 synthesis (Citation13, Citation14, Citation58). While Cox20ProtA and Mba1ProtA isolated Cox2 and ribosomes in wild-type mitochondria, Mba1ProtA isolated significantly less ribosome from pet111Δ mitochondria (, lane 8 versus lane 10). Moreover, in the absence of Pet111, Cox20ProtA did not purify ribosomes anymore (, lane 7 versus lane 9). More interestingly, the loss of Cox2 translation also abolished the interaction between Cox20 and Mba1 (). Accordingly, the Mba1-Cox20 interaction depends on Cox2 translation. This finding is in agreement with the observation that a loss of Mba1 did not disturb the interaction of newly synthesized mCox2 with Cox20 but that depletion of Cox20 leads to an accumulation of pCox2 with Mba1 (, lanes 9 and 10).
We next investigated the interaction of Cox20 and Mba1 with mitochondrial ribosomes in mitochondria from mba1Δ and cox20Δ strains. As seen before, Cox2 accumulated with Mba1 in mitochondria from the cox20Δ strain (, lanes 5 and 10). Surprisingly, the interaction of mitochondrial ribosomes with Mba1 was reduced when Cox20 was lacking (, lane 10). This finding suggests that during Cox2 maturation Mba1 interacts with Cox2 independently of the ribosome. We hypothesized that a disruption of Cox2 biogenesis at a later stage would lead to an accumulation of Cox2 and Cox20 with Mba1, while at the same time the ribosome would be dissociated. Hence, we isolated Mba1ProtA-containing complexes from a cox18Δ strain background (). As predicted, significantly more Cox20 and Cox2 were isolated with Mba1, while substantially fewer ribosomes were associated (, lane 8). These data suggest that the Cox20-Mba1 complex acts upstream of the Cox2–C-terminal tail translocase Cox18.
Previous work by Saracco and Fox (2002) (Citation34) reported that deletion of Cox18 leads to an accumulation of processed unassembled Cox2 (mCox2). To determine if the interaction of Mba1 with the mature Cox2 nascent chain is altered in the cox18Δ strain, we radiolabeled mitochondrial translation products and isolated Mba1ProtA-containing complexes. As expected, an accumulation of processed Cox2 (mCox2) with Mba1 was apparent in the absence of Cox18 (, lanes 6 and 8).
In summary, we found that Mba1 dynamically associates with Cox20 and early Cox2 assemblies at different stages of the biogenesis pathway. We conclude that Mba1 promotes Cox2 assembly by mediating the shuttling of newly synthesized Cox2 from the insertion machinery to the C-terminal export machinery in a Cox20-dependent manner.
DISCUSSION
Cox20 is a conserved protein that was previously defined to be a Cox2-specific chaperone. This classification is based on the observation that the majority of newly synthesized Cox2 associates with Cox20 in mutant mitochondria affected in Cox2 incorporation into assembly intermediates (Citation29, Citation59). Here, we defined the following to be Cox20-containing complexes: a trail of complexes at 200 kDa, the Cox20100 complex, and the Cox2065 complex. In agreement with the suggested Cox2 chaperone function of Cox20, all detected Cox20 assemblies contained Cox2, but they did not contain any other structural subunit of cytochrome c oxidase. Additional proteins implicated in Cox2 C-terminal processing, copper insertion, and protein export were identified in the Cox20 complexes at 200 kDa and in the Cox20100 complex. The Cox2 C-terminal export factor Cox18, together with Mss2 and Pnt1, were found in the trail of complexes at 200 kDa. This finding confirms the previously described interactions and suggests that these proteins act as a C-terminal export complex (Citation34, Citation60, Citation61). However, the association of Cox20 with Mss2, Pnt1, and Sco2 has not been reported before. In mammals, the homolog of Cox20 was found in complex with the copper chaperones SCO1 and SCO2 (both represent homologs of yeast Sco1) (Citation30). However, the molecular function of Cox20 in the copper insertion process remains elusive. In summary, these observations implicate Cox20 in several different processes during Cox2 biogenesis and agree with a role of Cox20 as a scaffold protein.
The translocation of the Cox2 C terminus is considered to occur posttranslationally (Citation62). Indeed, an association between Cox18 and the ribosome or Cox18 and the mitochondrial ribosome-associated Mba1 has not been detected. The identification of Mba1 in the Cox20100 complex was an unexpected finding. Previous publications implicated Mba1 in the cotranslational export of mitochondrion-encoded proteins of complexes III and IV and stabilization of Cox2 (Citation20, Citation22, Citation24). However, a mechanism by which Mba1 assists with Cox2 maturation has not yet been defined. Here we found an Mba1 interaction with Cox2 assembly intermediates (). In contrast to Cox20, we did not detect interactions between Mba1 and the copper chaperones Sco1 and Sco2, supporting the hypothesis that different pools of Cox20 exist. In addition, this finding indicates that the Cox20-Mba1 interaction is upstream of copper insertion into Cox2. The cooperation of Mba1 with Cox20 in the maturation of Cox2 can be suggested on the basis of the finding of a Cox20-Mba1 complex. In the absence of Cox20 and Cox18, Mba1 isolates with newly synthesized Cox2, suggesting that Mba1 escorts Cox2 during assembly. However, at steady state an association of Mba1 with newly synthesized Cox2 could not be identified. It appears likely that the Mba1-Cox2 interaction is transient but can be stabilized only in the absence of downstream assembly factors, such as Cox20 or Cox18. In agreement with this hypothesis, Mba1 is required for the interaction of pCox2 with Cox20. In contrast, the association of Cox20 with mature Cox2 is Mba1 independent. This is supported by the weak phenotype of an mba1Δ mutant, which displayed only a weak Cox2 assembly defect, indicative of Mba1-independent pathways (Citation22, Citation24).
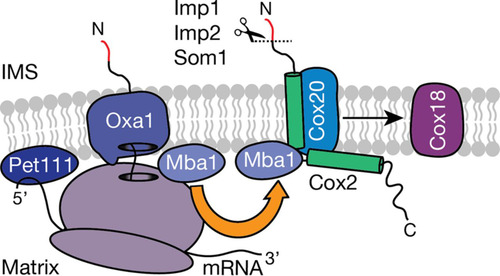
FIG 6 Ribosome-associated Mba1 forms a specific complex with Cox20. COX2 mRNA is translated with the help of the membrane-bound translational activator Pet111. Subsequently, the N-terminal tail is cotranslationally exported into the intermembrane space (IMS) via Oxa1. Mba1 shuttles newly synthesized Cox2 from the export machinery (Oxa1) to the Cox20 protein, promoting its maturation, which requires downstream assembly intermediates like the Cox2 C-terminal tail translocase, Cox18. Mba1 therefore links the cotranslational export of the N-terminal tail of Cox2 with its C-tail translocation.
Recent cryo-electron tomography studies localized Mba1 in proximity to the ribosomal exit tunnel (Citation22, Citation23, Citation63). However, unlike its human counterpart, MRPL45, Mba1 is not a structural component of the ribosome. Accordingly, Mba1 might be dynamically associated with the ribosome. In support of this, we found that Cox2 assembly defects affected the association of Mba1 with the ribosome. A similar phenotype could also be observed in the pet111Δ mutant, in which the Mba1 interaction with the ribosome was decreased but not abolished, indicating that Mba1 preferentially binds to ribosomes translating Cox2 (Citation13, Citation14). Our analyses unexpectedly revealed an association between Cox20 and the translating mitochondrial ribosome. This interaction was dependent on Cox2 translation. Since Cox20 lacks a significant matrix-exposed domain, the Cox20-ribosome interaction is unlikely to be direct. We speculate that a third factor could mediate this association. The nascent Cox2 would in fact be an attractive candidate for this.
The formation of the Cox20-Mba1 complex appears to be dynamic. Since the Cox20-Mba1 interaction requires Cox2, we suggest that Cox2 links the association between these proteins. Notably, in the Cox20-null mutant, Mba1 is sequestered with Cox2 and components of the C-terminal translocation complex. Instead, in the absence of Cox18, Mba1 accumulates with Cox20. These findings sustain a dynamic distribution of Mba1 between different Cox2 maturing assemblies. Collectively, the results of these analyses suggest that at the ribosome Mba1 shuttles newly synthesized Cox2 to Cox20 to support Cox2 maturation. Subsequently, Mba1 remains associated with Cox2 to facilitate transfer of the polypeptide to the tail export machinery in the inner membrane.
tmcb_a_12276092_sm0001.pdf
Download PDF (589.4 KB)tmcb_a_12276092_sm0002.xlsx
Download MS Excel (102.5 KB)tmcb_a_12276092_sm0003.xlsx
Download MS Excel (21.6 KB)ACKNOWLEDGMENTS
We are indebted to S. Callegari for critical reading of the manuscript and to A. Barrientos, R. Richter-Dennerlein, M. Deckers, and J. Dudek for discussion. We thank B. Knapp for excellent technical assistance.
This study was supported by the Deutsche Forschungsgemeinschaft (to B.W. and P.R.), GGNB, EXC 294 BIOSS (to B.W.), ERC (ERCAdG no. 339580) to P.R., MWK FoP 88b (to P.R.), and the Max Planck Society (to P.R.).
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00361-16.
REFERENCES
- Carr HS, Winge DR. 2003. Assembly of cytochrome c oxidase within the mitochondrion. Acc Chem Res 36:309–316. http://dx.doi.org/10.1021/ar0200807.
- Mick DU, Fox TD, Rehling P. 2011. Inventory control: cytochrome c oxidase assembly regulates mitochondrial translation. Nat Rev Mol Cell Biol 12:14–20. http://dx.doi.org/10.1038/nrm3029.
- Soto IC, Fontanesi F, Liu J, Barrientos A. 2012. Biogenesis and assembly of eukaryotic cytochrome c oxidase catalytic core. Biochim Biophys Acta 1817:883–897. http://dx.doi.org/10.1016/j.bbabio.2011.09.005.
- Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S. 1995. Structures of metal sites of oxidized bovine heart cytochrome c oxidase at 2.8 Å. Science 269:1069–1074. http://dx.doi.org/10.1126/science.7652554.
- Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S. 1996. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 Å. Science 272:1136–1144. http://dx.doi.org/10.1126/science.272.5265.1136.
- Herrmann JM, Woellhaf MW, Bonnefoy N. 2013. Control of protein synthesis in yeast mitochondria: the concept of translational activators. Biochim Biophys Acta 1833:286–294. http://dx.doi.org/10.1016/j.bbamcr.2012.03.007.
- Ott M, Amunts A, Brown A. 2016. Organization and regulation of mitochondrial protein synthesis. Biochemistry 85:77–101. http://dx.doi.org/10.1146/annurev-biochem-060815-014334.
- Barrientos A, Gouget K, Horn D, Soto IC, Fontanesi F. 2009. Suppression mechanisms of COX assembly defects in yeast and human: insights into the COX assembly process. Biochim Biophys Acta 1793:97–107. http://dx.doi.org/10.1016/j.bbamcr.2008.05.003.
- McStay GP, Su C-H, Tzagoloff A. 2013. Modular assembly of yeast cytochrome oxidase. Mol Biol Cell 24:440–452. http://dx.doi.org/10.1091/mbc.E12-10-0749.
- Ott M, Herrmann JM. 2010. Co-translational membrane insertion of mitochondrially encoded proteins. Biochim Biophys Acta 1803:767–775. http://dx.doi.org/10.1016/j.bbamcr.2009.11.010.
- Daley DO, Adams KL, Clifton R, Qualmann S, Millar AH, Palmer JD, Pratje E, Whelan J. 2002. Gene transfer from mitochondrion to nucleus: novel mechanisms for gene activation from Cox2. Plant J 30:11–21. http://dx.doi.org/10.1046/j.1365-313X.2002.01263.x.
- Elliott LE, Saracco SA, Fox TD. 2012. Multiple roles of the Cox20 chaperone in assembly of Saccharomyces cerevisiae cytochrome c oxidase. Genetics 190:559–567. http://dx.doi.org/10.1534/genetics.111.135665.
- Green-Willms NS, Butler CA, Dunstan HM, Fox TD. 2001. Pet111p, an inner membrane-bound translational activator that limits expression of the Saccharomyces cerevisiae mitochondrial gene COX2. J Biol Chem 276:6392–6397. http://dx.doi.org/10.1074/jbc.M009856200.
- Mulero JJ, Fox TD. 1993. PET111 acts in the 5′-leader of the Saccharomyces cerevisiae mitochondrial COX2 mRNA to promote its translation. Genetics 133:509–516.
- Poutre CG, Fox TD. 1987. PET111, a Saccharomyces cerevisiae nuclear gene required for translation of the mitochondrial mRNA encoding cytochrome c oxidase subunit II. Genetics 115:637–647.
- Sanchirico ME, Fox TD, Mason TL. 1998. Accumulation of mitochondrially synthesized Saccharomyces cerevisiae Cox2p and Cox3p depends on targeting information in untranslated portions of their mRNAs. EMBO J 17:5796–5804. http://dx.doi.org/10.1093/emboj/17.19.5796.
- Jia L, Kaur J, Stuart RA. 2009. Mapping of the Saccharomyces cerevisiae Oxa1-mitochondrial ribosome interface and identification of MrpL40, a ribosomal protein in close proximity to Oxa1 and critical for oxidative phosphorylation complex assembly. Eukaryot Cell 8:1792–1802. http://dx.doi.org/10.1128/EC.00219-09.
- Kruger V, Deckers M, Hildenbeutel M, van der Laan M, Hellmers M, Dreker C, Preuss M, Herrmann JM, Rehling P, Wagner R, Meinecke M. 2012. The mitochondrial oxidase assembly protein1 (Oxa1) insertase forms a membrane pore in lipid bilayers. J Biol Chem 287:33314–33326. http://dx.doi.org/10.1074/jbc.M112.387563.
- Szyrach G, Ott M, Bonnefoy N, Neupert W, Herrmann JM. 2003. Ribosome binding to the Oxa1 complex facilitates co-translational protein insertion in mitochondria. EMBO J 22:6448–6457. http://dx.doi.org/10.1093/emboj/cdg623.
- Bauerschmitt H, Mick DU, Deckers M, Vollmer C, Funes S, Kehrein K, Ott M, Rehling P, Herrmann JM. 2010. Ribosome-binding proteins Mdm38 and Mba1 display overlapping functions for regulation of mitochondrial translation. Mol Biol Cell 21:1937–1944. http://dx.doi.org/10.1091/mbc.E10-02-0101.
- Hell K, Herrmann J, Pratje E, Neupert W, Stuart RA. 1997. Oxalp mediates the export of the N- and C-termini of pCoxII from the mitochondrial matrix to the intermembrane space. FEBS Lett 418:367–370. http://dx.doi.org/10.1016/S0014-5793(97)01412-9.
- Ott M, Prestele M, Bauerschmitt H, Funes S, Bonnefoy N, Herrmann JM. 2006. Mba1, a membrane-associated ribosome receptor in mitochondria. EMBO J 25:1603–1610. http://dx.doi.org/10.1038/sj.emboj.7601070.
- Pfeffer S, Woellhaf MW, Herrmann JM, Förster F. 2015. Organization of the mitochondrial translation machinery studied in situ by cryoelectron tomography. Nat Commun 6:6019. http://dx.doi.org/10.1038/ncomms7019.
- Preuss M, Leonhard K, Hell K, Stuart RA, Neupert W, Herrmann JM. 2001. Mba1, a novel component of the mitochondrial protein export machinery of the yeast Saccharomyces cerevisiae. J Cell Biol 153:1085–1096. http://dx.doi.org/10.1083/jcb.153.5.1085.
- Rep M, Grivell LA. 1996. MBA1 encodes a mitochondrial membrane-associated protein required for biogenesis of the respiratory chain. FEBS Lett 388:185–188. http://dx.doi.org/10.1016/0014-5793(96)00543-1.
- Schneider A, Behrens M, Scherer P, Pratje E, Michaelis G, Schatz G. 1991. Inner membrane protease I, an enzyme mediating intramitochondrial protein sorting in yeast. EMBO J 10:247–254.
- Nunnari J, Fox TD, Walter P. 1993. A mitochondrial protease with two catalytic subunits of nonoverlapping specificities. Science 262:1997–2004. http://dx.doi.org/10.1126/science.8266095.
- Jan PS, Esser K, Pratje E, Michaelis G. 2000. Som1, a third component of the yeast mitochondrial inner membrane peptidase complex that contains Imp1 and Imp2. Mol Gen Genet 263:483–491. http://dx.doi.org/10.1007/s004380051192.
- Hell K, Tzagoloff A, Neupert W, Stuart RA. 2000. Identification of Cox20p, a novel protein involved in the maturation and assembly of cytochrome oxidase subunit 2. J Biol Chem 275:4571–4578. http://dx.doi.org/10.1074/jbc.275.7.4571.
- Bourens M, Boulet A, Leary SC, Barrientos A. 2014. Human COX20 cooperates with SCO1 and SCO2 to mature COX2 and promote the assembly of cytochrome c oxidase. Hum Mol Genet 23:2901–2913. http://dx.doi.org/10.1093/hmg/ddu003.
- Szklarczyk R, Wanschers BFJ, Nijtmans LG, Rodenburg RJ, Zschocke J, Dikow N, van den Brand MAM, Hendriks-Franssen MGM, Gilissen C, Veltman JA, Nooteboom M, Koopman WJH, Willems PHGM, Smeitink JAM, Huynen MA, van den Heuvel LP. 2013. A mutation in the FAM36A gene, the human ortholog of COX20, impairs cytochrome c oxidase assembly and is associated with ataxia and muscle hypotonia. Hum Mol Genet 22:656–667. http://dx.doi.org/10.1093/hmg/dds473.
- Fiumera HL, Dunham MJ, Saracco SA, Butler CA, Kelly JA, Fox TD. 2009. Translocation and assembly of mitochondrially coded Saccharomyces cerevisiae cytochrome c oxidase subunit Cox2 by Oxa1 and Yme1 in the absence of Cox18. Genetics 182:519–528. http://dx.doi.org/10.1534/genetics.109.101196.
- Fiumera HL, Broadley SA, Fox TD. 2007. Translocation of mitochondrially synthesized Cox2 domains from the matrix to the intermembrane space. Mol Cell Biol 27:4664–4673. http://dx.doi.org/10.1128/MCB.01955-06.
- Saracco SA, Fox TD. 2002. Cox18p is required for export of the mitochondrially encoded Saccharomyces cerevisiae Cox2p C-tail and interacts with Pnt1p and Mss2p in the inner membrane. Mol Biol Cell 13:1122–1131. http://dx.doi.org/10.1091/mbc.01-12-0580.
- Souza RL. 2000. Cloning and characterization of COX18, a Saccharomyces cerevisiae PET gene required for the assembly of cytochrome oxidase. J Biol Chem 275:14898–14902. http://dx.doi.org/10.1074/jbc.275.20.14898.
- Doss S, Lohmann K, Seibler P, Arns B, Klopstock T, Zühlke C, Freimann K, Winkler S, Lohnau T, Drungowski M, Nürnberg P, Wiegers K, Lohmann E, Naz S, Kasten M, Bohner G, Ramirez A, Endres M, Klein C. 2014. Recessive dystonia-ataxia syndrome in a Turkish family caused by a COX20 (FAM36A) mutation. J Neurol 261:207–212. http://dx.doi.org/10.1007/s00415-013-7177-7.
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115–132. http://dx.doi.org/10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2.
- Sikorski RS, Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19–27.
- Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, Knop M. 2004. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21:947–962. http://dx.doi.org/10.1002/yea.1142.
- Meisinger C, Pfanner N, Truscott KN. 2006. Isolation of yeast mitochondria. Methods Mol Biol 313:33–39.
- Mick DU, Vukotic M, Piechura H, Meyer HE, Warscheid B, Deckers M, Rehling P. 2010. Coa3 and Cox14 are essential for negative feedback regulation of COX1 translation in mitochondria. J Cell Biol 191:141–154. http://dx.doi.org/10.1083/jcb.201007026.
- Westermann B, Herrmann JM, Neupert W. 2001. Analysis of mitochondrial translation products in vivo and in organello in yeast. Methods Cell Biol 65:429–438. http://dx.doi.org/10.1016/S0091-679X(01)65025-8.
- Schägger H, von Jagow G. 1991. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem 199:223–231. http://dx.doi.org/10.1016/0003-2697(91)90094-A.
- Alkhaja AK, Jans DC, Nikolov M, Vukotic M, Lytovchenko O, Ludewig F, Schliebs W, Riedel D, Urlaub H, Jakobs S, Deckers M. 2012. MINOS1 is a conserved component of mitofilin complexes and required for mitochondrial function and cristae organization. Mol Biol Cell 23:247–257. http://dx.doi.org/10.1091/mbc.E11-09-0774.
- Cristodero M, Mani J, Oeljeklaus S, Aeberhard L, Hashimi H, Ramrath DJF, Lukeš J, Warscheid B, Schneider A. 2013. Mitochondrial translation factors of Trypanosoma brucei: elongation factor-Tu has a unique subdomain that is essential for its function. Mol Microbiol 90:744–755. http://dx.doi.org/10.1111/mmi.12397.
- Böttinger L, Oeljeklaus S, Guiard B, Rospert S, Warscheid B, Becker T. 2015. Mitochondrial heat shock protein (Hsp) 70 and Hsp10 cooperate in the formation of Hsp60 complexes. J Biol Chem 290:11611–11622. http://dx.doi.org/10.1074/jbc.M115.642017.
- Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. 2011. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res 10:1794–1805. http://dx.doi.org/10.1021/pr101065j.
- Cox J, Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26:1367–1372. http://dx.doi.org/10.1038/nbt.1511.
- Herrmann JM, Funes S. 2005. Biogenesis of cytochrome oxidase—sophisticated assembly lines in the mitochondrial inner membrane. Gene 354:43–52. http://dx.doi.org/10.1016/j.gene.2005.03.017.
- Fontanesi F, Clemente P, Barrientos A. 2011. Cox25 teams up with Mss51, Ssc1, and Cox14 to regulate mitochondrial cytochrome c oxidase subunit 1 expression and assembly in Saccharomyces cerevisiae. J Biol Chem 286:555–566. http://dx.doi.org/10.1074/jbc.M110.188805.
- McStay GP, Su C-H, Tzagoloff A. 2013. Stabilization of Cox1p intermediates by the Cox14p-Coa3p complex. FEBS Lett 587:943–949. http://dx.doi.org/10.1016/j.febslet.2013.02.022.
- Graef M, Seewald G, Langer T. 2007. Substrate recognition by AAA+ ATPases: distinct substrate binding modes in ATP-dependent protease Yme1 of the mitochondrial intermembrane space. Mol Cell Biol 27:2476–2485. http://dx.doi.org/10.1128/MCB.01721-06.
- Esser K, Jan PS, Pratje E, Michaelis G. 2004. The mitochondrial IMP peptidase of yeast: functional analysis of domains and identification of Gut2 as a new natural substrate. Mol Genet Genomics 271:616–626. http://dx.doi.org/10.1007/s00438-004-1011-y.
- Ong S-E, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. 2002. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics 1:376–386. http://dx.doi.org/10.1074/mcp.M200025-MCP200.
- Bareth B, Dennerlein S, Mick DU, Nikolov M, Urlaub H, Rehling P. 2013. The heme A synthase Cox15 associates with cytochrome c oxidase assembly intermediates during Cox1 maturation. Mol Cell Biol 33:4128–4137. http://dx.doi.org/10.1128/MCB.00747-13.
- Barros MH, Carlson CG, Glerum DM, Tzagoloff A. 2001. Involvement of mitochondrial ferredoxin and Cox15p in hydroxylation of heme O. FEBS Lett 492:133–138. http://dx.doi.org/10.1016/S0014-5793(01)02249-9.
- Hederstedt L. 2012. Heme A biosynthesis. Biochim Biophys Acta 1817:920–927. http://dx.doi.org/10.1016/j.bbabio.2012.03.025.
- Green-Willms NS, Fox TD, Costanzo MC. 1998. Functional interactions between yeast mitochondrial ribosomes and mRNA 5′ untranslated leaders. Mol Cell Biol 18:1826–1834. http://dx.doi.org/10.1128/MCB.18.4.1826.
- Keil M, Bareth B, Woellhaf MW, Peleh V, Prestele M, Rehling P, Herrmann JM. 2012. Oxa1-ribosome complexes coordinate the assembly of cytochrome c oxidase in mitochondria. J Biol Chem 287:34484–34493. http://dx.doi.org/10.1074/jbc.M112.382630.
- Broadley SA, Demlow CM, Fox TD. 2001. Peripheral mitochondrial inner membrane protein, Mss2p, required for export of the mitochondrially coded Cox2p C tail in Saccharomyces cerevisiae. Mol Cell Biol 21:7663–7672. http://dx.doi.org/10.1128/MCB.21.22.7663-7672.2001.
- He S, Fox TD. 1999. Mutations affecting a yeast mitochondrial inner membrane protein, Pnt1p, block export of a mitochondrially synthesized fusion protein from the matrix. Mol Cell Biol 19:6598–6607. http://dx.doi.org/10.1128/MCB.19.10.6598.
- Preuss M, Ott M, Funes S, Luirink J, Herrmann JM. 2005. Evolution of mitochondrial Oxa proteins from bacterial YidC. Inherited and acquired functions of a conserved protein insertion machinery. J Biol Chem 280:13004–13011.
- Amunts A, Brown A, Bai X-C, Llácer JL, Hussain T, Emsley P, Long F, Murshudov G, Scheres SHW, Ramakrishnan V. 2014. Structure of the yeast mitochondrial large ribosomal subunit. Science 343:1485–1489. http://dx.doi.org/10.1126/science.1249410.
